Abstract
Tumor suppressor p53-binding protein 1 (53BP1) regulates the repair of dysfunctional telomeres lacking the shelterin protein TRF2 by promoting their mobility, their nonhomologous end-joining (NHEJ), and, as we show here, by blocking 5′ resection by CtIP. We report that these functions of 53BP1 required its N-terminal ATM/ATR target sites and its association with H4K20diMe, but not the BRCT domain, the GAR domain, or the binding of 53BP1 to dynein. A mutant lacking the oligomerization domain (53BP1oligo) was only modestly impaired in promoting NHEJ of dysfunctional telomeres and showed no defect with regard to the repression of CtIP. This 53BP1oligo allele was previously found to be unable to support class switch recombination or to promote radial chromosome formation in PARP1 inhibitor-treated Brca1-deficient cells. The data therefore support two conclusions. First, the requirements for 53BP1 in mediating NHEJ at dysfunctional telomeres and in class switch recombination are not identical. Second, 53BP1-dependent repression of CtIP at double-strand breaks (DSBs) is unlikely to be sufficient for the generation of radial chromosomes in PARP1 inhibitor-treated Brca1-deficient cells.
The DNA damage response factor 53BP1 is a key regulator of the processing and repair of double-strand breaks (DSBs) (reviewed in refs. 1–3). Accumulation of 53BP1 at sites of DNA damage depends on phosphorylation of H2AX by the ATM and/or ATR kinases, binding of MDC1 to phosphorylated H2AX (γ-H2AX), and ubiquitylation of H2A and/or H2AX by MDC1-dependent ubiquitin ligases. Despite its dependence on these ATM/ATR-initiated events, 53BP1 does not bind H2AX, H2A, MDC1, or their interacting factors. Instead, 53BP1 interacts with histone H4 through an association of its tandem Tudor domain with the dimethylated form of H4 lysine 20 (H4K20Me2) (4). H4K20Me2 is a constitutive modification that has been proposed to become more accessible near sites of DNA damage due to ubiquitin-dependent removal of H4K20Me2 binding proteins, thus explaining the dependence of 53BP1 accumulation on γ-H2AX, MDC1, and ubiquitin ligases (5–7).
The role of 53BP1 in DNA repair surfaced in the context of Ig class switch recombination (CSR) in which 53BP1 is essential for nonhomologous end-joining (NHEJ) of activation-induced (cytidine) deaminase-induced DSBs and has been implicated in the synapsis of DNA ends separated by as much as 200 kb (8, 9). 53BP1 also promotes NHEJ during V(D)J recombination, in particular when the recombining ends are far apart, suggesting that 53BP1 can mediate the juxtaposition of distant DNA ends and facilitate their joining (10). Similarly, 53BP1 promotes fusions of dysfunctional telomeres generated through deletion of the shelterin protein TRF2 (11). Such deprotected telomeres undergo classical-NHEJ (c-NHEJ) in G1, forming trains of fused chromosomes that can be visualized in the following metaphase (12, 13). Live-cell imaging showed that, before their joining, dysfunctional telomeres become more mobile and sample larger territories (11), as was recently also shown for other DSBs (14–16). Because this change in mobility depends on 53BP1, it was proposed that 53BP1 stimulates the fusion of telomeres by improving the chance of telomere–telomere encounters (11). How 53BP1 might promote the synapsis and/or mobility of DSBs has not been established.
53BP1 also affects DNA repair through regulating DSB resection. During V(D)J recombination, the unrepaired coding ends are degraded when 53BP1 is absent (10), and 53BP1 deficiency results in frequent resection of DNA ends generated by I-SceI (17, 18). End resection is also unleashed in the absence of 53BP1 at telomeres that are deprived of all shelterin components (19). The 53BP1-controlled resection is dependent on ATM signaling and involves the CtIP nuclease (17–19).
In cells lacking BRCA1 function, PARP1 inhibitors (PARPi) induce lethal radial chromosomes that are thought to result from mis-rejoined DSBs (20, 21). Deletion of 53BP1 prevents the formation of these aberrant chromosomes and rescues the lethality of Brca1 deficiency in the mouse (18, 22, 23). It has been proposed that the effect of 53BP1 in this context is due to its propensity to block resection of DSBs thus preventing formation of the 3′ extensions needed to initiate homologous recombination and favoring NHEJ.
Using dysfunctional telomeres as an experimental setting, we determined which domains of 53BP1 are involved in repressing the CtIP-dependent 5′ end resection taking place in S/G2 and the induction of chromatin mobility and NHEJ in G1. This analysis revealed that oligomerization of 53BP1, although crucial for CSR, plays a lesser role in the joining of telomeres, indicating mechanistic differences between these two 53BP1-dependent NHEJ pathways. Furthermore, we find that oligomerization of 53BP1 is not required for the repression of CtIP-mediated 5′ end resection at telomeres. Because the oligomerization mutant was previously shown to be defective in radial chromosome formation in PARPi-treated Brca1 null cells (24), we infer that it is unlikely that CtIP inhibition is the sole determinant of this attribute of 53BP1.
Results
53BP1 Blocks ATM- and CtIP-Dependent Resection at Deprotected Telomeres.
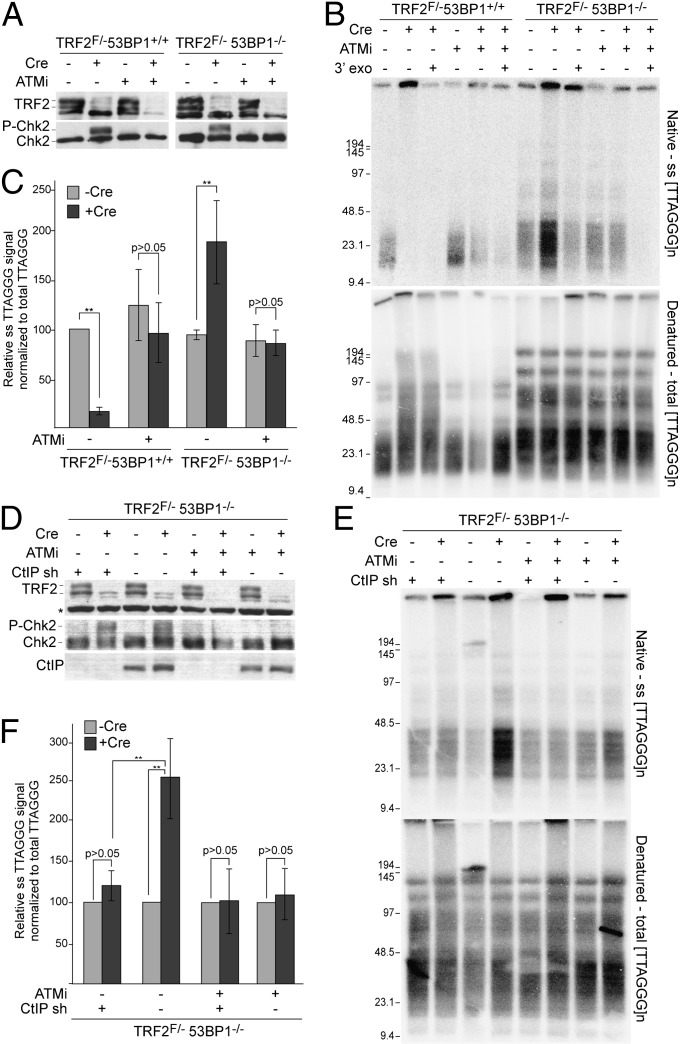
To test whether 53BP1 controls aberrant end-resection at telomeres lacking TRF2, we used a retroviral hit-and-run (H&R) Cre recombinase to delete TRF2 from SV40 large T antigen (SV40LT) immortalized TRF2F/-53BP1+/+ and TRF2F/-53BP1−/− mouse embryo fibroblasts (MEFs). As expected, TRF2 deletion elicited a DNA damage response in both 53BP1-proficient and -deficient MEFs, as evidenced by the phosphorylation of Chk2 (Fig. 1A). End resection was analyzed based on quantification of the single-stranded (ss) telomeric TTAGGG signal by in-gel hybridization to native DNA and normalization to the total telomeric signals obtained after DNA denaturation (Fig. 1 B and C). In the absence of 53BP1, TRF2 deletion results in a twofold increase in the normalized overhang signal. The ssTTAGGG repeat signal was derived from 3′ terminal sequences because it was sensitive to the Escherichia coli 3′ exonuclease ExoI (Fig. 1B). Parallel experiments with NHEJ-deficient Ku70−/− and Lig4−/− cells only showed a slight (<20%) increase in the telomeric overhang signal after TRF2 deletion (Fig. S1 A and B), indicating that the increased telomeric overhang signal in the 53BP1-null cells was not merely due to the diminished telomere fusions. Thus, 53BP1 protects telomeres lacking TRF2 from aberrant 5′ end resection. However, the extent of resection is less than that observed in the absence of the whole shelterin complex (19), indicating that, in addition to TRF2, other shelterin proteins protect telomeres from nucleolytic attack.
Fig. 1.
53BP1 protects dysfunctional telomeres from ATM/CtIP-dependent resection. (A) TRF2 and P-Chk2 immunoblots in TRF2F/-53BP1+/+(ATM+/−) and TRF2F/-53BP1−/− MEFs treated with KU55933 (ATMi) and H&R-Cre (72-h time point). (B) Telomeric overhang assay on the cells in A at 96 h after Cre. E. coli exonuclease ExoI was used to test for 3′ terminal ssDNA. (Upper) In-gel hybridization with 32P-(AACCCT)4 to native MboI/AluI-digested genomic DNA. (Lower) DNA was denatured in situ, and the gel was rehybridized with the same probe to determine the total telomeric signal. The ssTTAGGG signal was normalized to the total telomeric DNA in the same lane. (C) Quantification of normalized ssTTAGGG signals. Values represent means of three independent experiments with SDs. The normalized value was set at 100 for TRF2F/-53BP1+/+ cells not treated with Cre, and the other values are given relative to this value. (D) Immunoblots for CtIP, TRF2, and P-Chk2 in TRF2F/-53BP1−/− MEFs after treatment with CtIPsh, ATMi, and Cre (72 h) as indicated. (E) Representative telomeric overhang assays on TRF2F/-53BP1−/− MEFs treated as in D and assayed as in B. (F) Quantification of relative ss telomeric signal in TRF2F/-53BP1−/− MEFs. Values are means of three independent experiments with SDs. **P < 0.05 (unpaired Student t test).
We next asked whether the increased overhang signal was due to ATM/CtIP-mediated resection. Inhibition of ATM signaling prevented the overhang signal increase in response to deletion of TRF2 from 53BP1−/− cells (Fig. 1 B and C), in parallel with its expected effect on Chk2 phosphorylation and telomere fusions (Fig. 1 A–C and Fig. S1 A–C). The ssTTAGGG repeat signal was also significantly attenuated by a CtIP shRNA (Fig. 1 D–F). Consistent with ATM and CtIP acting in the same pathway, ATM inhibition did not further affect the overhang signal in CtIPsh-treated cells (Fig. 1 D–F). These data establish that ATM stimulates CtIP-dependent 5′ resection at dysfunctional telomeres, as it does at other sites of DNA damage (25–28), and that this resection is inhibited by 53BP1.
53BP1 Prevents Resection of Deprotected Leading-End Telomeres.
Two lines of evidence indicated that the ATM- and CtIP-dependent resection at dysfunctional telomeres in cells lacking 53BP1 preferentially affected the telomeres generated by leading-end DNA synthesis. First, chromosome orientation FISH (CO-FISH) showed that inhibition of ATM and/or CtIP induced frequent leading-end telomere fusions when TRF2 is deleted from 53BP1−/− cells (Fig. 2 A and B), whereas chromosome-type fusions were unaffected by ATM inhibition (Fig. 2B and Fig. S1 A–C). These leading-end telomere fusions were dependent on Ku70 and Lig4 (Fig. S1C), indicating that they are generated by c-NHEJ. Interestingly, these S/G2 c-NHEJ events do not require 53BP1, whereas those taking place in G1 are strongly diminished by the absence of 53BP1 (11).
Fig. 2.
53BP1 affects resection of leading-end telomeres. (A) CO-FISH to detect leading- and lagging-end telomeres in metaphases of TRF2F/-53BP1−/− MEFs with the indicated treatments at 96 h after Cre. Red, leading-end telomeres; green, lagging-end telomeres; arrows, leading-end telomere fusions. (B) Quantification of chromosome- and chromatid-type leading-end telomere fusions from metaphase analysis as shown in A. Values represent means of two experiments (>1,000 chromosomes per experiment) and SEMs. (C) Overhang assay of leading- and lagging-end telomeres of TRF2F/-53BP1−/− MEFs 96 h after TRF2 deletion in the absence or presence of ATMi as in Fig. 1A. Leading- and lagging-end telomeres were separated on CsCl gradients (Fig. S2B). (D) Quantification of relative overhang signals detected in C. Values represent means for three independent experiments and SDs. **P < 0.05 (unpaired Student t test).
A role for 53BP1 in blocking resection at leading-end telomeres also emerged from the analysis of separated leading- and lagging-end telomeres. Telomeres synthesized by leading- and lagging-strand DNA synthesis can be separated on CsCl density gradients based on their differential incorporation of BrdU (refs. 29 and 30; Fig. S2 A and B), and the ssTTAGGG repeat signal in fractions containing the newly synthesized leading- or lagging-end telomeres can then be evaluated by quantitative in-gel hybridization. TRF2 deletion in the absence of 53BP1 causes a threefold increase of the overhang of the leading-end telomeres, whereas the lagging-end telomeres were not affected (Fig. 2 C and D). The increase in the leading-end overhang signal was abolished upon inhibition of ATM (Fig. 2 C and D). Thus, dysfunctional telomeres that are replicated by leading-strand DNA synthesis are threatened by ATM-dependent 5′ resection, and this resection is inhibited by 53BP1.
Paradoxically, the ATM-dependent resection of the leading-end telomeres can be protective at telomeres in that it inhibits NHEJ, which is a major threat to chromosome ends (schematic in Fig. S3). We note that not all leading-end telomeres undergo fusion when CtIP fails to resect them. Whether these telomeres can be processed through a different pathway or persist without any 3′ overhang—and if the latter, how such blunt-ended telomeres might escape fusion—is not yet clear.
Based on our prior analysis of telomere fusions in TRF2/ATM double-knockout cells, we had previously speculated that the activation of ATM at TRF2-depleted telomeres might result in generation of a 3′ telomeric overhang, which can protect the telomeres from NHEJ (ref. 31 and Fig. S3 Top). Consistent with the data presented here, the protective effect of ATM-dependent resection appeared to be specific to leading-end telomeres because TRF2−/−ATM−/− cells showed leading-end telomere fusions (31). We attribute the leading-end specific effects of ATM, CtIP, and 53BP1 to the fact that leading-strand DNA synthesis is expected to generate a (nearly) blunt end that is vulnerable to NHEJ, whereas lagging-strand DNA synthesis would be expected to generate a product with a 3′ overhang. The presence of a short 3′ overhang may allow the POT1 proteins in shelterin to bind, thereby preventing NHEJ at the lagging-end telomeres (Fig. S3).
Functional Dissection of 53BP1.
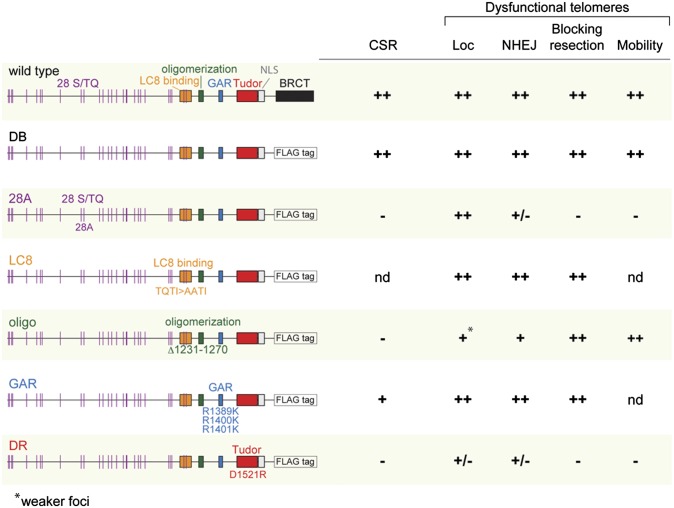
The data shown above establish telomeres deprived of TRF2 as an appropriate system to evaluate the role of 53BP1 in c-NHEJ, the associated increase in telomere mobility, as well as in the repression of CtIP-mediated end resection. We therefore used this system to test a panel of deletion and substitution mutants of 53BP1 (24) (Fig. 3 and Fig. S4 A and B). Mutation of all 28 N-terminal S/TQ sites that are potential targets for phosphatidylinositol 3-kinase-related kinase-mediated phosphorylation to AQ gave rise to an allele (53BP128A) that is deficient for CSR and fails to restore radial chromosome formation in Brca1/53BP1 double-mutant cells (24). In contrast, no strong CSR defects or other phenotypes were associated with three arginine-to-lysine substitutions in the GAR domain (referred to here as 53BP1GAR) (24). Deletion of amino acids 1,231–1,270, representing the oligomerization domain (referred to here as 53BP1oligo), resulted in a profound CSR defect and a strongly diminished ability to rescue radial fusions in Brca1/53BP1 mutant B cells (24). We mutated the LC8 binding domain of 53BP1 and showed the resulting mutant (1,171 TQTI>AATI) to be defective in interacting with dynein (Fig. S4C). Finally, the 53BP1DR mutant with a D1521R mutation in the tandem Tudor domain, which abrogates the association of 53BP1 with chromatin, did not support CSR and also failed to restore radial structure formation in 53BP1/Brca1 mutant B cells (24). Each of the mutations was created in the context of a human 53BP1 allele lacking the C-terminal BRCT motif. This truncated version of 53BP1 (53BP1DB) is indistinguishable from the wild-type protein in the context of CSR (24) and functions well at dysfunctional telomeres (Fig. S4 D–F) but is deficient for the repair of DSBs in heterochromatin (32, 33).
Fig. 3.
Summary of the phenotypes of 53BP1 mutants in CSR and at dysfunctional telomeres. 53BP1 mutants and their role in CSR were analyzed as the percentage of IgG1 switched cells obtained after stimulation of B cells (24). At dysfunctional telomeres, 53BP1 mutants were analyzed for TIF formation as shown in Fig. 4 (Loc), promotion of telomere fusion (NHEJ; Fig. 5), inhibition of hyperresection (blocking resection; Fig. 5), and promotion of telomere mobility (mobility; Fig. 5). For each column, the ability of the different mutants to perform the indicated specific function was compared with 53BP1DB: ++, fully functional; +, partially impaired; ±, strongly impaired; −, completely defective; nd, not determined.
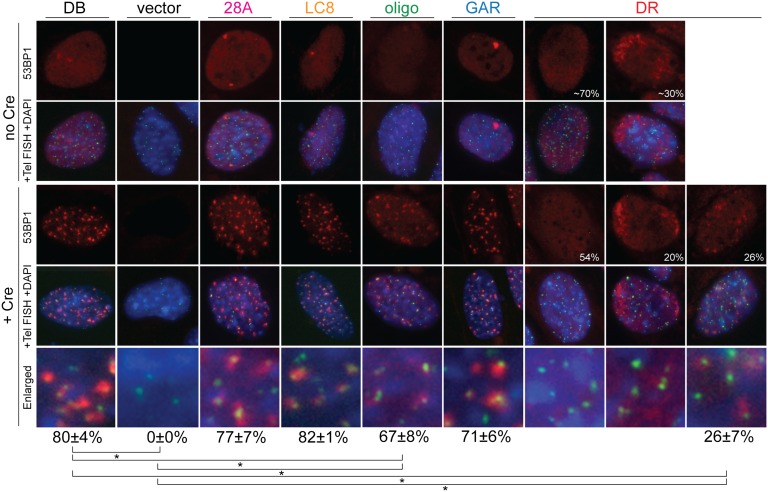
The mutant forms of 53BP1 were expressed at similar levels and did not affect the activation of Chk2 after TRF2 deletion from TRF2F/-53BP1−/− cells (Fig. S4B). With the exception 53BP1DR (see below), each of the 53BP1 alleles showed the anticipated diffuse nuclear localization in cells with functional telomeres (Fig. 4 Upper) and was readily detectable in the DNA damage foci formed at dysfunctional telomeres [referred to as telomere dysfunction induced foci (TIFs)] (ref. 34; Fig. 4 Lower). Quantification of 53BP1-containing TIFs indicated that the oligomerization mutant was slightly impaired with regard to its accumulation at deprotected telomeres (Fig. 4 and Fig. S5A). The TIFs formed by 53BP1oligo were slightly reduced in frequency, and the 53BP1 IF signal intensity in the foci appeared lower. Previous work with a different 53BP1 oligomerization mutant (D1256A) showed a dramatic reduction (∼10-fold) in its localization to dysfunctional telomeres and a commensurate reduction in its function (35). Given that the complete deletion of the oligomerization domain in 53BP1oligo does not have a strong effect on the localization to sites of DNA damage (Fig. 4) or chromatin (24), we suggest that the D1256A mutation may have affected additional aspects of 53BP1 (35).
Fig. 4.
Accumulation of 53BP1 alleles at dysfunctional telomeres. Shown is TIF assay on TRF2F/-53BP1−/− MEFs expressing the indicated 53BP1 alleles before (Upper) and 72 h after (Lower) Cre. 53BP1 mutants detected with an antibody to 53BP1 amino acids 350–400 (present in all alleles) (red) and telomeres detected with FITC-[CCCTAA]3 (green) are shown. DNA was stained with DAPI. Numbers in the DR images indicate percent of cells with the localization pattern shown. Numbers below the images indicate quantified TIF response for each allele scored based on localization of 53BP1 at five or more telomeres per cell [means of five independent experiments (>50 cells per experiment) and SDs]. *P < 0.05 (paired Student t test).
As expected, the 53BP1DR mutation in the tandem Tudor domain had the strongest effect on TIF formation by 53BP1, with only 26% of the cells showing detectable TIFs after TRF2 deletion and the few TIFs that formed showing very faint IF signals (Fig. 4 and Fig. S5B). Another 20% of the 53BP1DR expressing cells showed very strong and diffuse signals at one or both poles of the nuclei in cells that appeared to be undergoing division, but this pattern was independent of TRF2 deletion (Fig. 4 Upper).
Our findings on the diminished localization of 53BP1DR at dysfunctional telomeres are in agreement with our published data (11). However, another study on the localization of 53BP1DR reported that the Tudor domain mutation completely abrogated, rather than diminished, the ability of 53BP1 to associate with telomeres lacking TRF2 (35). We suggest that the discrepancy between this current study and the prior one may be due to differing detection levels of 53BP1, presumably due to differences in imaging protocols.
BRCT, LC8, and GAR Mutations Have Minimal Effects.
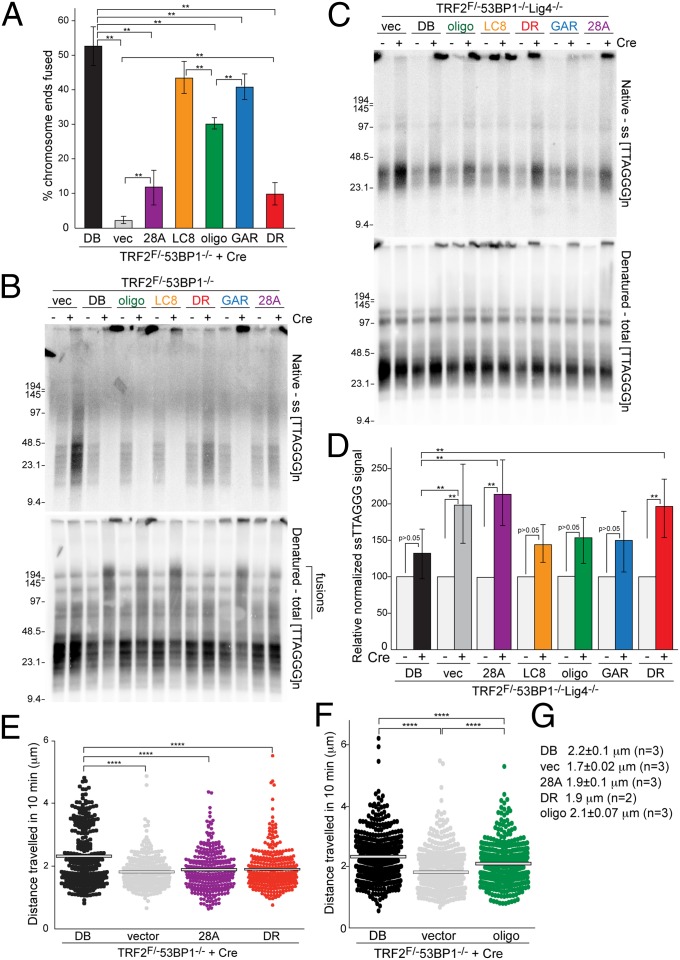
Deletion of the BRCT domain, mutation of the LC8 binding site, or mutation of the GAR motif did not strongly affect the processing of dysfunctional telomeres. Analysis of metaphase chromosomes showed a slight reduction (∼20%) in the rate of telomere fusions for the LC8 and GAR mutations, whereas absence of 53BP1 diminished the fusions by >90% (Fig. 5A and Fig. S6A). The telomere fusions promoted by the mutant alleles of 53BP1 were also readily detectable as larger telomeric restriction fragments in pulsed-field gel electrophoresis (PFGE) gels (Fig. 5B). In addition, a prior study showed that 53BP1 alleles with mutations in the GAR or BRCT domains were proficient for the fusion of dysfunctional telomeres (35).
Fig. 5.
53BP1 domains required for NHEJ, blocking resection, and promoting mobility. (A) Quantification of chromosome-type telomere fusions in TRF2F/-53BP1−/− MEFs with the indicated 53BP1 alleles 96 h after Cre. Means of five independent experiments (n > 1,000 chromosomes per experiment) and SDs are shown. **P < 0.05 (paired Student t test). (B) PFGE analysis of telomeric DNA of TRF2F/-53BP1−/− MEFs expressing the indicated 53BP1 alleles at 96 h after Cre. Quantification of the telomeric overhangs is shown in Fig. S6B. (C) Telomeric overhang assay on TRF2F/F53BP1−/−Lig4−/− MEFs with the indicated 53BP1 alleles. (D) Quantification of the telomeric overhang based on assays as shown in C. Values represent means of five independent experiments and SDs. The normalized value was set at 100 for TRF2F/F53BP1−/−Lig4−/− cells expressing 53BP1DB without Cre and all other values are given relative to this value. **P < 0.05 (paired Student t test). (E and F) Distribution of the cumulative distances traveled by all mCherry-BP1-2 foci per cell (average 40 foci cell; >10 cells per allele) in TRF2F/-53BP1−/− cells infected with the indicated 53BP1 alleles in one representative experiment. ****P < 0.05 (two-tailed Mann-Whitney test). The difference in the distances traveled in the presence of 53BP1DB allele in E and F is not significant. (G) Summary of data obtained with experiments shown as in E and F. Median distances traveled and SDs are given. The difference in distance traveled in cells expressing the DB and oligo alleles of 53BP1 is not statistically significant.
Because the telomere fusions in cells expressing 53BP1LC8, 53BP1DB, or 53BP1GAR masked effects on end resection (Fig. 5B and Fig. S6B), we introduced these alleles as well as 53BP1DB into TRF2F/F53BP1−/−Lig4−/− MEFs, in which telomeric fusions do not take place due to the absence of DNA ligase IV (12). In this setting, 53BP1LC8, 53BP1GAR, and 53BP1DB behaved similarly and prevented the increase in the ss telomeric DNA signal that is normally observed when TRF2 is deleted from 53BP1-deficient cells (Fig. 5 C and D). Therefore, we conclude that the BRCT domain, LC8 dynein binding domain, and the GAR motif are not required to protect the dysfunctional telomeres from 5′ end resection.
Requirement for the Tandem Tudor Domain and N-Terminal S/TQ Sites.
Different results were obtained with 53PB128A and 53BP1DR. Cells expressing these alleles showed a nearly fivefold reduction in telomere fusions after TRF2 deletion (Fig. 5A and Fig. S6A), and their genomic DNA showed scant evidence for joined telomeres when examined by PFGE (Fig. 5B). This result is consistent with our previous analysis of a different Tudor domain mutant made in the context of full-length 53BP1, which showed strongly diminished, but not absent, NHEJ of dysfunctional telomeres (11). Our finding of residual NHEJ of dysfunctional telomeres in cells expressing 53PB128A or 53BP1DR departs from findings by others who found no residual NHEJ of dysfunctional telomeres in cells expressing either a Tudor mutant of 53BP1 or an allele of 53BP1 mutated for 15 N-terminal ST/Q sites (35).
Neither 53BP1DR nor 53BP128A were capable of protecting telomeres from 5′ end resection. When telomere fusions were prevented in TRF2F/F53BP1−/−Lig4−/− MEFs, these alleles did not block the increase in the overhang signal after TRF2 deletion (Fig. 5 C and D).
We then tested the ability of these mutants to increase the mobility of the telomeric chromatin, which was previously shown to be associated with efficient telomere fusion (11). To this end, the dysfunctional telomeres were identified in living cells based on their decoration with an mCherry fusion protein containing the tandem Tudor domain of 53BP1, which localizes to sites of DNA damage (11). Time-lapse microscopy was performed at 72 h after introduction of Cre, and individual mCherry foci were tracked so that the cumulative distance traveled in 10 min could be calculated for each dysfunctional telomere in the cell. The results indicated that, although dysfunctional telomeres in cells expressing 53BP1DB showed the expected increase in mobility compared with 53BP1-deficient controls, neither 53BP1DR nor 53BP128A were capable of promoting the mobility of dysfunctional telomeres (Fig. 5 E and G).
Oligomerization of 53BP1 Mildly Affects Telomeric NHEJ but Not End Resection.
The 53BP1oligo mutant also did not behave as a wild-type allele, but its defects in the setting of dysfunctional telomeres were subtle. The frequency of telomere fusions in cells expressing the 53BP1oligo mutant was reduced by 30–40% compared with the control, and this reduction was mostly due to the absence of cells with the longest multichromosome fusion products (Fig. 5 A and B and Fig. S6 A and C).
Interestingly, although 53BP1oligo was partially defective in promoting NHEJ at dysfunctional telomeres, the allele appeared fully functional with regard to blocking 5′ end resection. This attribute was most obvious in the context of Lig4-deficient cells from which TRF2 was deleted (Fig. 5 C and D). The change in the 3′ telomeric overhang observed after TRF2 loss was minimal in cells expressing 53BP1oligo similar to that observed for cells expressing 53BP1DB, 53BP1LC8, or 53BP1GAR.
Given that 53BP1oligo was proficient in repressing CtIP yet had a modest NHEJ defect, we tested its ability to promote the increased mobility of dysfunctional telomeres. Interestingly, the mutant resulted in an increase in telomere mobility that was very close to that for 53BP1DB (Fig. 5 F and G). Thus, the diminished NHEJ of the dysfunctional telomeres observed with the 53BP1oligo mutant is unlikely to be due to a deficiency in telomere mobility.
Discussion
Dysfunctional telomeres proved a versatile system for the rapid dissection of the function of 53BP1 in the context of its ability to promote NHEJ and protect DNA ends from resection by CtIP. These two functions do not require the GAR, LC8, or BRCT domains, although it is possible that redundancies exist that hide contributions of these parts of 53BP1. The tandem Tudor domain of 53BP1 is paramount for its localization to dysfunctional telomeres (11), but even without the full engagement of this domain, 53BP1 has some ability to promote NHEJ of dysfunctional telomeres. This result suggests that once 53BP1 has arrived at a site of damage, perhaps through a secondary interaction, it can promote telomere fusions in a manner that is largely independent of the tandem Tudor domain. Despite the normal accumulation of 53BP128A at dysfunctional telomeres, this allele is functionally inactive with regard to promoting telomere mobility, NHEJ, and blocking the ATM-dependent resection of leading-end telomeres by CtIP (Fig. 3). The phenotypes observed in the 53BP128A mutant strongly suggest that ATM-dependent phosphorylation of 53BP1, although not required for its localization, is essential for its function, likely serving as recognition sites for other factors involved in blocking resection and promoting mobility. The behavior of this set of mutants closely correlates with their activity in the context of CSR where the tandem Tudor domain and the N-terminal ST/Q sites are required, whereas the GAR and BRCT domains are not.
The most informative mutant analyzed in this study is the allele lacking the oligomerization domain, 53BP1oligo. This mutant shows a moderate defect for the fusion of dysfunctional telomeres. Because the residual activity of 53BP1oligo in the NHEJ of dysfunctional telomeres is substantial, it highlights a contrast with its performance in the context of CSR in which this mutant behaves as a null allele (24). The simplest interpretation of this difference is that the activities that mediate CSR and joining of dysfunctional telomeres are only partially overlapping. Specifically, as proposed previously, CSR may require a synapsis of DSBs that is mediated by oligomerization of 53BP1, whereas the NHEJ of dysfunctional telomeres may be more reliant of an increase in their mobility, which is less influenced by the oligomerization state of 53BP1. However, because CSR is analyzed in B cells, whereas telomere fusions are best studied in fibroblasts, it is also possible that the distinct outcomes with 53BP1oligo are due to unforeseen cell-type-specific aspects of 53BP1.
The ability of 53BP1oligo to repress CtIP at dysfunctional telomeres appears equivalent to wild-type 53BP1. This observation, which we assume extends to other sites of DNA damage, may be informative with regard to the role of 53BP1 in the Brca1-deficient cells. When Brca1/53BP1-null cells are complemented with 53BP1oligo instead of wild-type 53BP1, PARPi treatment does not induce radial chromosomes (24). If its action at dysfunctional telomeres indeed reflects the behavior of 53BP1 at other sites of DNA damage, our results would suggest that inhibition of CtIP is not sufficient for radial chromosome formation in Brca1-deficient cells. Rather, the formation of radial chromosomes may be due to a combination of other NHEJ-promoting aspects of 53BP1 in addition to its ability to repress CtIP and other nucleases.
Materials and Methods
Cell Lines, Plasmids, and shRNA.
TRF2F/-ATM+/−, TRF2 F/-ATM−/−, TRF2F/-Lig4−/−, TRF2F/-Ku70−/−, TRF2F/-53BP1−/−, and TRF2F/F MEFs have been described (11, 12). The TRF2F/F53BP1−/−Lig4−/− MEFs were isolated from embryonic day (E)12.5 embryos from TRF2F/F53BP1+/−Lig4+/− intercrosses, immortalized with pBabeSV40LargeT (a gift from G. Hannon, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) at passage P2, and cultured as described (11, 12). MEFs were infected two or three times at 12-h intervals with pMMP H&R Cre retrovirus as described (12) with experimental time point 0 set at 12 h after the first infection. ATM was inhibited with 2.5 μM KU55933 (TOCRIS Bioscience). Retroviruses carrying CtIP shRNA and 53BP1 mutant alleles (18, 24) were introduced into MEFs by using six infections at 6- or 12-h intervals, followed by 3–5 d of selection in 2–3 μM puromycin before Cre infections.
Immunoblotting, IF, and IF-FISH.
Immunoblotting was performed as described (12) with the following primary antibodies: TRF2 (1254, rabbit polyclonal); Chk2 (mouse monoclonal; BD Biosciences); CtIP (rabbit polyclonal; Santa Cruz H-300); 53BP1(100-304A, rabbit polyclonal; Novus Biologicals); myc (9B11, mouse monoclonal; Cell Signaling); flag (F1804-M2, mouse monoclonal; Sigma). For IF-FISH, cells on coverslips were fixed for 10 min in 3% (wt/vol) paraformaldehyde and 2% (wt/vol) sucrose at room temperature and IF-FISH was carried out as described (12), using primary 53BP1 antibody (see above) and FITC-OO-[CCCTAA]3 PNA telomere probe (Applied Biosystems). DNA was counterstained with DAPI, and slides were mounted with ProLong Gold antifade (Sigma).
FISH, CO-FISH, Overhang Assay.
FISH and CO-FISH were performed as described (12) by using Tamra-OO-[TTAGGG]3 and FITC-OO-[CCCTAA]3 PNA probes. MEFs were harvested at 96 h after Cre following 2 h in 0.2 μg/mL colcemid (Sigma). For CO-FISH, cells were grown in 10 μM BrdU:BrdC (3:1) for 13–14 h before colcemid was added. Telomeric overhangs and telomeric restriction fragment patterns were analyzed 96 h after Cre by in-gel hybridization with a γ-[32P]ATP end-labeled [AACCCT]4 probe as described (12). ImageQuant software was used to quantify the single-stranded telomere overhang signals and the signal from total telomeric DNA in the denatured gel.
Live-Cell Imaging.
Dysfunctional telomeres were visualized based on their association with mCherry-BP1-2 (11). TRF2F/-53BP1−/− MEFs with 53BP1 mutant alleles were plated 24 h after Cre treatment onto MatTek glass-bottom plates and imaged for 10 min at 66–72 h after Cre. The tracking analysis of mCherry-BP1-2 foci was performed with ImageJ software for at least 10 cells for each genotype with the following parameters for particle detection and tracking: radius = 1–2 pixels; cutoff = 1–2 pixels; percentile = 1–6; link range = 1; displacement = 5 pixels. Only mCherry-BP1-2 foci continuously tracked for at least 18 out of 20 frames were considered.
Supplementary Material
Acknowledgments
We thank Devon White for expert mouse husbandry; Peng Wu for instruction on separation of leading- and lagging-end telomeres; Nadya Dimitrova for instruction on live-cell imaging of dysfunctional telomeres; and members of the T.d.L. and M.C.N. laboratories for helpful comments on the manuscript. This work was supported by grants from the National Institutes of Health (to T.d.L. and M.C.N.) and by a grant from Breast Cancer Research Foundation (to T.d.L.). M.C.N. is a Howard Hughes Medical Institute investigator. T.d.L. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222617110/-/DCSupplemental.
References
- 1.Noon AT, Goodarzi AA. 53BP1-mediated DNA double strand break repair: Insert bad pun here. DNA Repair (Amst) 2011;10(10):1071–1076. doi: 10.1016/j.dnarep.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13(10):1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 4.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acs K, et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18(12):1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- 6.Mallette FA, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31(8):1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart GS. The demise of a TUDOR under stress opens a chromatin link to 53BP1. EMBO J. 2012;31(8):1847–1849. doi: 10.1038/emboj.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165(4):459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5(5):481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 10.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456(7221):529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456(7221):524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7(7):712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 13.Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22(9):1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krawczyk PM, et al. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci. 2012;125(Pt 9):2127–2133. doi: 10.1242/jcs.089847. [DOI] [PubMed] [Google Scholar]
- 15.Miné-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14(5):510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 16.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol. 2012;14(5):502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 17.Bothmer A, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207(4):855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336(6081):593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 21.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 22.Bunting SF, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46(2):125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17(6):688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42(3):319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280(46):38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 26.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28(1):134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell. 2006;21(3):427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Wu P, Takai H, de Lange T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150(1):39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrova N, de Lange T. Cell cycle dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of NHEJ in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol. 2009;29:5552–5563. doi: 10.1128/MCB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, Goodarzi AA, Jeggo PA, Paull TT. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J. 2010;29(3):574–585. doi: 10.1038/emboj.2009.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noon AT, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12(2):177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 34.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13(17):1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 35.Rai R, et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010;29(15):2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.