Abstract
Developmental transcription factors important in early neuron specification and differentiation often remain expressed in the adult brain. However, how these transcription factors function to mantain appropriate neuronal identities in adult neurons and how transcription factor dysregulation may contribute to disease remain largely unknown. The transcription factor Nurr1 has been associated with Parkinson's disease and is essential for the development of ventral midbrain dopamine (DA) neurons. We used conditional Nurr1 gene-targeted mice in which Nurr1 is ablated selectively in mature DA neurons by treatment with tamoxifen. We show that Nurr1 ablation results in a progressive pathology associated with reduced striatal DA, impaired motor behaviors, and dystrophic axons and dendrites. We used laser-microdissected DA neurons for RNA extraction and next-generation mRNA sequencing to identify Nurr1-regulated genes. This analysis revealed that Nurr1 functions mainly in transcriptional activation to regulate a battery of genes expressed in DA neurons. Importantly, nuclear-encoded mitochondrial genes were identified as the major functional category of Nurr1-regulated target genes. These studies indicate that Nurr1 has a key function in sustaining high respiratory function in these cells, and that Nurr1 ablation in mice recapitulates early features of Parkinson's disease.
Keywords: NR4A2, nuclear receptor, laser capture microdissection, RNA sequencing, orphan receptor
Under experimental conditions, somatic differentiated cells can undergo reprogramming into other cell types or induced pluripotent stem cells (1). This remarkable plasticity raises questions of how the differentiated cellular identity is maintained for extended periods in normal life (2). Of particular relevance is how neurons, which should retain their specific functions for decades in a human brain, stably maintain their unique differentiated properties, and how disrupted maintenance of the correct differentiated identity may be related to disease. Under embryonic development, signaling events induce the expression of transcription factors that combinatorially function to specify appropriate identities and differentiation of specific neuron types. Many of these transcription factors continue to be expressed in adult neurons as well; however, little is known of their functions in the adult brain, or the extent to which they contribute to the stability of the differentiated state (3).
Degeneration of ventral midbrain (VMB) dopamine (DA) neurons, particularly neurons of the substantia nigra compacta (SNc), causes many of the characteristic symptoms in patients with Parkinson's disease (PD). PD is characterized by a progressive pathology involving the appearance of insoluble protein inclusions known as Lewy bodies and eventually the death of neurons. Several studies have indicated that loss of striatal DA and other dopaminergic properties cause symptoms in PD long before cell bodies within the SNc actually die (4). Thus, PD cell pathology may influence differentiated neuronal properties, and from this perspective, it is of interest to explore how developmental transcription factors contribute to DA neuron function in the adult brain.
The transcription factor Nurr1 is one of a family of nuclear receptors critical for DA neuron development. Structural studies have found that this protein is distinct from many other ligand-binding nuclear receptors, lacking a ligand-binding cavity, and thus may function as a ligand-independent transcription factor (5). During the development of DA neurons, Nurr1 expression is induced in early postmitotic neurons and is then maintained under differentiation and in the adult brain (6). In Nurr1 null gene-targeted mice, DA neurons fail to differentiate, and DA neuron markers are absent at birth (7–9). Several recent studies have suggested that disrupted Nurr1 function in adult DA neurons may contribute to the cellular pathology in PD; Nurr1 gene polymorphisms have been associated with PD, and Nurr1 heterozygous mutant mice show increased vulnerability to DA neuron toxic insults (10–13). Moreover, Nurr1 is down-regulated in peripheral lymphocytes of PD patients, and postmortem studies have found that in PD, Nurr1 is down-regulated in the remaining DA neurons showing signs of neuropathology (14, 15).
Because Nurr1 null mice die soon after birth owing to deficiencies in nondopaminergic cells, these animals cannot be used to demonstrate how Nurr1 functions in maintaining mature DA neuron identity in the adult brain. Moreover, DA neurons in heterozygous Nurr1 null mice have developed with subthreshhold levels of Nurr1 and thus may have defects acquired under development and neuronal maturation. Previous studies of mice with heterozygous null mutations in other transcription factor genes have also elucidated deficiencies in adult DA neurons, but whether or not these observations reflect developmental or adult functions remains unclear (16, 17). For these reasons, in previous work, we used mice with a floxed Nurr1 allele, allowing conditional Nurr1 gene targeting in adult brains through stereotaxic injection of adeno-associated virus vectors harboring a Cre-encoding gene (18). The results indicated an association between Nurr1 ablation in mature DA neurons decreased levels of some, but not all, dopaminergic markers. However, for technical reasons that strategy yielded a relatively high degree of variability and did not provide a comprehensive view of the gene expression programs regulated by Nurr1 in adult DA neurons. Moreover, because not all DA neurons were targeted by Cre expression, it was difficult to assess whether Nurr1 ablation resulted in a DA neuron pathology related to PD.
In the present study, Nurr1 was conditionally ablated by tamoxifen treatment of conditional Nurr1 gene-targeted mice expressing the CreERT2 enzyme under the dopamine transporter (DAT) gene regulatory sequences (19). Thus, complete Nurr1 ablation in all DA neurons was achieved in a tamoxifen-controlled manner. This allowed more extensive histological and behavioral analyses and, importantly, the use of laser capture microdissection (LCM) followed by next-generation mRNA sequencing to provide unbiased analyses of differential gene expression resulting from Nurr1 ablation in adult DA neurons. Taken together, our data show that Nurr1 regulates key genes, including a battery of nuclear-encoded mitochondrial genes, and that adult Nurr1 ablation recapitulates early pathological features of PD.
Results
Reduced Levels of Striatal DA and Impaired Behavior in Nurr1-Ablated Mice.
For Nurr1 ablation in mature DA neurons, we used Nurr1 floxed mice and a mouse strain harboring a tamoxifen-inducible Cre (CreERT2) linked to the DAT gene regulatory sequences in a bacterial artificial chromosome (BAC-DAT-CreERT2 mice) (18, 19). We generated homozygous Nurr1 floxed mice harboring either no copies (cNurr1Ctrl) or a single copy of the BAC-DAT-CreERT2 transgene (cNurr1DatCreER). As described previously, Cre-mediated ablation results in removal of the first coding exon of Nurr1. All animals used for conditional ablation were treated with tamoxifen at the indicated time points.
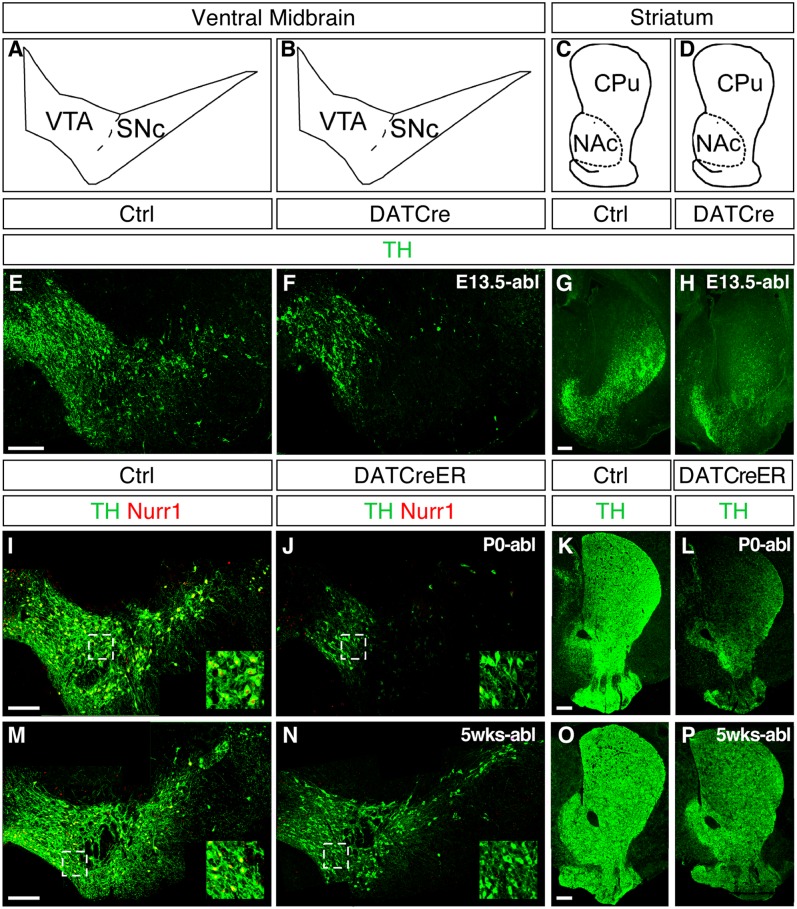
We first analyzed the consequences of Nurr1 ablation at different developmental stages. Nurr1 was ablated in developing DA neurons at embryonic day (E) 13.5, by crossing with mice harboring Cre targeted to the DAT locus (DAT-Cre) (18), or by tamoxifen treatment of cNurr1DatCreER mice at birth (P0) or at age 5 wk. Tissue was collected at 1 wk after Nurr1 ablation and used for combined tyrosine hydroxylase (TH) and Nurr1 immunostaining (Fig. 1). Ablation during embryogenesis resulted in a nearly complete loss of TH within the striatum and DA neuron cell bodies within the SNc (Fig. 1 E–H). Ablation at birth also resulted in a drastic loss of TH (Fig. 1 I–L) within both the midbrain and the striatum, whereas ablation at age 5 wk resulted in a more modest reduction of TH immunostaining and no apparent loss of cell bodies (Fig. 1 M–P). As expected, nuclear Nurr1 immunostaining was not detected in the VMB from Nurr1-ablated mice (Fig. 1 F, J, and N). A more comprehensive analysis of markers in animals treated with tamoxifen at P0 demonstrated drastic down-regulation of DAT and vesicular monoamine transporter 2 (VMAT2) as well, whereas aromatic amino acid decarbocylase (AADC) and Pitx3 were less affected (Fig. S1). These results demonstrate that DA neurons become progressively more resistant to Nurr1 ablation with increasing age.
Fig. 1.
Nurr1 ablation in developing and adult DA neurons. (A–D) Cross-sections at the level of VMB showing the VTA and SNc and at the level of striatum showing the CPu and NAc. (E–P) Immunostaining of TH (green) and Nurr1 (red) at the level of the VMB and in the striatum of cNurr1Ctrl (Ctrl), cNurr1DatCre (DatCre), and cNurr1DatCreER (DatCreER) mice. Nurr1 was ablated by DAT-Cre crosses (cNurr1DatCre mice) at approximately E13.5 and analyzed at P0 (E–H), or treated with tamoxifen at P0 (I–L) or at 5 wk after birth (M–P). In I–P, mice were killed at 1 wk after tamoxifen treatment. Nurr1 immunoreactivity was almost completely lost in TH-positive cells in the cNurr1DatCre and cNurr1DatCreER mice (compare the high-magnification insets in I, J, M, and N).
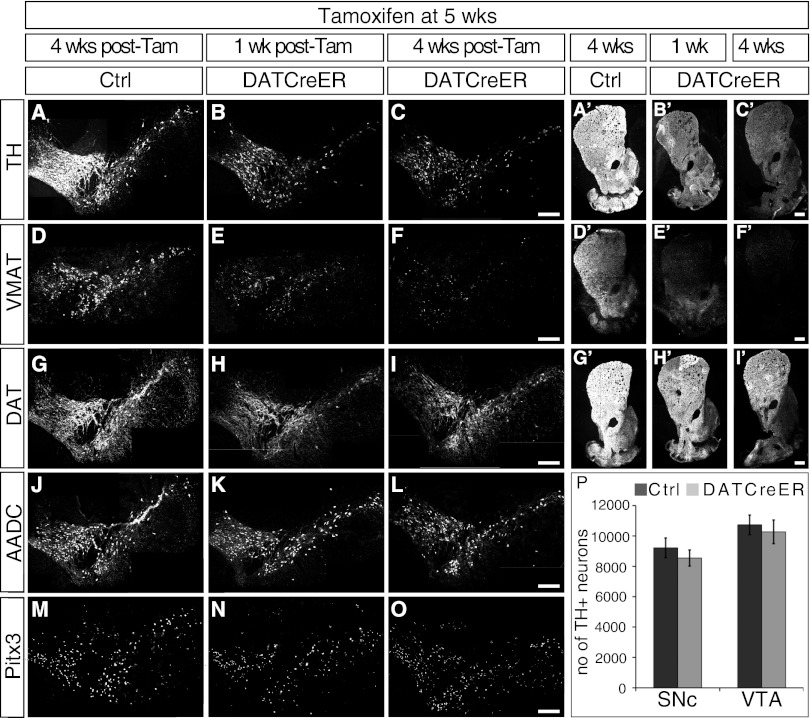
We wished to analyze the consequences of Nurr1 ablation in mature DA neurons in more depth. In all of the following experiments cNurr1DatCreER mice were treated with tamoxifen at 5 wk after birth and analyzed at the indicated time points. At the level of the VMB, a marked reduction of TH and VMAT2 immunoreactivity was noted at 1 wk after tamoxifen treatment in cNurr1DatCreER and was further exaggerated at 4 wk (Fig. 2 A–F). In contrast, DAT, AADC, and Pitx3 were decreased only modestly (Fig. 2 G–O) at both time points anayzed. At the level of the striatum, TH, VMAT2, and DAT were decreased at 1 wk and 4 wk after Nurr1 ablation (Fig. 2 A′–I′). Moreover, the number of TH-positive cell bodies was not decreased significantly at 2 mo after tamoxifen treatment in Nurr1-ablated VMB, as determined by stereology (Fig. 2P).
Fig. 2.
Expression analysis of DA neuron markers and cell counting in Nurr1-ablated mice. Nurr1 was ablated at age 5 wk by tamoxifen treatment. Analyses were performed in cNurr1Ctrl (Ctrl) and cNurr1DatCreER (DatCreER) mice at either 1 wk or 4 wk after tamoxifen treatment. (A–O) Marker expression at the level of the VMB. (A′–I′) Marker expression at the level of the striatum. (P) Stereology of TH-positive neurons in the SNc and VTA in cNurr1Ctrl (Ctrl) and cNurr1DatCreER (DatCreER) mice at 2 mo after tamoxifen treatment.
Similar severe decreases in TH and DAT expression were also apparent within the VMB at 4 mo and 11 mo after tamoxifen treatment (Fig. S2). Of note, at these advanced stages, AADC also remained robustly expressed within DA neuron cell bodies. Thus, Nurr1 ablation in mature DA neurons is associated with decreased expression of TH, VMAT2, and DAT; however, the overall cellular integrity of DA neuron cell bodies within the VMB remains intact in Nurr1-ablated mice.
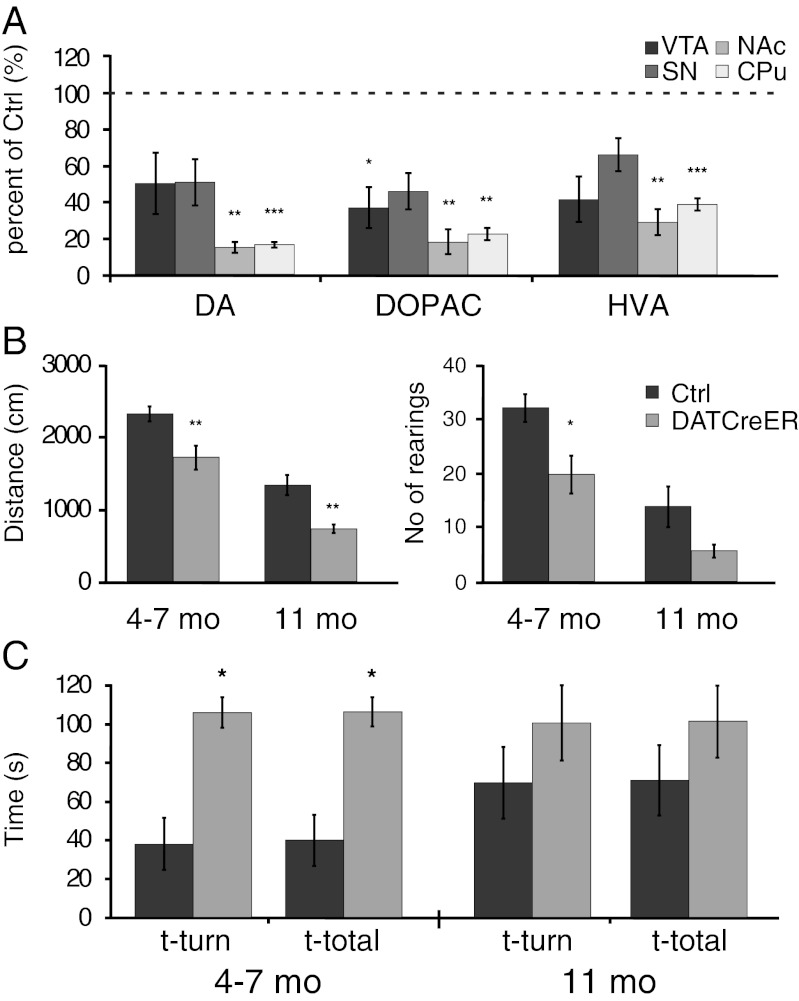
Levels of DA and DA metabolites were measured in the striatum by HPLC of dissected tissue extracts from cNurr1DatCreER at 11 mo after tamoxifen treatment. DA was dramatically reduced in both the caudate putamen (CPu) and the nucleus accumbens (NAc) (Fig. 3A). Of note, DA levels were reduced more significantly within the striatum than within the SNc and VTA (Fig. 3A). Consistent with these findings, motor behaviors were affected in cNurr1DatCreER mice. Open-field locomotion tests performed at 3–5 mo and 11 mo after tamoxifen treatment demonstrated significant reductions in both horizontal and vertical (i.e., number of rearings) locomotor activity in mutant mice (Fig. 3B). Moreover, the vertical pole test indicated prolonged turning time and total task time in cNurr1DatCreER mice (Fig. 3C). Taken together, our findings indicate that ablation of Nurr1 in mature DA neurons results in a severe decrease in DA neuron markers, reduced striatal DA, and significant motor impairment.
Fig. 3.
Analysis of catecholamines and motor behavior in Nurr1-ablated mice. (A) Striatal levels of DA, 3,4-Dihydroxyphenylacetic acid (DOPAC), and homovanillic acid (HVA) in cNurr1Ctrl (Ctrl) and cNurr1DatCreER (DatCreER) mice at 11 mo after tamoxifen treatment in 5-wk-old mice. Separate analyses were performed on tissue extracts from the VTA, SNc, NAc, and CPu, as indicated. (B and C) Open-field measures of horizontal (distance in cm over a 5-min measurement) and vertical (number of rearings during a 5-min measurement) locomotion (B) and vertical pole test of posture control (C) of cNurr1Ctrl (Ctrl) and cNurr1DatCreER (DatCreER) mice at 4–7 mo and 11 mo after tamoxifen treatment in 5-wk-old mice. Here t-turn is the time required to orient downward, and t-total is the total time taken to turn and descend the pole. Note that overall performance was also poor in controls at 11 mo.
Fiber Pathology in Nurr1-Ablated Mice.
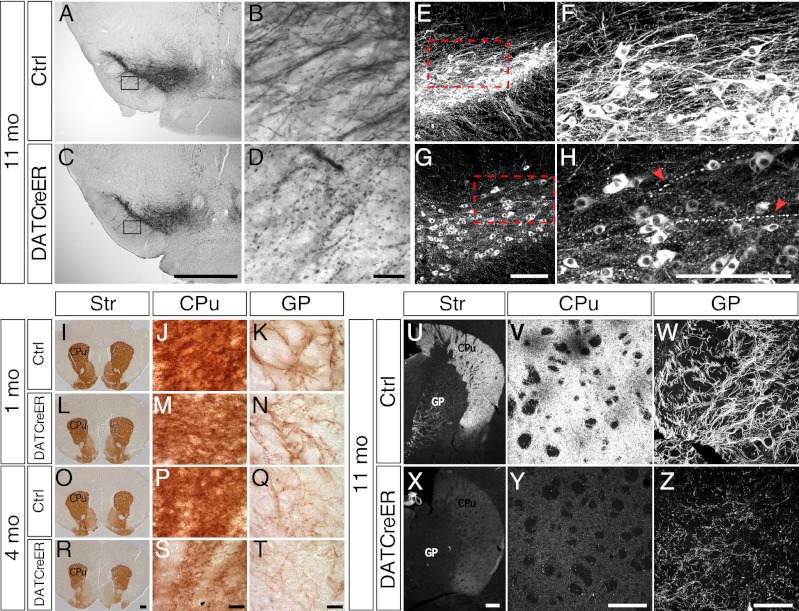
Although no major loss of cell bodies was apparent after Nurr1 ablation (Fig. 2P and Fig. S2 I–L), progressive DA neuron fiber pathology became evident in these animals. TH-positive dendrites extending into the substantia nigra pars reticulata appeared normal in terms of fiber density (but not in TH intensity, as reported earlier) at 4 wk after Nurr1 ablation, but progressively reduced fiber density was seen at 4 mo and 11 mo, and examination at high magnification revealed clear signs of fiber pathology, characterized by fragmented dendrites frequently interrupted by varicosities (Fig. 4 A–D). Similar abnormalities in fibers extending rostrally toward the striatum were also apparent in cNurr1DatCreER mice at 11 mo after tamoxifen treatment (Fig. 4 E–H).
Fig. 4.
Fiber integrity in Nurr1-ablated mice. (A–D) TH immunostaining by horseradish peroxidase/diaminobenzidine (DAB) staining in either low magnification (4×; A and C) or high magnification (40×; B and D) showing dendrites extending into the substantia nigra reticulata in cNurr1Ctrl (Ctrl) and cNurr1DatCreER (DatCreER) mice at 12 mo after tamoxifen treatment in 5-wk-old mice. Boxed regions in A and C are shown in B and D. (E–H) TH immunofluorescence in either low magnification (20×) or high magnification (35×) showing fibers extending dorsorostrally from the substantia nigra cell bodies in the VMB. Arrowheads in H indicate abnormal fibers. (I–T) DAT immunostaining (DAB) of the striatum in cNurr1Ctrl (Ctrl) and cNurr1DatCreER (DatCreER) mice at 1 mo and 4 mo after tamoxifen treatment in 5-wk-old mice. Images of the striatum (Str) in I, L, O, and R are at low magnification (2×). Images at high magnification (100×) (J, K, M, N, P, Q, S, and T) show fibers in either the CPu or in the GP, as indicated. (U–Z) TH immunofluorescence of the striatum in cNurr1Ctrl (Ctrl) and cNurr1DatCreER (DatCreER) mice at 11 mo after tamoxifen treatment in 5-wk-old mice. Images at low magnification (U and X) and high magnification (V–Z) show the Str, CPu, or GP, as indicated.
An overall decreased density of DAT-positive fibers was also seen in the striatum and globus pallidus (GP) of these animals at 4 mo after tamoxifen treatment (Fig. 4 I–T; compare O, P, and Q with R, S, and T). Axon bundles within the medial forebrain bundle appeared normal at 1 mo after tamoxifen treatment but were significantly decreased at 4 mo after tamoxifen treatment (Fig. S3 I–L). Moreover, at 11 months after tamoxifen treatment, TH-positive axons within the GP appeared fragmented and also contained frequent varicosities in cNurr1DatCreER mice, but not in cNurr1Ctrl mice (Fig. 4 U–Z). DAT-positive axons within the medial forebrain bundle appeared normal at 1 mo after Nurr1 ablation, but were detected at decreased density at 4 mo after Nurr1 ablation (Fig. S3 M–T).
Profiling Nurr1-Regulated Gene Expression by RNA Sequencing.
The possibility of targeting Nurr1 in mature DA neurons prompted us to analyze global gene expression changes occurring as a consequence of Nurr1 ablation. LCM was used to isolate SNc and VTA TH-stained neurons from adult mice (Fig. S4), with 200 microdissected cells pooled from each animal and used for RNA isolation and synthesis of a cDNA library suitable for mRNA sequencing. We used a recently developed method for next-generation mRNA sequencing (Smart-Seq), designed for mRNA transcriptomics using low amounts of total RNA down to what can be isolated even from single cells (20).
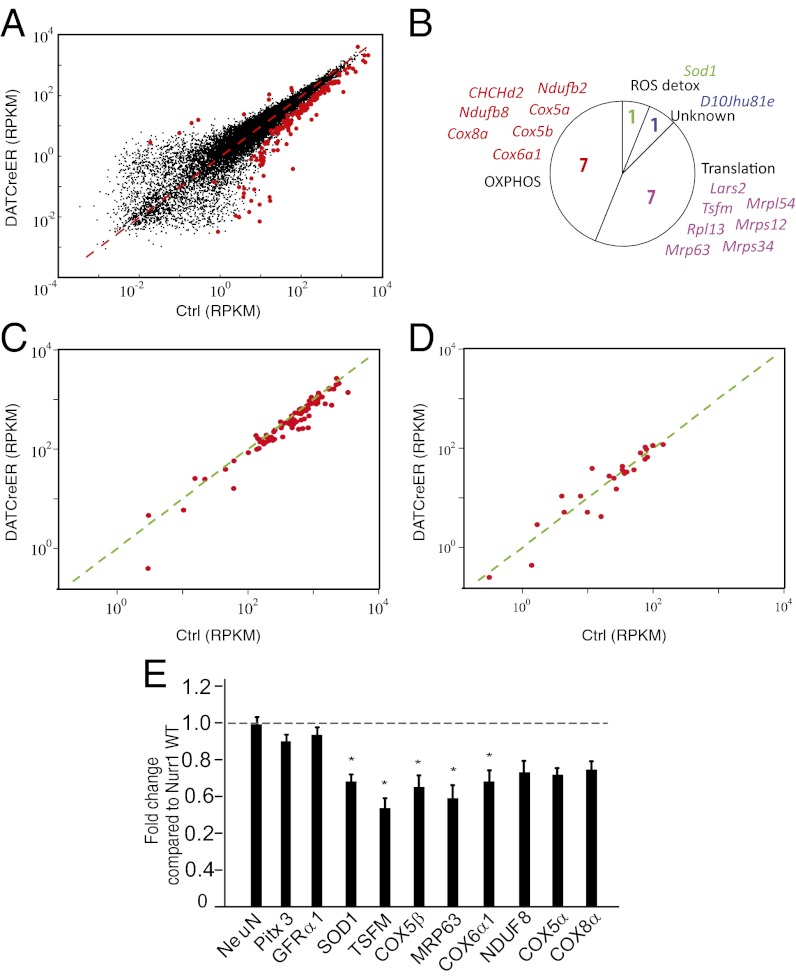
We used the Smart-Seq protocol to analyze mRNA gene expression in DA neurons from cNurr1ctrl (n = 6) and cNurr1DatCreER (n = 9) mice. To avoid detection of gene expression changes resulting from a Nurr1 ablation-induced progressive pathology over an extended period, LCM was performed early, at 1 wk after the completion of tamoxifen treatment. The Spearman correlation coefficient between biological replicates averaged 0.86 and was not below 0.81 for any sample, demonstrating high reproducibility between biological replicates. DEseq statistical analysis based on a negative binomial distribution (21) detected 168 differentially expressed genes in samples from cNurr1DatCreER and cNurr1ctrl DA neurons (Table S1). Several important observations were derived from this analysis. First, a vast majority (94%; P < 1 × 10−11, binomial test) of the 168 differentially expressed genes were expressed at a lower level in cNurr1DatCreER DA neurons, indicating that Nurr1 functions primarily as an activator of gene expression (Fig. 5A). Moreover, of the genes encoding proteins involved in regulating the DA neurotransmitter phenotype, only TH (4.2-fold) and VMAT2 (1.9-fold) were differentially expressed at this early time point (Table S1). In contrast, and consistent with immunostaining and in situ hybridization analysis, AADC and DAT were not differentially expressed at 1 wk after Nurr1 ablation (Fig. 2 J–O).
Fig. 5.
RNA sequencing of control and Nurr1-ablated DA neurons. (A) Scatter diagram showing all detected genes. Differentially expressed genes are highlighted by red dots. The x-axis shows mRNA sequencing RPKM values from control cells; the y-axis, mRNA sequencing RPKM values from Nurr1-ablated cells. (B) Circle diagram showing functional categories and gene names of significantly regulated nuclear-encoded mitochondrial genes. The majority of these genes encode either proteins involved in oxidative phosphorylation or mitochondrial translation. One gene (Sod1) is implicated in radical oxygen species (ROS) detoxification. (C) Scatter diagram highlighting genes encoding proteins involved in oxidative phosphorylation. The x-axis shows mRNA sequencing RPKM values from control cells; the y-axis, mRNA sequencing RPKM values from Nurr1-ablated cells. (D) Scatter diagram highlighting genes encoding proteins involved in fatty acid metabolism. The x-axis shows mRNA sequencing RPKM values from control cells; the y-axis, mRNA sequencing RPKM values from Nurr1-ablated cells. (E) qPCR analysis in Nurr1 ablated and control mice 8–10 wk after tamoxifen treatment. Data are expressed as mean ± SEM (n = 5 per group). *P < 0.05 compared with Nurr1WT/WT group.
We also found that other transcription factors previously identified as essential for the generation of DA neurons, including Pitx3, Lmx1a, Lmx1b, Otx2, and Engrailed1/2, were not differentially expressed in cNurr1DatCreER DA neurons. In Nurr1-ablated cells, an aberrant truncated transcript remained expressed, because the promoter and regulatory sequences of Nurr1 remained intact. The expected loss of excised coding sequences was confirmed by the absence of reads in deleted sequences in cells from cNurr1DatCreER neurons, but not from cNurr1Ctrl neurons. Interestingly, the aberrant Nurr1 mRNA transcribed from the Cre-deleted nr4a2 gene locus (Table S1) was up-regulated (by 3.0-fold). Although this finding suggests that Nurr1 may function in autoregulatory negative feedback, we cannot exclude the possibility that the abnormal floxed Nurr1 mRNA may be more stable and thus result in apparent up-regulation in Nurr1-ablated neurons.
Gene Ontology and pathway enrichment analyses were performed using ToppGene (22) to identify functional categories of differentially expressed genes. This analysis showed that nuclear-encoded mitochondrial genes were strikingly overrepresented among the gene sets expressed at significantly lower levels in Nurr1-ablated DA neurons (P < 0.01). Most of these genes encode proteins with functions in either oxidative phosphorylation or mitochondrial ribosomes (Fig. 5B). We decided to look at different functional categories of nuclear-encoded mitochondrial genes in an attempt to identify regulatory trends extending beyond the genes identified as statistically significantly regulated by DeSeq analysis. Indeed, a highly significant proportion (90%; P < 1.3 × 10−15, binomial test) of all nuclear genes encoding respiratory chain proteins (n = 90), including those that are significantly deregulated as determined by DeSeq, show lower reads per kilobase per million mapped reads (RPKM) values in Nurr1-ablated DA neurons compared with controls (Fig. 5C). In contrast, only 58% of nuclear-encoded mitochondrial genes (n = 24; P = 0.54, binomial test) involved in fatty acid metabolism showed lower RPKM values in Nurr1-ablated DA neurons (Fig. 5D). Additional categories also did not appear to be lower in Nurr1-ablated cells, including those involved in apoptosis (57%; n = 6) and glyconeogenesis (44%; n = 9).
To confirm the dysregulated expression of mitochondrial genes, we performed quantitative PCR analysis of independent mRNA extracted from the VMB of adult control and cNurr1DatCreER mice. Sod1, TSFM, Cox5b, Mrp63, and Cox6a1 were expressed at significantly lower levels in cNurr1DatCreER mice, and nonsignificant trends toward lower expression was seen for Nduf8, Cox5a, and Cox8a. Of note, dissection of VMB tissue is also expected to include a substantial number of non-DA neurons, suggesting that the actual level of dysregulation is underestimated by quantitative PCR analysis. In addition, the pan-neuronal marker NeuN and genes not identified as dysregulated by RNAseq (Pitx3 and Gfra1) did not differ from controls in samples obtained from dissected VMB (Fig. 5E). Taken together, our findings indicate that Nurr1 selectively contributes to the maintenance of normal expression levels of nuclear-encoded genes involved in oxidative respiration, but not of any other functional categories of nuclear-encoded mitochondrial genes.
Discussion
Under certain experimental conditions, particularly after forced expression of developmental transcription factors, somatic differentiated cells can be reprogrammed into pluripotency or transdifferentiate into other cell types. How differentiated cells stably maintain their identities under normal conditions remains largely unclear, however. It seems likely that transcription factors that are important for establishing the differentiated neuronal identity are also involved in its maintenance. We addressed this question by analyzing how the DA neuron transcription factor Nurr1 contributes to the maintenance of DA neurons. It seems intuitive that maintenance of identity is critical for the function of healthy neurons that should persist for many decades in the human brain, and we have been particularly interested in examining whether pathological abnormalities in PD may be related to a failure of cells to sustain transcription factor function in mature DA neurons. The results reported here indeed support that adult Nurr1 ablation recapitulates features of PD.
Our findings showing that the loss of striatal DA and other phenotypic changes occur long before neurons actually die are consistent with the idea that dysfunctional transcription factor function may contribute to PD (4). Several developmental transcription factors, including Nurr1, Lmx1a/b, Engrailed 1, and Pitx3, remain expressed in mature DA neurons, and nucleotide polymorphisms in human genes encoding these factors have been associated with PD (11–13, 23–26). Moreover, analysis of postmortem brain tissue has demonstrated down-regulation of Nurr1 and other key transcription factors in remaining DA neurons in PD, and significantly reduced Nurr1 and Pitx3 mRNA expression levels in peripheral blood cells in PD patients (14, 15). Thus, it seems likely that PD-associated pathological changes will influence transcription factor expression and consequently result in down-regulation of DA neuron-specific genes and accelerated pathology. It is also interesting to note that α-synuclein, the main protein constituent in Lewy bodies, is expressed in both the cytoplasm and in the nucleus, where it may exert at least some of its toxicity in PD by interfering with transcription (27–29).
If abnormal transcription factor function is indeed contributing to PD, then genetic disruption of transcription factors in adult neurons in mice should recapitulate aspects of the degenerative disease process. Conditional gene targeting of transcription factor genes in mature DA neurons provides a rational strategy for testing this hypothesis. Indeed, the phenotype of adult Nurr1-ablated mice reported here resembles early features of PD, apparently supporting the possibility that diminished Nurr1 activity may contribute to the disease. Accordingly, in both PD and the gene-targeted mice studied here, striatal DA loss is accompanied by motor deficiencies that become apparent before DA neurons actually die. Reduced expression of VMAT2, as seen in Nurr1-ablated mice, has been noted in PD as well (30). Nurr1 ablation also results in a distinct fiber pathology that affects both dendrites and axons, which is interesting in light of PD studies identifying axon degeneration as an early event in disease progression (4). It should be noted that defects were always uniform on both sides of the VMB and striatum. Thus, taken together, our observations identify abnormalities that resemble significant aspects of presymptomatic and early stages of PD, and suggest that Nurr1 conditional KO mice can serve as a relevant PD model.
Central to understanding how transcription factors may contribute to the function and disease in DA neurons is the identification of regulated target genes. Examining the transcriptome in specific populations of adult neurons presents a formidable challenge, given that specific neuron types are intermingled with other cell types and thus are difficult to isolate. Using LCM for isolation of specific neuron populations provides a powerful strategy; however, low RNA yields is a common obstacle that makes comprehensive analysis of differential gene expression pattern technically challenging. The Smart-Seq method used in the present study was developed for mRNA sequencing of very small amounts of RNA, down to what can be extracted even from single cells (20), and thus has potential for isolating RNA from limited populations of brain cells isolated by LCM. Our data indicate that comprehensive transcriptome data can be derived from relatively small populations of neurons; we found that the amount of RNA extracted from 200 laser-microdissected DA neurons per analyzed animal was more than sufficient for comprehensive Smart-Seq analysis.
Cells were captured already at 1 wk after tamoxifen treatment with the aim of primarily identifying direct regulatory targets of Nurr1. It is impossible to exclude the possibility that some of the differentially expressed genes are indirectly regulated via other transcription factors; however, it is notable that none of the other known DA neuron developmental transcription factors, including Pitx3, Engrailed 1/2, FoxA2, and Otx2, were differentially expressed in Nurr1-ablated neurons. The finding that Nurr1 seems to function in transcriptional activation in DA neurons (>90% of differentially expressed genes were higher in controls) is also consistent with previous in vitro data showing that Nurr1 functions as a potent activator of transcription (6). Although regulation of DAT and Ret by Nurr1 has been suggested, these genes were not significantly dysregulated, as determined by RNAseq analysis. Through in situ hybridization of RNA expression, our analysis confirmed that these two genes indeed are not down-regulated or are only modestly down-regulated at 1 wk after ablation. However, Nurr1 ablation is likely to result in pathology that leads to indirect down-regulation of numerous additional genes at later time points.
Analysis of differential gene expression has provided a unique fingerprint of early gene expression changes occurring as a result of Nurr1 ablation in mature DA neurons. A remarkable outcome of this analysis is the finding that Nurr1 regulates a large number of nuclear-encoded mitochondrial genes. Of interest, genes encoding components of the respiratory chain seem to be particularly affected. Seven genes encoding respiratory chain subunits were significantly dysregulated, but analysis of the entire battery of genes involved in oxidative phosphorylation revealed that >90% of those genes had lower RPKM values in Nurr1-ablated DA neurons compared with controls (Fig. 5). Midbrain DA neurons are autonomous pacemakers and constantly fire with moderate (i.e., tonic firing) or very high (i.e., burst firing) frequencies to control a sustained and adaptive release of DA in innervated target areas (31). This unique behavior of DA neurons is highly energy-demanding and linked to a high requirement for ATP-generating oxidative phosphorylation. Of note, the SNc and VTA contain significantly higher levels of mitochondrial DNA compared with other brain regions, reflecting the need for efficient oxidative phosphorylation in these cells (32). Our findings suggest that Nurr1 is an important transcription factors for sustaining this distinguishing trait of DA neurons by positively regulating a large number nuclear-encoded mitochondrial genes. How Nurr1 regulates these genes is an interesting avenue of study. It seems likely that Nurr1 functionally interacts with other factors, such as PGC-1α or nuclear respiratory factors, to regulate the battery of nuclear-encoded mitochondrial genes.
Numerous studies have implicated mitochondrial failure in the pathogenesis of PD, and it seems plausible that insufficient ATP production as a result of Nurr1 down-regulation can contribute to pathology, including the appearance of dystrophic axons and dendrites seen in Nurr1-deficient animals and in PD patients. Other genes that appear to be regulated by Nurr1 are of interest in terms of neurodegeneration, including Sod1, which is essential in protection against radical oxygen species, and ApoE, which has been implicated as a risk factor in Alzheimers's disease (Table S1). It may be possible to pharmacologically target the nuclear receptor Nurr1 via its heterodimerization partner RXR. Whether or not Nurr1 functions alone or together with RXR to regulate key DA neuron genes remains unknown, but an evaluation of this action will be critical to the attempt to develop Nurr1 as a PD drug target.
Materials and Methods
Animals.
Generation of conditional Nurr1 gene-targeted mice and mice expressing the CreERT2 enzyme under the DAT gene regulatory sequences in a bacterial artificial chromosome (BAC-DAT-CreERT2 mice) has been described previously (18, 19). Crosses between these transgenic lines facilitates inducible Nurr1 gene ablation exclusively in mDA neurons by generating mice homozygous for the conditional targeted Nurr1 allele and heterozygous for the BAC-DAT-CreERT2 allele (cNurr1DATCreER). Littermates homozygous for Nurr1 floxed allel harboring no copy of the BAC-DAT-CreERT2 transgene served as controls (cNurr1Ctrl). BAC-DAT-CreERT2 mice were also crossed with reporter mice in which the LacZ gene was introduced under control of the ROA26 promoter that harbors a loxP-flanked stop cassette (33). Mice were kept in rooms with controlled 12-h light/dark cycles, temperature, and humidity, with food and water provided ad libitum. All animal experiments were performed with permission from the local Animal Ethics Committee.
Histological Analyses.
At early neonatal stages, mouse brains were dissected and fixed for 24 h in 4% phosphate-buffered paraformaldehyde (PFA). For the isolation of adult brains, animals were anesthetized with tribromoethanol (0.5 mg/g) and perfused through the left ventricle with body-temperature PBS, followed by ice-cold 4% PFA. The brains were dissected and postfixed overnight in 4% PFA, then cryoprotected for 24–48 h in 30% sucrose at 4 °C before being cut into 30-µm sections on a sliding microtome or embedded in optimum cutting temperature compound (Sakura Finetek). The embedded brains were cryosectioned at 14 μm (midbrain) or 20 μm (forebrain) onto slides (SuperFrostPlus; Menzel Gläser). Littermates were used in all comparative experiments. Detailed information on immunohistochemistry and in situ hybridization protocols is provided in SI Materials and Methods.
Cell Counting and Optical Densitometry Analysis.
The total number of TH-positive neurons in the VMB was measured according to the optical fractionator principle, using the Olympus CAST-Grid stereology system. Every sixth section covering the entire extent of the VTA and SN was included in the counting procedure. A coefficient of error of <0.10 was accepted.
LCM and RNA Sequencing.
Mice were euthanized at 1 wk after the completion of tamoxifen treatment using CO2 asphyxiation, after which brains were quickly removed and snap-frozen in dry ice-cooled isopentane. Then 10-µm-thick cryostat coronal sections of the midbrain were cut and mounted on membrane glass slides (Zeiss 415101–4401-000) at −20 °C. Before LCM, rapid TH staining was performed to visualize the midbrain DA neurons to be captured. LCM was performed immediately after staining using a Leica laser microdissection system. A total of 200 TH-positive neurons were captured unilaterally throughout the midbrain from every 14th section per animal, and total RNA was extracted using the Pico RNA Isolation Kit (Arcturus Engineering). The eluted volume was decreased to approximately 3 µL by vacuum spinning. Synthesis and amplification (18 cycles) of the cDNA library were performed using Smart-SEq (20). These protocols are described in more detail in SI Materials and Methods.
Behavioral Tests.
Adult (3-5 mo after tamoxifen treatment) and old (11 mo after tamoxifen treatment) mice were used to assess the motor phenotype. A single cohort of mice was tested at each age, with a 6-d interval between behavioral tests. Mice were randomized according to genotype. At each test, mice were allowed to habituate to the experiment room for at least 1 h. Details of the open-field and pole tests are provided in SI Materials and Methods.
Measurement of DA and DA Metabolites.
Twelve-month-old mice were killed by decapitation. Brains were rapidly removed and frozen in dry ice-cooled isopentane. Regions of interest were then collected using a tissue punch, weighed, and kept at −80 °C until processing with HPLC.
Supplementary Material
Acknowledgments
We thank Günther Schütz, David Engblom, Pierre Chambon, and Daniel Metzger for mouse strains, and members of the T.P. laboratory for discussions and advice. This work was supported by grants from the Swedish Strategic Science Foundation (to T.P., A.B., and P.S.) and from the European Union, Seventh Framework Programme under grant agreement mdDANeurodev (Grant 222999, to T.P.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE42912).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221077110/-/DCSupplemental.
References
- 1.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 2.Holmberg J, Perlmann T. Maintaining differentiated cellular identity. Nat Rev Genet. 2012;13(6):429–439. doi: 10.1038/nrg3209. [DOI] [PubMed] [Google Scholar]
- 3.Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- 4.Cheng H-C, Ulane CM, Burke RE. Clinical progression in Parkinson disease and the neurobiology of axons. Ann Neurol. 2010;67(6):715–725. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423(6939):555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 6.Perlmann T, Wallén-Mackenzie A. Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 2004;318(1):45–52. doi: 10.1007/s00441-004-0974-7. [DOI] [PubMed] [Google Scholar]
- 7.Zetterström RH, et al. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276(5310):248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 8.Saucedo-Cardenas O, et al. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA. 1998;95(7):4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo SO, et al. Dopamine biosynthesis is selectively abolished in substantia nigra/ventral tegmental area but not in hypothalamic neurons in mice with targeted disruption of the Nurr1 gene. Mol Cell Neurosci. 1998;11(1-2):36–46. doi: 10.1006/mcne.1998.0673. [DOI] [PubMed] [Google Scholar]
- 10.Le W, Conneely OM, He Y, Jankovic J, Appel SH. Reduced Nurr1 expression increases the vulnerability of mesencephalic dopamine neurons to MPTP-induced injury. J Neurochem. 1999;73(5):2218–2221. [PubMed] [Google Scholar]
- 11.Grimes DA, et al. Translated mutation in the Nurr1 gene as a cause for Parkinson’s disease. Mov Disord. 2006;21(7):906–909. doi: 10.1002/mds.20820. [DOI] [PubMed] [Google Scholar]
- 12.Xu P-Y, et al. Association of homozygous 7048G7049 variant in the intron six of Nurr1 gene with Parkinson’s disease. Neurology. 2002;58(6):881–884. doi: 10.1212/wnl.58.6.881. [DOI] [PubMed] [Google Scholar]
- 13.Zheng K, Heydari B, Simon DK. A common NURR1 polymorphism associated with Parkinson disease and diffuse Lewy body disease. Arch Neurol. 2003;60(5):722–725. doi: 10.1001/archneur.60.5.722. [DOI] [PubMed] [Google Scholar]
- 14.Chu Y, et al. Nurr1 in Parkinson’s disease and related disorders. J Comp Neurol. 2006;494(3):495–514. doi: 10.1002/cne.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, et al. Decreased NURR1 and PITX3 gene expression in Chinese patients with Parkinson’s disease. Eur J Neurol. 2012;19(6):870–875. doi: 10.1111/j.1468-1331.2011.03644.x. [DOI] [PubMed] [Google Scholar]
- 16.Albéri L, Sgadò P, Simon HH. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development. 2004;131(13):3229–3236. doi: 10.1242/dev.01128. [DOI] [PubMed] [Google Scholar]
- 17.Kittappa R, Chang WW, Awatramani RB, McKay RDG. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5(12):e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadkhodaei B, et al. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci. 2009;29(50):15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engblom D, et al. Glutamate receptors on dopamine neurons control the persistence of cocaine seeking. Neuron. 2008;59(3):497–508. doi: 10.1016/j.neuron.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305-11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergman O, et al. Do polymorphisms in transcription factors LMX1A and LMX1B influence the risk for Parkinson’s disease? J Neural Transm. 2009;116(3):333–338. doi: 10.1007/s00702-009-0187-z. [DOI] [PubMed] [Google Scholar]
- 24.Bergman O, et al. PITX3 polymorphism is associated with early-onset Parkinson’s disease. Neurobiol Aging. 2010;31(1):114–117. doi: 10.1016/j.neurobiolaging.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Haubenberger D, et al. Association of transcription factor polymorphisms PITX3 and EN1 with Parkinson’s disease. Neurobiol Aging. 2011;32(2):302–307. doi: 10.1016/j.neurobiolaging.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Tang L, et al. Meta-analysis of association between PITX3 gene polymorphism and Parkinson’s disease. J Neurol Sci. 2012;317(1-2):80–86. doi: 10.1016/j.jns.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 27.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15(20):3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 28.Goers J, et al. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry. 2003;42(28):8465–8471. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- 29.Schell H, Hasegawa T, Neumann M, Kahle PJ. Nuclear and neuritic distribution of serine-129 phosphorylated alpha-synuclein in transgenic mice. Neuroscience. 2009;160(4):796–804. doi: 10.1016/j.neuroscience.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Scherman D, et al. Striatal dopamine deficiency in Parkinson’s disease: role of aging. Ann Neurol. 1989;26(4):551–557. doi: 10.1002/ana.410260409. [DOI] [PubMed] [Google Scholar]
- 31.Grace AA, Bunney BS. The control of firing pattern in nigral dopamine neurons: Single spike firing. J Neurosci. 1984;4(11):2866–2876. doi: 10.1523/JNEUROSCI.04-11-02866.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuke S, Kubota-Sakashita M, Kasahara T, Shigeyoshi Y, Kato T. Regional variation in mitochondrial DNA copy number in mouse brain. BBA Bioenergetics. 2011;1807(3):270–274. doi: 10.1016/j.bbabio.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.