Abstract
Huntington disease (HD) is a devastating, late-onset, inherited neurodegenerative disorder that manifests with personality changes, movement disorders, and cognitive decline. It is caused by a CAG repeat expansion in exon 1 of the HTT gene that translates to a polyglutamine tract in the huntingtin protein (HTT). The formation of HTT fragments has been implicated as an essential step in the molecular pathogenesis of HD and several proteases that cleave HTT have been identified. However, the importance of smaller N-terminal fragments has been highlighted by their presence in HD postmortem brains and by the fact that nuclear inclusions are only detected by antibodies to the N terminus of HTT. Despite an intense research effort, the precise length of these fragments and the mechanism by which they are generated remains unknown. Here we show that CAG repeat length–dependent aberrant splicing of exon 1 HTT results in a short polyadenylated mRNA that is translated into an exon 1 HTT protein. Given that mutant exon 1 HTT proteins have consistently been shown to be highly pathogenic in HD mouse models, the aberrant splicing of HTT mRNA provides a mechanistic basis for the molecular pathogenesis of HD. RNA-targeted therapeutic strategies designed to lower the levels of HTT are under development. Many of these approaches would not prevent the production of exon 1 HTT and should be reviewed in light of our findings.
Keywords: exon 1 huntingtin, huntingtin fragment, mis-splicing, SRSF6
Huntington disease (HD) is a devastating, late-onset, inherited neurodegenerative disease caused by a CAG repeat expansion in exon 1 of the HTT gene that translates to a polyglutamine tract in huntingtin protein (HTT). Individuals with (CAG)40 and above will develop HD within a normal lifespan, whereas repeats above (CAG)70 will invariably cause childhood onset (1). The hypothesis that an N-terminal fragment of mutant HTT represents the “toxic” species is well-established (2), and the search for the proteases that generate HTT fragments has identified caspase-3 (3), caspase-6 (3), calpains (4, 5), and matrix metalloproteinase 10 (6). However, the potential importance of smaller N-terminal fragments is highlighted by their presence in HD postmortem brains (7), their release by formic acid solubilization from brain tissue (8), and that nuclear inclusions are only detected by antibodies to N-terminal HTT epitopes (7, 9). In cell models, HTT can be cleaved into small fragments, namely cp-A and cp-B (8) or cp-1 and cp-2 (10, 11), but the proteases responsible for this have not been identified. We have recently shown that the smallest fragment present in the brains of HdhQ150 knock-in mice is an exon 1 HTT protein (12). Analysis of the R6 transgenic mouse lines has previously shown an exon 1 HTT protein is produced by translation of an exon 1 HTT transcript (13). This led us to investigate whether the exon 1 HTT protein in the HdhQ150 knock-in mice might have been generated by aberrant splicing rather than proteolytic cleavage.
Results
Aberrant Splicing of Mouse Htt Exon 1 to Exon 2 Results in a Small Polyadenylated mRNA.
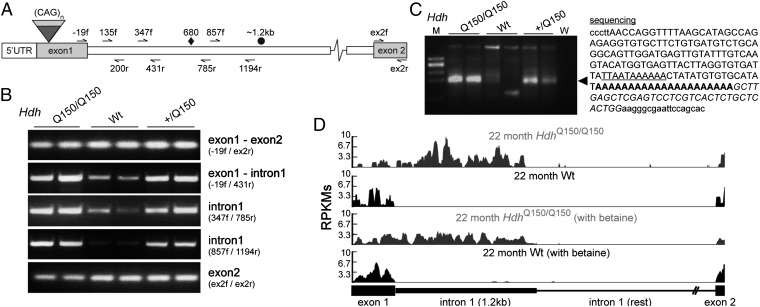
The HdhQ150 knock-in mouse model was generated by the insertion of ∼150 CAGs into exon 1 of the mouse Htt gene (14). To determine whether exon 1 of Htt mRNA had spliced to exon 2, we used a series of RT-PCR assays (Fig. 1A) on cDNA prepared from the brains of homozygous (HdhQ150/Q150), heterozygous (Hdh+/Q150) and WT animals at 2 mo of age (Fig. 1B). Nonquantitative end-point RT-PCR products were comparable between all three genotypes for assays that amplified exon 2 and spanned the exon 1–exon 2 junction. In contrast, the levels of the RT-PCR products obtained for the exon 1–intron 1 boundary and for intron 1 sequences upstream of ∼1.2 kb were more intense in cDNA prepared from the brains of HdhQ150/Q150 compared with Hdh+/Q150 animals, which were in turn more intense than those obtained from their WT littermates (Fig. 1B). Examination of the genomic sequence for Htt intron 1 identified a cryptic polyadenylation signal at position 677–685 bp and 3′RACE showed the presence of a polyadenylated short mRNA in HdhQ150/Q150 and Hdh+/Q150 but not WT brains (Fig. 1C). This unspliced exon 1–intron 1 transcript was present throughout the brain and in a range of peripheral tissues (Figs. S1 and S2). To independently verify the presence of the exon 1–intron 1 transcript, we used total RNA sequencing (RNA-Seq) on RNA prepared from the cortex of HdhQ150/Q150 and WT mice. Considerable read densities mapping to the first 1.2 kb of intron 1 were observed in HdhQ150/Q150, but not in WT littermate samples (Fig. 1D and Fig. S3). To approximate the percentage of nonspliced transcripts, we compared the 3′UTR read densities by the Mixture-of-Isoforms (MISO) software and found a 20% reduction in the full-length mRNA produced in the HdhQ150/Q150 samples (Fig. S3). Finally, we failed to detect any evidence for higher intronic reads in homozygotes compared with WT at any other position along the mouse Htt gene (Fig. S3C).
Fig. 1.
Aberrant splicing of Htt exon 1 to exon 2 results in a short polyadenylated mRNA in the HdhQ150 knock-in mouse model. (A) Schematic representation of the mouse Htt gene and primers used for the RT-PCR analysis. ♦, cryptic polyadenylation signal; ●, end of overrepresented intronic sequences. (B) RT-PCR analysis of exon 1, the exon 1–intron 1 boundary, intron 1, and exon 2. (C) 3′RACE product was generated from HdhQ150/Q150 and Hdh+/Q150 brain RNA (◄), but not from WT controls, and contained a polyA tail located ∼700 bp into intron 1. The cryptic polyadenylation signal is underlined, the polyA tail is shown in bold, the primer sequence is in italics, and vector sequence is in lowercase. (D) RNA-Seq reads from cortex of 22 mo HdhQ150/Q150 and WT mice mapping to the Htt exon 1–exon 2 region. M, low-molecular-weight marker (New England Biolabs); W, water.
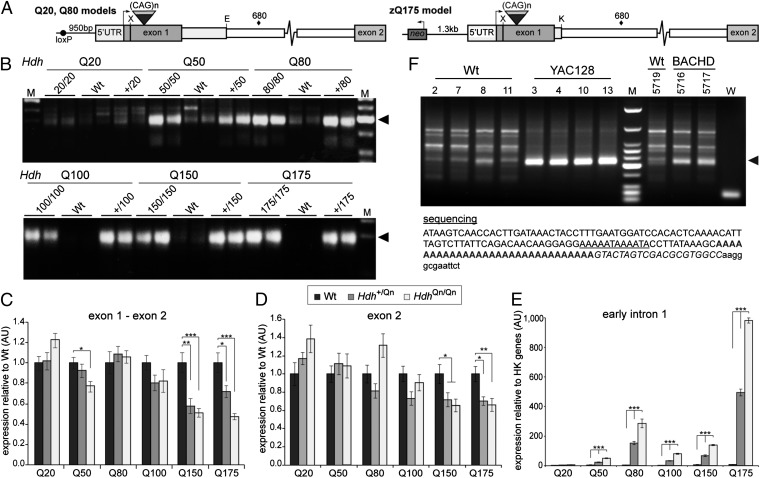
Aberrant Splicing Occurs in All HD Knock-In Mouse Models and Is CAG Repeat Length–Dependent.
As stated previously, individuals with mutations of (CAG)40 and above will develop HD within a normal lifespan, whereas repeats above (CAG)70 will invariably cause childhood onset (1). Therefore, we used a series of knock-in mouse lines: HdhQ20, Q50, Q80, Q100, Q150, and zQ175 (Figs. 1A and 2A) to determine whether aberrant splicing occurred in the context of a wide range of CAG repeats (Table S1 shows CAG repeats lengths). We analyzed RNA at 2 mo of age for each of the knock-in lines and were able to detect the same 3′RACE product in all cases except for HdhQ20 (Fig. 2B). Quantitative RT-PCR (indicated that the spliced full-length Htt mRNA levels, as determined by amplifying the exon 1–exon 2 junction and exon 2 sequences, were largely comparable between genotypes for Q20, Q50, Q80, and Q100. However, the full-length Htt transcripts were underrepresented in the Q150 and zQ175 mice (Fig. 2 C and D), consistent with the RNA-Seq data (Fig. S3B) and a previous report (15). It was not possible to normalize the levels of any intronic sequences to WT because in these mice, intron 1 sequences in processed mRNA have been removed by splicing and any signal represents noise (Fig. 2E). However, quantitative RT-PCR showed that transcripts containing early intron 1 sequences were highly increased in a gene–dose-dependent manner for all lines except HdhQ20 (Fig. 2E). The level of these transcripts also increased in a polyQ length–dependent manner when comparing the Q50, Q100, and Q150 lines (mouse Htt sequences only) or the Q80 and zQ175 lines (human-mouse chimeric Htt) (Fig. 2 A and E).
Fig. 2.
Aberrant splicing is CAG repeat length–dependent and occurs in the context of both mouse Htt and human HTT. (A) Schematic representations of chimeric human/mouse Htt genes in the HdhQ20, HdhQ80, and zQ175 lines. The 5′UTR and first 28 bp of exon 1 are always of mouse origin, whereas the remaining exon 1 sequence is human. The HdhQ20 and HdhQ80 lines contain 268 bp of human intron 1, 124 bp of mouse intron 1 is deleted, and there is a loxP site 5′ to the ATG. Line zQ175 contains 10 bp of human intron 1 with 94 bp of mouse intron 1 deleted and an intact neo-cassette 1.3-kb 5′ to the ATG. X, XmnI; E, EcoRV; K, KpnI; ♦, cryptic polyadenylation signal. (B) 3′RACE indicates the presence of the identical polyadenylated transcript (◄) in all lines except HdhQ20. (C and D) The relative levels of the spliced exon 1–exon 2 and exon 2 transcripts are shown relative to WT. (E) The expression level of early intron 1 transcripts is shown relative to the geometrical mean of three housekeeping (HK) genes (Atp5b, Eif4a3, Sdha). Primers for quantitative RT-PCR are specified in Table S3. Exon 1–exon 2, 19f-ex2r; exon 2, ex2f-ex2r; early intron 1, 135f–200r. n = 8/genotype; *P < 0.05; **P < 0.01; ***P < 0.001. Data are mean ± SEM. (F) A 3′RACE product was generated from YAC128 and BACHD brain RNA (◄), but not from WT controls, and contained a polyA tail, ∼7.3 kb into intron 1. The cryptic polyadenylation signal is underlined, the polyA tail is in bold, the primer sequence (UAPdT18) is shown in italics, and the vector sequence is in lowercase letters. M is HaeIII-digested ΦX174 (B) and low-molecular-weight markers (D) (New England Biolabs).
Aberrant Splicing Occurs in the Context of Human HTT.
To extrapolate these findings to human HTT, we performed in silico analysis and identified nine predicted polyadenylation signals in intron 1 (Fig. S4). 3′RACE on brain RNA from 2-mo-old YAC128 (16) and BACHD (17) mice, both of which are transgenic for human HTT, showed that the signal with the highest predictive score (∼7.3 kb) results in a polyadenylated transcript (Fig. 2F). A second polyadenylated transcript, resulting from a signal at ∼2.7 kb, was also found in the YAC128 mice (Fig. S5).
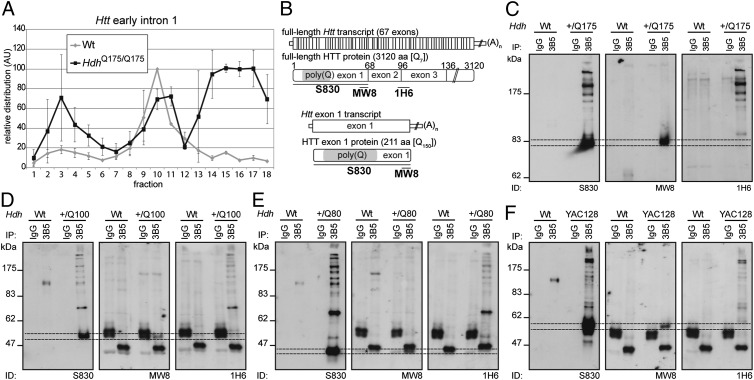
Next we sought to determine whether the aberrantly spliced mRNA was translated. We performed RT-PCR on RNA isolated from polysome gradients prepared from homozygous zQ175 and WT brains at 2 mo of age (Fig. 3A and Fig. S6). RT-PCR with primers to Hsf1 and Atp5b demonstrated that the polysomes isolated from both genotypes were intact (Fig. S6 A–C). RNA isolated from the polysome gradients from WT mice showed the expected profiles: Htt exonic sequences, but not early intron 1 sequences were associated with polysomes (fractions 12–18) (Fig. 3A and Fig. S6D). In contrast, early intron 1 sequences from zQ175 mice were present in the polysome fractions (Fig. 3A), which is consistent with the nonspliced RNA being translated.
Fig. 3.
The aberrantly spliced Htt transcript is translated and produces an exon 1 HTT protein. (A) Polysome gradients showing the relative distribution of early Htt intron 1 (135f/200r) transcripts (n = 2). Data are mean ± SEM. (B) Schematic shows the position of the HTT antibody epitopes. (C–F) HTT proteins were immunoprecipitated with 3B5H10-coupled magnetic beads from WT and (C) zQ175, (D) HdhQ100, (E) HdhQ80, and (F) YAC128 brain lysates, and Western immunoblots were immunoprobed with S830, MW8, and 1H6 antibodies. Dotted lines indicate the gel migration of the exon 1 HTT proteins, which are retarded by the polyglutamine tract and do not migrate as would be predicted by their molecular weight. To resolve exon HTT proteins with different polyQ tracts, the gel depicted in (C) was run for a longer time than those in (D–F). On the HdhQ80 blots, exon 1 HTT comigrates with a nonspecific band (*). 3B5 = 3B5H10.
Aberrantly Spliced Htt Transcript Is Translated and Produces an Exon 1 HTT Protein.
An unspliced exon 1 is followed by an evolutionarily conserved stop codon (Fig. S7), resulting in the production of an exon 1 protein that, in all vertebrates, terminates in a proline residue. We have previously shown that the smallest N-terminal fragment generated in the HdhQ150 lines corresponds to exon 1 HTT (12). To determine whether this is also present in the other HD models, we performed immunoprecipitation with 3B5H10, and Western blots were immunoprobed with S830, MW8 [exon 1 HTT C-terminal neo-epitope (12)], and 1H6 (Fig. 3B). Comparison of the S830 and MW8 blots (which only detect an exon 1 HTT protein on Western blots) reveals that an exon 1 HTT protein (dotted lines) is present in the zQ175, HdhQ150, HdhQ100, HdhQ80, and YAC128 brains but not in those from their WT littermates or in the IgG controls (Fig. 3 C–F and Fig. S8), as would be predicted from the presence of the nonspliced short transcript.
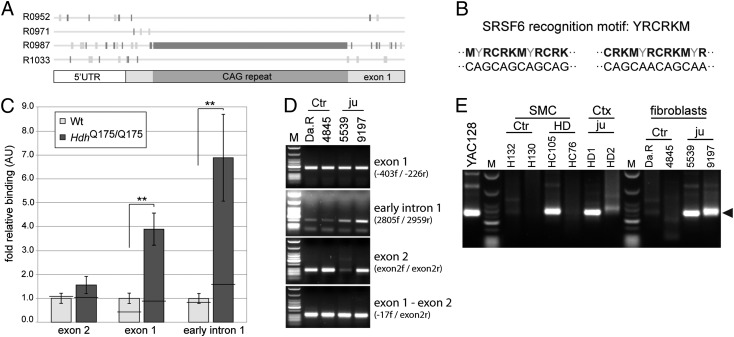
Splicing Factor SRSF6 Binds to Expanded CAG Repeats in Htt Transcripts.
To investigate the underlying mechanism, we used bioinformatics to predict regulatory motifs in exon 1 of Htt (Fig. 4A) and mapped the binding site of SRSF6 to a CAG or CAGCAA repeat (Fig. 4B). RNA coimmunoprecipitation with an antibody to SRSF6 captured Htt 5′UTR and early intron 1 sequences from homozygous zQ175 mice with a much higher efficiency than those from WT brain lysates (Fig. 4C). Exon 2 transcripts were not immunoprecipitated from either zQ175 or WT lysates (Fig. 4C), which is consistent with SRSF6 binding to the expanded CAG repeat in the zQ175 mice and inhibiting the splicing of exon 1 to exon 2.
Fig. 4.
The splicing factor SRSF6 binds to expanded CAG repeats in Htt transcripts and the aberrantly spliced transcript is present in HD patient tissues. (A) RegRNA predicts a cluster of R0987 regulatory motifs (CTGN) in the expanded CAG repeat of Htt. Using ESEfinder 3.0, this motif was mapped to the binding site of SRSF6. (B) The RNA recognition motif of SRSF6 is YRCRKM, closely resembling a CAG or CAGCAA repeat. Nucleotide abbreviations: Y, T or C; R, A or G; K, G or T; M, A or C. (C) RNA-IP of SRSF6 from zQ175 brain resulted in the coprecipitation of higher levels of early intronic (135f-200r) and exon 1 (5′UTR 1f-5′UTR 1r) transcripts compared with WT. In contrast, the coprecipitated levels of zQ175 and WT exon 2 transcripts were not significantly different. n = 6/genotype. Horizontal bar, IgG control immunoprecipitation. Data are mean ± SEM; **P < 0.01. (D) RT-PCR of human samples. Human fibroblast lines can be found in Table S2. (E) 3′RACE of human samples. ◄, the expected RACE product size of about 260 bp; Ctr, control subject; Ctx, cortex; HD, adult onset HD; ju, juvenile-onset HD; M, low-molecular-weight markers (New England Biolabs); SMC, sensory motor cortex; tg, transgenic; W, water.
The Aberrantly Spliced Transcript Is Present in HD Patient Tissues.
To determine whether the aberrant splicing also occurs in humans, we performed RT-PCR on RNA from two juvenile HD and two control fibroblast lines, peripheral blood mononuclear cells from HD and control subjects, postmortem sensory motor cortex from two HD and two control subjects, and cortical tissue from two individuals with juvenile HD (Table S2). In the fibroblast lines, the signals were relatively comparable for exon 1, the exon 1–exon 2 junction, and exon 2 transcripts. In contrast, the early intron 1 transcripts were elevated in the HD fibroblast compared with the control lines (Fig. 4D). However, the pattern was more difficult to interpret in the postmortem brain samples, probably reflecting differences in the extent to which the RNA had degraded in these tissues. We performed 3′RACE for the same cryptic polyadenylation signal as used in the YAC128 and BACHD mice. The polyadenylated transcript was clearly apparent in the two fibroblast lines as well as in the sensory motor cortex of an HD patient with 42 CAGs and the cortex of a juvenile HD individual with 72 CAGs (Fig. 4E). We were only able to detect a 3′RACE product in one of eight peripheral blood mononuclear cells samples, suggesting that the missplicing events are very rare in these cells.
Discussion
We have identified a small exon 1–intron 1 polyadenylated mRNA transcript in the brains of all HD mouse models expressing mutant Htt (mouse) or HTT (human). Furthermore, we have shown that the same transcript is also present in fibroblast lines derived from HD patients and in postmortem HD brains. We have shown that the SRSF6 splicing factor binds to the 5′end of the Htt gene with an expanded CAG repeat, consistent with its known recognition motif (18). SRSF6 regulates splicing and facilitates translation of partially spliced transcripts (19, 20). SR proteins might also sequester U1 small nuclear ribonucleoprotein complexes (21), a phenomenon that promotes polyadenylation from cryptic polyA signals within introns (22). Therefore, an increased association of SRSF6 with expanded CAG repeats could account for the CAG repeat length–dependent production of the exon 1–intron 1 transcript. Translation of this transcript produces an exon 1 HTT protein.
The pathological consequences of the expression of this aberrantly spliced product in mice have been demonstrated in multiple experiments. The R6/ mouse lines are transgenic for a genomic fragment that spans the 5′ end of the HTT gene, exon 1, and a portion of intron 1 (13). Therefore, the R6 lines can be considered to be models of aberrant splicing in HD and demonstrate that an exon 1 HTT protein is highly pathogenic, a result that was recapitulated in a set of independent experiments (23). Surprisingly, the late-stage phenotypes of R6/2 mice closely resemble those present in the HdhQ150 knock-in mice (24–27), the main difference between these two models being the age of phenotype onset and rate of disease progression. This suggests that the pathogenic process in HdhQ150 mice could be driven by the same exon 1 HTT fragment differing only in its abundance and accumulation over time.
Our discovery that exon 1 HTT is generated by aberrant splicing will provide the opportunity to test the extent to which an exon 1 protein contributes to disease pathogenesis in the knock-in lines and the extent to which full-length HTT and/or other N-terminal HTT fragments, generated by proteolytic cleavage, are also pathogenic. RNA-targeted therapeutic approaches designed to lower the levels of HTT through the use of antisense oligonucleotides, RNAi, or small hairpin RNAs are under development (28). Many of these approaches would not prevent the production of exon 1 HTT and should be reviewed in the light of our findings.
Materials and Methods
Mouse Maintenance and Breeding.
The HdhQ50 and HdhQ100 lines were generated by selective breeding for alterations in germ-line repeat size starting with a C57BL/6 congenic of the HdhQ150 lines (14). The Hdh+/Q20, Hdh+/Q80 (29, 30), and zQ175 (31) knock-in mice were supplied from CHDI colonies maintained at The Jackson Laboratory. The HdhQ20, HdhQ50, HdhQ80, HdhQ100, and zQ175 lines were maintained by backcrossing to C57BL/6J (Charles River). All experimental procedures were approved by the King's College London Ethical Review Committee and performed in accordance with United Kingdom Home Office regulations.
RNA Sequencing.
Samples were prepared using a modified, strand-specific version of the Illumina Tru-Seq protocol. Final PCR amplification was performed with KAPA HiFi polymerase and GC buffer (Kapa Biosystems). The paired-end, strand-specific cDNA libraries were multiplexed onto the Illumina HiSEq (40-bp reads). Read data were mapped to the mm9 build with the Bowtie alignment program using the “best” setting. For quality control statistics of the RNA-Seq libraries, see Table S4. Differential expression of the Htt gene was analyzed with the R DESeq package. Gene expression was calculated as reads per kilobase of exon per million mapped reads (RPKMs). Splicing was analyzed using the Python/C version of MISO.
Polysome Gradients.
A 10–40% (wt/vol) sucrose gradient was prepared in 50 mM Tris⋅Cl (pH 6.6), 140 mM NaCl, 12 mM magnesium chloride, 200 µg/mL cycloheximide, and 1 mM DTT. Mouse brain tissue was lysed in freshly prepared polysome buffer [10 mM Tris⋅Cl (pH 7.4), 140 mM NaCl, 12 mM magnesium chloride, 1% (wt/vol) Triton X-100, 1 mM DTT, 200 µg/mL cycloheximide, 0.5 U/µL RNAsin, and 10 mM ribonucleoside vanadyl complex]. A volume corresponding to 250 µg absorbance at 260 nm was layered on the 10–40% sucrose gradients. The gradients were centrifuged at 39,000 rpm at 4 °C for 1 h 40 min in a SW41-Ti swing out rotor. Fractions were collected and RNA was extracted.
SRSF6 RNA Coimmunoprecipitation.
Mouse brain tissue was lysed in freshly prepared Triton buffer [50 mM Hepes/NaOH pH 7.6, 160 mM NaCl, 7 mM magnesium chloride, 3 mM calcium chloride, 5 mM potassium chloride, 1% (wt/vol) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 0.5 U/µL RNAsin and complete protease and phosphatase inhibitor mixture]. Two milligrams of total protein was immunoprecipitated for 5 h on a rotating wheel at 4 °C with 9 μL of Dynabeads protein G and 3 μg of anti-SRSF6 antibody (LS-B5712; LifeSpan BioSciences). The beads were washed four times with 0.5 mL Triton buffer followed by RNA extraction.
Detailed descriptions of mouse maintenance, breeding, genotyping, and CAG repeat sizing, quantitative RT-PCR, mouse/human RT-PCR (see also Table S5) and 3′RACE, RNA sequencing, polysome gradients, antibodies, immunoprecipitation, Western blotting, SRSF6 RNA-IP, bioinformatics, and statistical analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jenny Morton for providing YAC128 mice, Yvon Trottier for the 1H6 antibody, the Massachusetts Institute of Technology BioMicro Center for RNA-Seq preparation and sequencing, and the Neurological Foundation of New Zealand for Huntington disease postmortem brain tissue. This work was supported by Grant G0801314 from the Medical Research Council and a grant from the CHDI Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221891110/-/DCSupplemental.
References
- 1. Bates GP, Harper PS, Jones AL, eds (2002) Huntington's Disease (Oxford University Press, Oxford), 3rd Ed.
- 2.Ross CA, Tabrizi SJ. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10(1):83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 3.Wellington CL, et al. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington’s disease. J Neurosci. 2002;22(18):7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YJ, et al. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington’s disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci USA. 2001;98(22):12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gafni J, et al. Inhibition of calpain cleavage of huntingtin reduces toxicity: Accumulation of calpain/caspase fragments in the nucleus. J Biol Chem. 2004;279(19):20211–20220. doi: 10.1074/jbc.M401267200. [DOI] [PubMed] [Google Scholar]
- 6.Miller JP, et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington’s disease. Neuron. 2010;67(2):199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Lunkes A, et al. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell. 2002;10(2):259–269. doi: 10.1016/s1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- 9.Schilling G, et al. Characterization of huntingtin pathologic fragments in human Huntington disease, transgenic mice, and cell models. J Neuropathol Exp Neurol. 2007;66(4):313–320. doi: 10.1097/nen.0b013e318040b2c8. [DOI] [PubMed] [Google Scholar]
- 10.Ratovitski T, et al. Mutant huntingtin N-terminal fragments of specific size mediate aggregation and toxicity in neuronal cells. J Biol Chem. 2009;284(16):10855–10867. doi: 10.1074/jbc.M804813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratovitski T, et al. N-terminal proteolysis of full-length mutant huntingtin in an inducible PC12 cell model of Huntington’s disease. Cell Cycle. 2007;6(23):2970–2981. doi: 10.4161/cc.6.23.4992. [DOI] [PubMed] [Google Scholar]
- 12.Landles C, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010;285(12):8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 14.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10(2):137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 15.Giles P, et al. Longitudinal analysis of gene expression and behaviour in the HdhQ150 mouse model of Huntington’s disease. Brain Res Bull. 2012;88(2-3):199–209. doi: 10.1016/j.brainresbull.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Slow EJ, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12(13):1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 17.Gray M, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28(24):6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009;10(3):R30. doi: 10.1186/gb-2009-10-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tranell A, Tingsborg S, Fenyö EM, Schwartz S. Inhibition of splicing by serine-arginine rich protein 55 (SRp55) causes the appearance of partially spliced HIV-1 mRNAs in the cytoplasm. Virus Res. 2011;157(1):82–91. doi: 10.1016/j.virusres.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Swanson CM, Sherer NM, Malim MH. SRp40 and SRp55 promote the translation of unspliced human immunodeficiency virus type 1 RNA. J Virol. 2010;84(13):6748–6759. doi: 10.1128/JVI.02526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labourier E, et al. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev. 1999;13(6):740–753. doi: 10.1101/gad.13.6.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150(1):53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benn CL, et al. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington’s disease. Hum Mol Genet. 2005;14(20):3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 24.Woodman B, et al. The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res Bull. 2007;72(2-3):83–97. doi: 10.1016/j.brainresbull.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Moffitt H, McPhail GD, Woodman B, Hobbs C, Bates GP. Formation of polyglutamine inclusions in a wide range of non-CNS tissues in the HdhQ150 knock-in mouse model of Huntington’s disease. PLoS ONE. 2009;4(11):e8025. doi: 10.1371/journal.pone.0008025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn A, et al. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16(15):1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 27.Labbadia J, et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Invest. 2011;121(8):3306–3319. doi: 10.1172/JCI57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121(2):500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum Mol Genet. 1999;8(1):115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 30.White JK, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17(4):404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 31.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465(1):11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.