Abstract
Photosynthesis uses chlorophylls for the conversion of light into chemical energy, the driving force of life on Earth. During chlorophyll biosynthesis in photosynthetic bacteria, cyanobacteria, green algae and gymnosperms, dark-operative protochlorophyllide oxidoreductase (DPOR), a nitrogenase-like metalloenzyme, catalyzes the chemically challenging two-electron reduction of the fully conjugated ring system of protochlorophyllide a. The reduction of the C-17=C-18 double bond results in the characteristic ring architecture of all chlorophylls, thereby altering the absorption properties of the molecule and providing the basis for light-capturing and energy-transduction processes of photosynthesis. We report the X-ray crystallographic structure of the substrate-bound, ADP-aluminium fluoride–stabilized (ADP·AlF3-stabilized) transition state complex between the DPOR components L2 and (NB)2 from the marine cyanobacterium Prochlorococcus marinus. Our analysis permits a thorough investigation of the dynamic interplay between L2 and (NB)2. Upon complex formation, substantial ATP-dependent conformational rearrangements of L2 trigger the protein–protein interactions with (NB)2 as well as the electron transduction via redox-active [4Fe–4S] clusters. We also present the identification of artificial “small-molecule substrates” of DPOR in correlation with those of nitrogenase. The catalytic differences and similarities between DPOR and nitrogenase have broad implications for the energy transduction mechanism of related multiprotein complexes that are involved in the reduction of chemically stable double and/or triple bonds.
Keywords: dynamic switch protein, electron transfer
The biosynthesis of chlorophylls is essential for the capture of global energy. This complex, multienzymatic process generates chlorophyllide a (Chlide) through the stereospecific reduction of the C-17=C-18 double bond of ring D in protochlorophyllide a (Pchlide) (Fig. 1A). Two completely unrelated enzymes have evolved for Pchlide reduction: a monomeric, light-dependent system (1), found in angiosperms and cyanobacteria, and the dark-operative protochlorophyllide oxidoreductase (DPOR), found in anoxygenic photosynthetic bacteria, cyanobacteria, algae, and gymnosperms (2). DPOR is a two-component metalloprotein comprising an ATP-dependent reductase (L2) and a catalytic unit [(NB)2], both sharing a substantial degree of structural and sequence identity with nitrogenase (Fig. 1B) (3, 4). As in nitrogenase, both components of DPOR carry redox active metallocenters (5–8), which mediate the ATP-driven electron transfer from L2 to the site of substrate reduction in (NB)2. L2 and (NB)2 are only transiently associated with each other during catalysis (9), and ATP hydrolysis triggers their association and dissociation, permitting control of the timing of the accompanying electron transfer process between the two proteins. Previously determined structures of L2 and (NB)2 (5–7) provided a static picture of DPOR catalysis. However, only the structural investigation of the “trapped” ternary transition state complex allows for the molecular understanding of DPOR protein dynamics and subcomplex interaction.
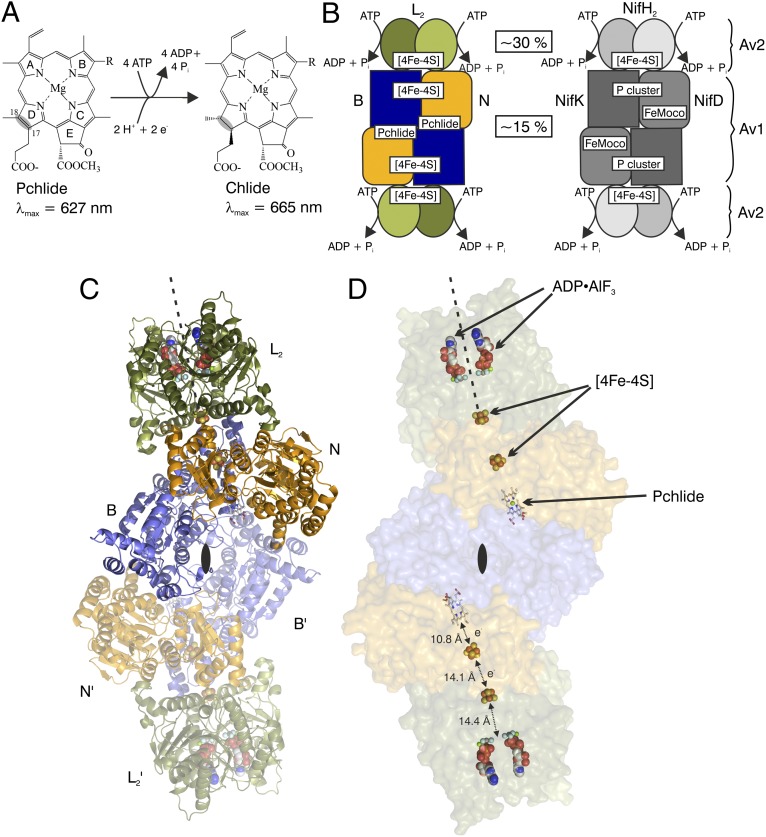
Fig. 1.
Catalysis and 3D structure of DPOR complex (L2)2(NB)2. (A) DPOR catalyzes the formation of Chlide through ATP-dependent, stereospecific reduction of the C-17=C-18 double bond of Pchlide ring D. (B) Schematic representation of DPOR (Left) and nitrogenase (Right) complexes. L2 and NifH2 (also named Av2) both contain a subunit-bridging [4Fe–4S] cluster, whereas the [4Fe–4S] cluster at the N/B subunit interface of (NB)2 is located in an analogous position as the [8Fe–7S] P-cluster at the NifD/NifK subunit interface of (NifDK)2 (Av1). Sequence identities between the subunits are shown in boxes. (C) Structure of the octameric DPOR complex. Subunits N and B (orange and blue, respectively) are responsible for Pchlide binding. Presence of the ATP analog ADP•AlF3 triggers the binding of two L2 protein dimers (green). The dyad axis of L2 is shown as a broken line. The overall (L2)2(NB)2 complex shows perfect symmetry as indicated by the twofold axis (black lenses); subunits of the symmetry-related protomer are marked L′, N′, and B′ and rendered transparent. (D) Cofactors and the substrate of DPOR are highlighted in a transparent surface representation of the octameric DPOR complex. Edge-to-edge distances are indicated.
Results and Discussion
The crystal structure determination of the 360 kDa DPOR complex from the marine cyanobacterium Prochlorococcus marinus (10) is summarized in Table S1, and the heterooctameric complex is depicted in Fig. 1. Subunits N and B are structurally homologous, generating a pseudo-twofold symmetry axis that is colinear with the molecular twofold axis of L2 (Fig. 1C). Both [4Fe–4S] clusters are centered around this extended axis: the L2 cluster is symmetrically ligated by four cysteinyl ligands between the two subunits, whereas the NB cluster is asymmetrically ligated by three cysteine residues from N and one aspartate residue from B. Apparently, the docking of L2 on NB induces a linear arrangement of the two redox-active clusters with the substrate, Pchlide (Fig. 1D). Moreover, upon complex formation, the two [4Fe–4S] clusters are brought to a distance of 14.1 Å, which is substantially shorter than the distance calculated on the basis of the noncomplexed structure of (NB)2 upon theoretical rigid-body docking of L2 (5). The distance between the NB cluster and the conjugated Pchlide ring system, by contrast, remains constant at ∼11 Å both in the absence and presence of L2. Clearly, electron transfer in DPOR is mediated through the spatial organization of the redox-active [4Fe–4S] centers and the substrate.
Biochemical studies identified L2 as a dynamic switch protein (9) that links ATP hydrolysis to conformational changes in the protein. Two different states of the L2 protein were characterized biochemically: the “on state” is induced by ATP analogs and has a high affinity for (NB)2; the “off state” is generated in the presence of ADP and does not form a complex with (NB)2. The existence of the two states of L2 is further supported by structural comparison of L2 within the octameric complex with the free form of a related L2 protein and the free or complexed form of NifH2 of nitrogenase (Table S2), which strongly indicates a parallelism between the switch mechanism of DPOR and nitrogenase.
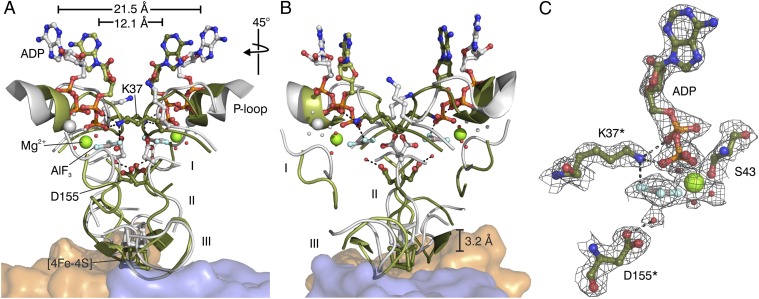
Formation of the DPOR complex leads to a more compact overall structure of L2. The underlying structural movements can be described as a rotation of the nucleotide-bound L monomers toward the dimer interface (Fig. 2), which reveals a striking degree of sequence conservation (Fig. S1). The pronounced intersubunit rearrangement is mainly triggered by critical interface residues in direct response to a status change of the bound nucleotides (Fig. 2C, Table S3). The AlF3-moiety of the nucleotide analog represents the trigonal bipyramidal γ-phosphate transition state during ATP hydrolysis. The adjacent activating Mg2+ ion is octahedrally coordinated. Amino acid residues Asp155* and Lys37* are provided by the second L monomer. Asp155* is responsible for positioning and/or activating a specific water molecule for the subsequent ATP hydrolysis, whereas Lys37* of the P-loop possibly assists the release of γ-phosphate upon ATP hydrolysis. In the ADP-bound state, both amino acids are involved in contacts within the L monomer [Protein Data Bank (PDB) ID code 3FWY]. In both the ADP•AlF3– and the ADP-bound structure, conserved contacts are responsible for binding of the adenosine moiety of the nucleotide. The nucleotide-dependent signal transduction involves several key regions of L2 (Fig. S1). First, there is a so-called “switch I region” (Gly64–Thr72). Located near the “terminal phosphate” of the nucleotide analog, this region shows a significant positional Cα deviation upon “conversion” of L2 from the “off state” to the “on state” (Fig. 2), indicating that it may play a key role in communicating the nucleotide state to the docking loop (Ile84–Glu96). Second, there is a so-called “switch II region” (Asp151–Phe161), where Asp155 and Cys158 (one ligand of the [4Fe–4S] cluster in L2) reside. It was demonstrated earlier that deletion of Leu153 in the switch II region resulted in a conformation of L2 protein that was “arrested” in a nondissociating complex with (NB)2 (9), suggesting that this region likely communicates the nucleotide status to the [4Fe–4S] cluster of L2. Both switch I and switch II are conserved in nucleotide-binding proteins that are more distantly related to L2, such as G proteins or myosin (11). Third, peptides Met79–Glu96 and Pro118–Gly126, including ligand Cys124 of the [4Fe–4S] cluster in L2 undergo major structural rearrangements to accommodate the dynamic repositioning of the [4Fe–4S] cluster of L2 upon complex formation, allowing for rapid electron transfer to the [4Fe–4S] cluster of NB (Fig. 2).
Fig. 2.
Dynamic switch mechanism of DPOR. (A) Superposition of the “on state” conformation of L2 in the ADP•AlF3–stablized complex (green) and the “off state” conformation of L2 in the ADP-bound state (gray, PDB ID code 3FWY). Nucleotide-dependent conformational rearrangements trigger the affinity of L2 for the (NB)2 core (orange and blue). The [4Fe–4S] cluster of L2 moves ∼3 Å toward the [4Fe–4S] cluster of (NB)2. Peptide segments undergoing large Cα-rearrangements are marked I (Asp66–Asp70 of switch I), II (Leu154–Cys158 of switch II), and III (loop region Pro118–Gly126). Regions not involved in significant conformational changes are omitted for clarity. The overall movement of each L subunit toward the L2 dimer interface becomes evident by a 9.4 Å decrease of the distance between the ADP molecules (measured between the N3-atoms of the two adenine bases in the “off state” and the “on state” of L2). Relevant water molecules are shown as red spheres. (B) Identical superposition after a 45° clockwise rotation. (C) Binding of ADP•AlF3 to L2 in the octameric DPOR complex. Residues provided by the second L monomer are indicated by asterisks. The 2Fo–Fc electron density is contoured at 1.5 σ.
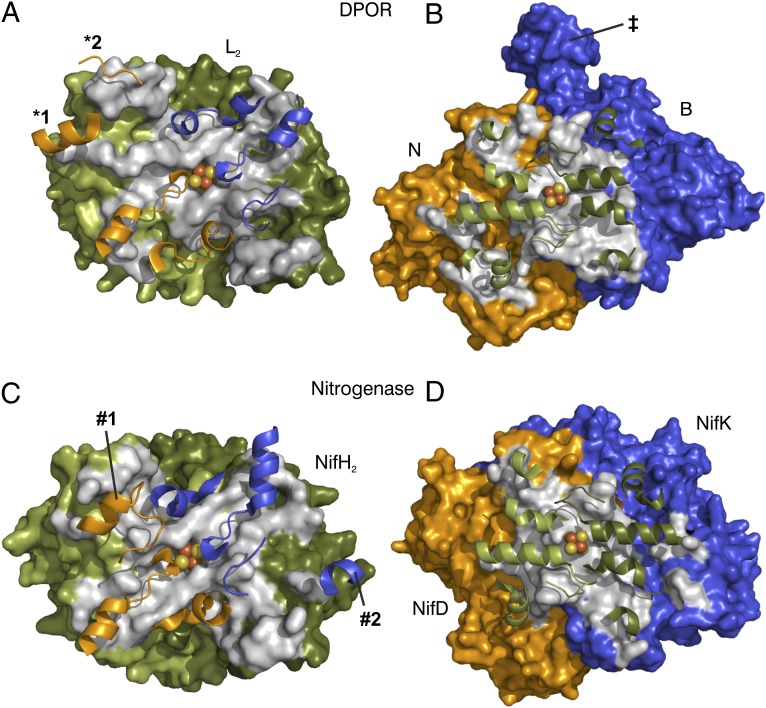
The L2/NB protein interface area of each DPOR half-octamer measures ∼1,900 Å2. A comparison of the docking interfaces of DPOR and an ADP•AlF4−–stabilized nitrogenase complex (12)—supported by structure-based sequence alignments—shows that residues of L2 (NifH2) that contact the surface of subunits N (NifD) and B (NifK) are conserved with respect to their positions both in the amino acid sequence and in the 3D structure (Fig. 3 B and D, Figs. S1–S3). Interestingly, the amino acid residues themselves are not conserved between the two enzymes, which may be nature’s design to prevent deleterious “cross-talks” between different biological systems (7, 13). The spatial distributions of the protein surfaces of L2 and NifH2 that are involved in the binding of (NB)2 and (NifDK)2, respectively, are largely conserved between DPOR and nitrogenase (Fig. 3 A and C). Conversely, only some central secondary structure elements of (NB)2 and (NifDK)2 are equivalently involved in binding L2 and NifH2. However, outside this core docking region, there are nonrelated secondary structure elements of (NB)2 that contribute to the complex formation, such as an additional helix (Thr218–Asp226) and an elongated loop (Tyr319–Glu323) in subunit N (*1 and *2, respectively, in Fig. 3A). Additionally, in nitrogenase, a loop followed by a helix in NifD (Arg182–His196) and a helical segment in NifK (Glu299–Lys303) (#1 and #2, respectively, in Fig. 3C) participate in NifH2 docking; equivalent elements are not found at the interface of the DPOR complex. It is important to note that the additional helix in DPOR subunit N (*1 in Fig. 3A) corresponds to the additional helix of nitrogenase subunit NifK (#2 in Fig. 3C) when the pseudo-twofold rotational symmetry of the complex is applied. Thus, as far as the docking interface is considered, subunit N would correspond to NifK (rather than to NifD) and vice versa. This observation seems to contradict the analyses based on the sequence alignment and the positioning of the subunits in the tetrameric (NB)2 core (Fig. 1 and ref. 5), both of which suggest the respective association of N to NifD and B to NifK. Taken together, these results imply that the subunits of both complexes could have diverged from a more symmetric predecessor.
Fig. 3.
Protein–protein interaction surfaces for transition state complexes of DPOR and nitrogenase (PDB ID code 1M34). (A) Van der Waals surface of L2 viewed along the pseudo-twofold rotational symmetry axis from (NB). (B) Van der Waals surface of (NB) viewed along the twofold rotational symmetry axis from L2. (C) NifH2 surface (chains E and F) viewed in the same orientation as in A. (D) (NifDK) surface (chains A and B) viewed in the same orientation as in B. Colors are as follows: subunit L chain A (NifH chain F), dark green; subunit L chain B (NifH chain E), light green; subunit N (NifD), orange; subunit B (NifK), blue. Surface areas of residues involved in protein–protein interactions are shown in gray. In all cases, only half of the core tetramer is depicted as one functional unit for clarity. Key secondary structure elements of the docking partner are displayed as cartoon. [4Fe–4S] clusters located on L2 and NifH2 are represented by van der Waals spheres. Secondary structural elements exclusive to the docking of DPOR (*1 and *2, both in subunit N) or nitrogenase (#1 in NifD and #2 NifK), as well as the C-terminal extension of DPOR subunit B (‡), are labeled.
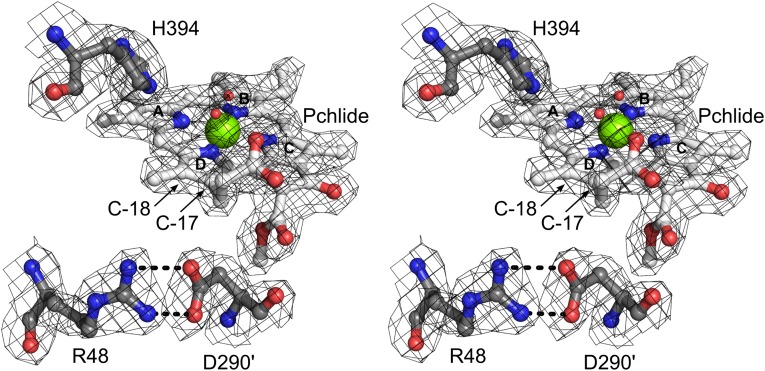
The binding of Pchlide to (NB)2 protein was recently demonstrated as an initial step toward efficient DPOR complex formation (9). The ring system of Pchlide is buried in a cavity formed mainly by hydrophobic amino acid residues. Therefore, significant conformational changes are required for the channeling of substrate into this pocket. Consistent with the earlier observation of a partial unwinding of a helical segment of chain B′ in the orthologous Rhodobacter capsulatus (NB)2 upon Pchlide binding (6), the same unwinding of a segment between residues Pro421 and Gly425 can be observed in the ADP•AlF3–stabilized DPOR complex. Moreover, a C-terminal domain of subunit B (His480–Phe528, Fig. 3B) closes the substrate-binding cavity at the NB interface, possibly preventing the organism from Pchlide-induced photodynamic damage. With regard to the regio- and stereo-specific protonation of Pchlide, structural and mutational analyses (Tables S4 and S5) suggest that residue Asp290′ of subunit B′ is directly responsible for the protonation at C-17, in agreement with a recent R. capsulatus study (6). The present investigation further reveals that the accurate positioning of Asp290 is ensured via salt bridge formation with Arg48 (Fig. 4). Based on the noncomplexed R. capsulatus structure, it was proposed that the C-17 propionate of the substrate can directly function as the proton donor in the trans-specific protonation at C-18 (6). However, the moderately retained activity upon His394Ala mutation points to a critical role of this residue in the specific protonation at C-18, probably by positioning a water molecule at a distance of 3.2 Å from C-18 above the ring (Fig. 4). These structural and biochemical data suggest a C-18 protonation mechanism via this water molecule, assisted by combined interaction with residue His394 and the C-17 propionate of Pchlide.
Fig. 4.
Stereoview of Pchlide binding in the ADP•AlF3–stabilized DPOR complex. Three polar amino acid residues are relevant for protonation at C-17 and C-18: residues His394 and Arg48 are located on one B subunit, whereas Asp290′ is provided by the other B subunit. The water molecule in close proximity to C-18 may act as a proton donor, whereas a second water molecule serves as an axial ligand to the central magnesium. The 2Fo–Fc electron density is contoured at 1.0 σ.
The Pchlide-binding site in DPOR can be considered as being loosely analogous to the FeMoco-binding site in nitrogenase, as the catalytic events at both sites involve the reductive protonation of double- and/or triple-bond substrates. Interestingly, the substrate-free DPOR can catalyze the two-electron reduction of N3− or N2H4 to NH3, which mirrors the ability of a FeMoco-deficient “apo nitrogenase” variant to catalyze the same reactions (Table 1). However, despite sharing the two “simple” nitrogenase substrates, substrate-free DPOR or “apo nitrogenase” is not capable of reducing “complex” nitrogenase substrates, such as N2 (14) and CO (15, 16), which require the transfer of more than two electrons (Table 1). The narrow substrate spectrum of these two systems likely originates from the presence of [4Fe–4S]–type clusters instead of a high-nuclearity [8Fe–7S] cluster at their respective “P-cluster” sites: DPOR has a [4Fe–4S] cluster at the N/B interface, whereas apo nitrogenase variant has a pair of [4Fe–4S]–like clusters at the NifD/NifK interface (17). Nevertheless, the striking similarities between the structure and function of DPOR and nitrogenase point to an evolutionary link between the two enzyme systems and suggest the possibility to use DPOR as an important platform to gain insights into the mechanisms of both chlorophyll biosynthesis and nitrogen fixation.
Table 1.
Specific substrate-reducing activities of substrate-free DPOR, apo nitrogenase variant, and holo nitrogenase
| Enzyme | Activities (nmol⋅min−1⋅mg−1) |
|||||
| Chlide formation from 0.02 mM Pchlide | NH3 formation from 40 mM N3− | NH3 formation from 40 mM N2H4 | NH3 formation from 100% N2 | C2H4 formation from 60% C2H2 | C2H4 formation from 100% CO | |
| Substrate-free DPOR | 1.1 ± 0.1 | 1.2 ± 0.1 | 1.07 ± 0.01 | 0 (0) | 0 (0) | 0 (0) |
| Apo nitrogenase variant | 0 (0) | 2.4 ± 0.3 | 0.4 ± 0.2 | 0 (0) | 3.3 ± 0.5 | 0 (0) |
| Holo nitrogenase | 0 (0) | 624 ± 16 | 263 ± 2 | 481 ± 19 | 2216 ± 145 | 0.013 ± 0.001 |
Specific substrate-reducing activities of substrate-free DPOR, apo nitrogenase variant, and holo nitrogenase are shown. Substrate-free DPOR consists of L2 and (NB)2, whereas apo nitrogenase variant consists of NifH2 and apo (NifDK)2 variant. Data are presented as mean ± SD (n = 4). The lower detection limits were 0.08, 0.0005, and 0.001 nmol⋅min−1⋅mg−1 of protein for Chlide-, C2H4-, and NH3-formation, respectively.
Materials and Methods
Protein Crystallization.
DPOR subcomplexes L2 and (NB)2 were purified under anoxic conditions as described earlier and the ternary complex was assembled (9). A total of 51 mg (431 nmol) of purified L2 protein fused at the N terminus to GST were immobilized on 21 mL glutathione agarose. Complex formation was initiated by adding 54 mg (259 nmol) pure (NB)2 and 1.3 µmol Pchlide in the presence of 40 mM NaF, 1.6 mM AlCl3, and 8 mM NaADP. After washing with a buffer containing 100 mM Hepes/NaOH (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 50 mM NaF, and 2 mM AlCl3, the green colored (L2)2(NB)2 complex was proteolytically released by PreScission protease (GE Healthcare) and concentrated to 9 mg/mL. DPOR crystals were prepared in sitting drops by vapor diffusion at 17 °C under anoxic conditions. A total of 480 conditions were screened. Crystallization drops contained 1 µl reservoir solution and 1 µl of the (L2)2(NB)2 complex solution (diluted to 7.5 mg/mL with washing buffer). Plates were stored in the dark. Green crystals appeared after 3–4 wk using a reservoir of 0.1 M KCl, 0.1 M Tris (pH 8.5), and 3% (wt/vol) PEG 6000. For cryoprotection, crystals were soaked in 15% (wt/vol) glycerol, 13% (wt/vol) PEG 8000, 5 mM DTT, 10 µM Pchlide, 62 mM NaCl, 4 mM MgCl2, 10 mM NaF, 208 µM AlCl3, and 0.17% (vol/vol) DMSO in 142 mM Hepes/NaOH (pH 7.5). Crystals were frozen in liquid nitrogen.
Structure Determination.
X-ray diffraction data were collected at beamline 14.2 of BESSY II of the Helmholtz–Zentrum Berlin (HZB), Germany, at a wavelength of 0.91841 Å at 100 K. Data were processed in space group C2 with XDS and scaled with XSCALE (18), whereas upper resolution limits were assessed through the observation of I/σI and Rmerge. Data were extended to 2.1 Å resolution along a* and c* but only to 2.8 Å along b*, and they were anisotropy corrected using the anisotropy correction server (19). The resulting dataset showed reasonable completeness to 2.6 Å resolution. However, incomplete data to 2.15 Å resolution were included in later calculations for more detailed maps. Molecular replacement (MR) with phenix.auto_mr (20) using monomers of L from Rhodobacter sphaeroides (PDB ID code 3FWY) and N and B from Thermosynechococcus elongatus (PDB ID code 2XDQ) as search models after pruning with chainsaw (21) yielded two copies of subunit L (chains A and B) and one of each N (chain C) and B (chain D) per asymmetric unit. The resulting electron density and the model were improved by alternate rebuilding and relaxation with phenix.mr_rosetta (22). The model was manually rebuilt in COOT (23), refined through rigid body, restrained, and TLS refinement with phenix.refine (20). Data collection and refinement statistics are listed in Table S1. A Ramachandran analysis of all residues showed that 97.6% have favored, 2.2% have allowed, and 0.2% have disallowed backbone dihedral angles.
Structure-Based Sequence Analysis.
Structure-based sequence alignments were calculated using the “MatchMaker” and “MatchAlign” subroutine of UCSF Chimera (24, 25). The implemented structure analysis tool was used for the calculation of ADP•AlF3, Mg2+ and peptide contacts, respectively. ClustalW (26, 27) was used for primary sequence analysis. Docking interfaces of DPOR and nitrogenase were analyzed with the PISA server (28). Molecular graphics and also the superposition of Cα atoms of DPOR and nitrogenase (“align” command) were computed with PyMOL (29).
DPOR Activity Assay.
The activity of mutant DPOR proteins was analyzed as shown earlier (3). For the investigation of artificial substrate-reducing activities, all DPOR and nitrogenase assays were carried out as described earlier (9, 13, 30, 31). The substrate concentrations are detailed in Table 1. Assays of Pchlide reduction by DPOR contained 0.04 mg (NB)2 and 0.06 mg L2 in a total volume of 0.25 mL; assays of N3−, N2H4, N2, and C2H2 reduction by DPOR contained 0.4 mg (NB)2 and 0.6 mg L2 in a total volume of 1 mL; and assays of CO reduction by DPOR contained 8 mg (NB)2 and 12 mg L2 in a total volume of 1 mL. Assays of Pchlide, N3−, N2H4, N2, and C2H2 reduction by nitrogenase contained 1.5 mg NifH2 and either 0.15 mg (NifDK)2 or 0.15 mg apo (NifDK)2 variant in a total volume of 1 mL, and assays of CO reduction by nitrogenase contained 20 mg NifH and either 3 mg (NifDK)2 or 3 mg apo (NifDK)2 variant in a total volume of 1 mL. The products of these assays were analyzed as described previously (13, 15, 30, 31). Apo (NifDK)2 variant was generated by deletion of nifH, which results in a (NifDK)2 conformation that contains a pair of [4Fe–4S]–like clusters in place of the [8Fe–7S] P-cluster (32).
Supplementary Material
Acknowledgments
We especially thank Simone Virus and Marion Schwietering for help with protein crystallization. This work was supported by the Deutsche Forschungsgemeinschaft (Grant JA470/9-2).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2YNM).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218303110/-/DCSupplemental.
References
- 1.Heyes DJ, Hunter CN. Making light work of enzyme catalysis: Protochlorophyllide oxidoreductase. Trends Biochem Sci. 2005;30(11):642–649. doi: 10.1016/j.tibs.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Fujita Y. Protochlorophyllide reduction: A key step in the greening of plants. Plant Cell Physiol. 1996;37(4):411–421. doi: 10.1093/oxfordjournals.pcp.a028962. [DOI] [PubMed] [Google Scholar]
- 3.Bröcker MJ, et al. Substrate recognition of nitrogenase-like dark operative protochlorophyllide oxidoreductase from Prochlorococcus marinus. J Biol Chem. 2008;283(44):29873–29881. doi: 10.1074/jbc.M805206200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita Y, Bauer CE. Reconstitution of light-independent protochlorophyllide reductase from purified bchl and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J Biol Chem. 2000;275(31):23583–23588. doi: 10.1074/jbc.M002904200. [DOI] [PubMed] [Google Scholar]
- 5.Bröcker MJ, et al. Crystal structure of the nitrogenase-like dark operative protochlorophyllide oxidoreductase catalytic complex (ChlN/ChlB)2. J Biol Chem. 2010;285(35):27336–27345. doi: 10.1074/jbc.M110.126698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muraki N, et al. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature. 2010;465(7294):110–114. doi: 10.1038/nature08950. [DOI] [PubMed] [Google Scholar]
- 7.Sarma R, et al. Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase with MgADP bound: A homologue of the nitrogenase Fe protein. Biochemistry. 2008;47(49):13004–13015. doi: 10.1021/bi801058r. [DOI] [PubMed] [Google Scholar]
- 8.Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387(6631):370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 9.Bröcker MJ, et al. Biosynthesis of (bacterio)chlorophylls: ATP-dependent transient subunit interaction and electron transfer of dark operative protochlorophyllide oxidoreductase. J Biol Chem. 2010;285(11):8268–8277. doi: 10.1074/jbc.M109.087874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63(1):106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sablin EP, Fletterick RJ. Nucleotide switches in molecular motors: Structural analysis of kinesins and myosins. Curr Opin Struct Biol. 2001;11(6):716–724. doi: 10.1016/s0959-440x(01)00265-2. [DOI] [PubMed] [Google Scholar]
- 12.Schmid B, et al. Biochemical and structural characterization of the cross-linked complex of nitrogenase: Comparison to the ADP-AlF4(-)-stabilized structure. Biochemistry. 2002;41(52):15557–15565. doi: 10.1021/bi026642b. [DOI] [PubMed] [Google Scholar]
- 13.Wätzlich D, et al. Chimeric nitrogenase-like enzymes of (bacterio)chlorophyll biosynthesis. J Biol Chem. 2009;284(23):15530–15540. doi: 10.1074/jbc.M901331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96(7):2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Lee CC, Ribbe MW. Extending the carbon chain: Hydrocarbon formation catalyzed by vanadium/molybdenum nitrogenases. Science. 2011;333(6043):753–755. doi: 10.1126/science.1206883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CC, Hu Y, Ribbe MW. Vanadium nitrogenase reduces CO. Science. 2010;329(5992):642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Ribbe MW. Biosynthesis of the metalloclusters of molybdenum nitrogenase. Microbiol Mol Biol Rev. 2011;75(4):664–677. doi: 10.1128/MMBR.05008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103(21):8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.DiMaio F, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473(7348):540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Goujon M, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server issue):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29. Schrödinger, LLC (2010) The PyMOL molecular graphics system, Version 1.3.
- 30.Corbin JL. Liquid chromatographic-fluorescence determination of ammonia from nitrogenase reactions: A 2-min assay. Appl Environ Microbiol. 1984;47(5):1027–1030. doi: 10.1128/aem.47.5.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavini N, Burgess BK. FeMo cofactor synthesis by a nifH mutant with altered MgATP reactivity. J Biol Chem. 1992;267(29):21179–21186. [PubMed] [Google Scholar]
- 32.Corbett MC, et al. Comparison of iron-molybdenum cofactor-deficient nitrogenase MoFe proteins by X-ray absorption spectroscopy: Implications for P-cluster biosynthesis. J Biol Chem. 2004;279(27):28276–28282. doi: 10.1074/jbc.M403156200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.