Abstract
The microbial cosmopolitan dispersion hypothesis often invoked to explain distribution patterns driven by high connectivity of oceanographic water masses and widespread dispersal ability has never been rigorously tested. By using a global marine bacterial dataset and iterative matrix randomization simulation, we show that marine bacteria exhibit a significantly greater dispersal limitation than predicted by our null model using the “everything is everywhere” tenet with no dispersal limitation scenario. Specifically, marine bacteria displayed bipolar distributions (i.e., species occurring exclusively at both poles and nowhere else) significantly less often than in the null model. Furthermore, we observed fewer taxa present in both hemispheres but more taxa present only in a single hemisphere than expected under the null model. Each of these trends diverged further from the null expectation as the compared habitats became more geographically distant but more environmentally similar. Our meta-analysis supported a latitudinal gradient in bacterial diversity with higher richness at lower latitudes, but decreased richness toward the poles. Bacteria in the tropics also demonstrated narrower latitudinal ranges at lower latitudes and relatively larger ranges in higher latitudes, conforming to the controversial macroecological pattern of the “Rapoport rule.” Collectively, our findings suggest that bacteria follow biogeographic patterns more typical of macroscopic organisms, and that dispersal limitation, not just environmental selection, likely plays an important role. Distributions of microbes that deliver critical ecosystem services, particularly those in polar regions, may be vulnerable to the same impacts that environmental stressors, climate warming, and degradation in habitat quality are having on biodiversity in animal and plant species.
Keywords: ICoMM, MIRADA-LTERS, microbial biogeography, macroecology
Bipolar distributions refer to the presence of identical taxa in the polar or subpolar provinces of the Northern and Southern hemispheres without evidence for connecting populations that span the tropics. The oceanographic literature first described bipolar-distributed fauna and flora, among the oldest of biogeographic patterns, more than 160 y ago during early voyages to polar provinces (1, 2). In the past decade, molecular approaches also confirmed bipolarity in the microbial eukaryotes (e.g., planktonic foraminifera and dinoflagellates) (3–5), whereby molecular evidence suggested that the same species recurrently migrated between both hemispheres through the equator (3). Bipolar species distributions as a biogeographic pattern hypothesize that a historically cosmopolitan-distributed species experienced an event that limited its range to both polar provinces or that a species restricted to one pole dispersed to the other. The marine fossil record supports a cosmopolitan distribution of diatoms (6), but suggests limited dispersal-driven biogeography near polar provinces for this group.
The Baas-Becking (BB) tenet, “everything is everywhere, but the environment selects” (ref. 7, p. 15), may describe the current biogeography of microorganisms. The first part of the BB tenet, “everything is everywhere” explains the seemingly cosmopolitan distribution of microorganisms (8) by invoking a lack of dispersal limitation. The second component of the BB tenet accounts for regional distinctions by invoking environmental selection in a process equivalent to “species sorting” in metacommunity theory (9). Arguments for “everything is everywhere” cite enormous microbial population sizes (10) or the presence of strong passive dispersal forces (i.e., thermohaline circulation and oceanic currents) (11) or their ability to hitchhike on larger species (12). However, recent molecular evidence describes microbial biogeographical patterns that suggest limited gene flow (13, 14) and indicate the potential for limited species dispersal for “cosmopolitan” diatoms (15).
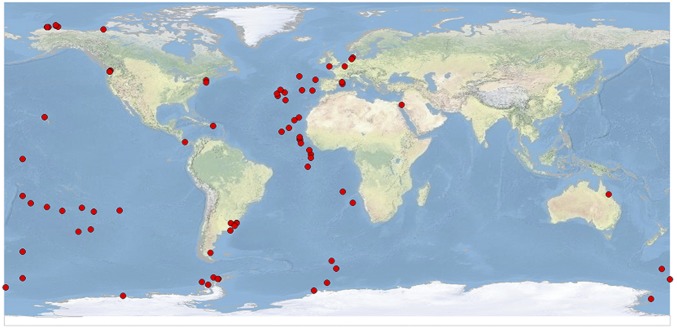
The recent application of deep sequencing to determine microbial community structures from many latitudinal zones in the world’s ocean offers an opportunity to examine bacterial macroecological distribution patterns (16, 17). In a synthesis of data from the first marine microbial census, we first observed the pattern of shared taxa common to samples from opposite poles (16). However, this preliminary investigation showed that the most abundant taxa in the polar regions were also found elsewhere and thus failed to conclusively demonstrate the classical “bipolar” species distribution. In the present study, we investigated the distribution of bacteria over latitudinal zones by analyzing 277 epipelagic bacterial communities from the Arctic, Atlantic, Pacific, and Southern Oceans spanning latitudes 72.4°N to 75.6°S (Fig. 1 and Dataset S1). Our dataset consisted of 4.23 million bacterial small-subunit (SSU) rRNA gene V6 hypervariable pyrotag sequences that clustered into 65,545 operational taxonomic units (OTUs). We conservatively treated OTUs as taxa, approximately equivalent to the smallest unit of bacterial diversity for testing biogeographical patterns (18).
Fig. 1.
Sampling locations of 277 marine epipelagic bacterial communities from the Arctic, Atlantic, Pacific, and Southern oceans spanning latitudes 72.4°N to 75.6°S. Samples include ones from the International Census of Marine Microbes and Palmer Station in Antarctica as part of the Microbial Inventory Research Across Diverse Aquatic Long Term Ecological Research Sites project.
We hypothesized that species sorting coupled with some degree of dispersal limitation best explains current microbial distributions. We compared global-scale observations vs. a null model that assumes no limitation on dispersal (i.e., everything can be everywhere) and no environmental selection (i.e., the environment does not favor the growth of specific microbes). Our null model is similar to Hubbell’s neutral theory (19) in that it lacks environmental forcing; however, it is distinct in that it assumes no limits on taxon dispersal. Our approach offers a quantitative means of assessing classical biogeographic patterns in nature on the microbial scale.
Results and Discussion
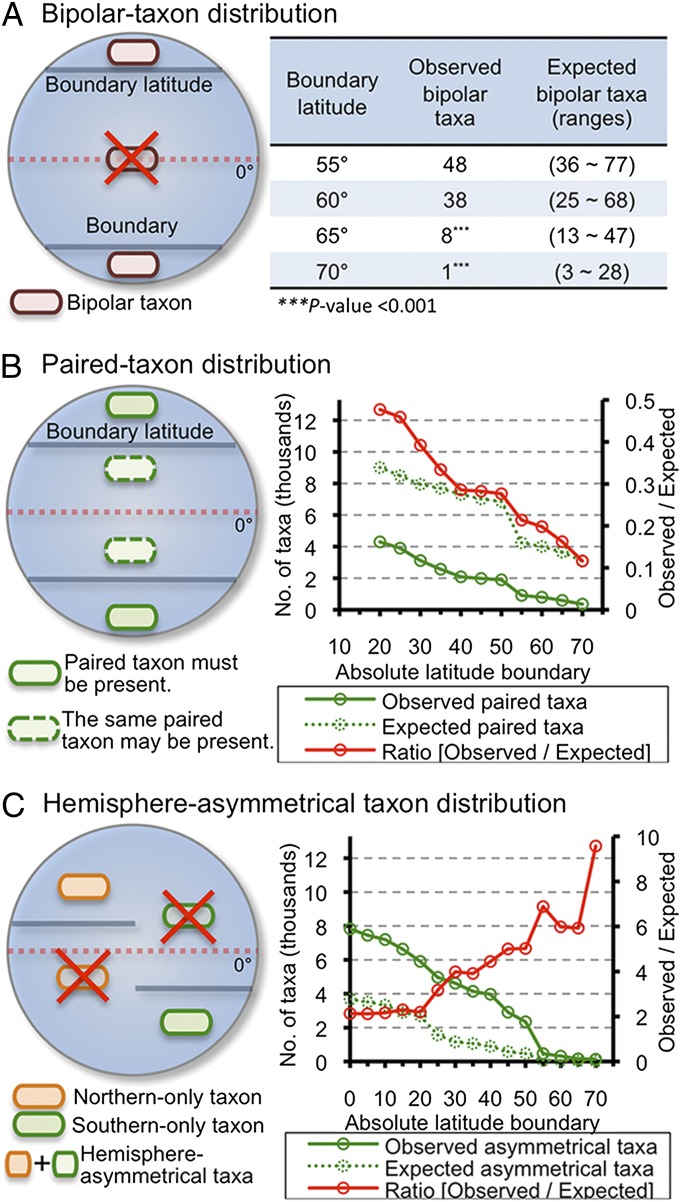
A bipolar species is a taxon that occurs in both poles and does not occur in intervening latitudes (Fig. 2A). We compared numbers of observed bipolar-distributed taxa vs. the numbers of expected bipolar-distributed taxa based on a null model. To assess the null model, we used an iterative matrix randomization process to obtain the expected number of bipolar-distributed taxa. Starting from a taxon by sample matrix of instances of the occurrence of a taxon in a sample (i.e., counts), the randomization process shuffled counts among taxa and samples without replacement, while holding constant the number of counts per sample and the absolute abundance of each taxon across all samples. This process controlled for the sampling design and taxon abundance, and returned results expected if populations had unlimited dispersal and were free of selection pressures. It is likely that environmental selection does play an important role in structuring marine microbial populations, but, by strategically comparing null expectations and observations across environmental gradients, we can use this minimal null expectation to highlight the relative importance of dispersal limitation and selection. By comparing populations in increasingly selectively similar environments at increasingly large distances one can discriminate between distributions generated under the BB tenet, and a model in which limited dispersal plays a role (i.e., everything is not everywhere, and the environment selects).
Fig. 2.
Nonrandom global biogeographical patterns of marine epipelagic bacteria. (A) A schematic illustration of the bipolar taxon distribution, where taxa occur only above the subtropics on both sides of the equator. Table (Right) compares observed bipolar-distributed taxa vs. expected ones. (B) A schematic illustration of a paired-taxon distribution. The numbers of observed, expected, and the ratio of observed-to-expected paired taxa are shown in the graph (Right). (C) A schematic illustration of a hemisphere-asymmetrical taxon distribution, whereby taxa occur only above a boundary latitude in a selected hemisphere. The numbers of observed, expected, and the ratio of observed-to-expected hemisphere-asymmetrical taxa are shown in the graph (Right). All expected numbers of taxa and P values were obtained from null-simulation (i.e., random-distribution) based matrix randomization processes with 1,000 iterations.
We first summarized the distributions of bipolar distributed taxa in a collection of 1,000 shuffled matrices at latitudinal increments of 5° above 55°. The numbers of observed bipolar distributed taxa at absolute values of 55° and 60° fell within the expected bipolar distributed taxon values from the null model (Fig. 2A). In contrast, we observed fewer bipolar-distributed taxa (Table S1 provides their taxonomy) at absolute latitudes 65° or 70° than expected, consistent with a biogeographical pattern characterized by limited dispersal (Fig. 2A). This pattern of increasing divergence from expectation as environments became more distant and more selectively similar was a consistent feature in our results.
Our observed bacterial bipolarity led us to measure whether “paired-taxon” distributions also deviated from the null model. Under our definition, a paired taxon is one that occurred in both hemispheres at or higher than an absolute boundary latitude, without regard to the taxon’s occurrence between these latitudes. By this definition, all bipolar taxa at each boundary latitude are paired taxa, but many paired taxa are not bipolar taxa. First, we counted the numbers of observed paired taxa at latitudinal increments of 5° above 20°, and then compared this number to the number of expected paired taxa by using the same iterative matrix randomization process described earlier. At all examined latitudes, we observed significantly fewer paired taxa than expected under the null model (Fig. 2B). The ratios of observed to expected paired taxa decreased sharply from 0.35 to 0.11 toward higher boundary latitudes (Fig. 2B, red line). This incremental divergence from 1:1 in the ratio of observed to expected paired taxa as habitats became further apart and more selectively similar suggested an increasing barrier to dispersal over increasing latitudinal distance.
We also explored a hemisphere-asymmetrical taxon distribution (Fig. 2C), whereby taxa only occurred poleward of an absolute boundary latitude in one hemisphere and were excluded from other latitudes. After using the same iterative matrix randomization process, we observed more hemisphere-asymmetrical taxa than expected under the null hypothesis. If one assumes that environmental characteristics of the two hemispheres at similar latitudes are comparable (Fig. S1), this result suggested that tropical/subtropical provinces may serve as barriers to dispersal of marine microbial taxa. Also, we noted that the ratio of observed to expected hemisphere-asymmetrical taxa diverged strongly from one with increasing boundary latitude (Fig. 2C). This pattern diverged from expectations under the BB tenet and further supported the notion that as distances between selectively similar habitats increase, dispersal between them is increasingly rare.
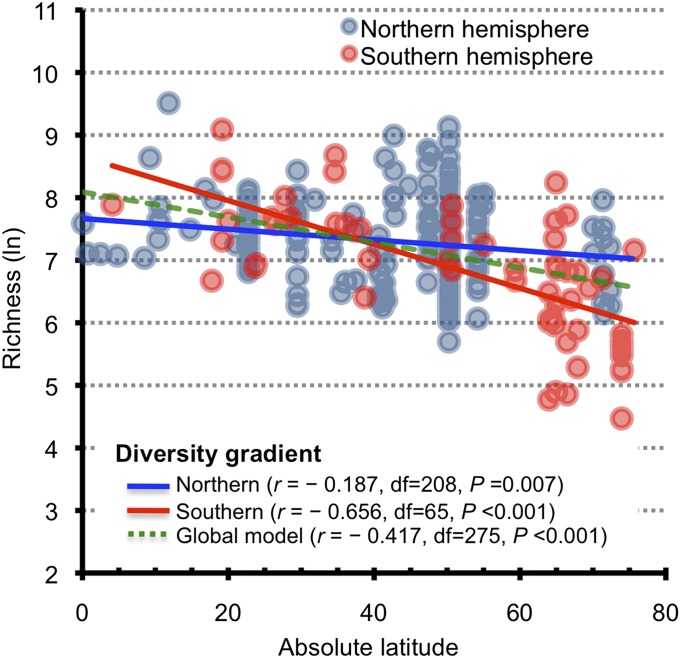
Our study is one of few (20–23) that address large-scale gradients in diversity. The observations of a latitudinal gradient in bacterial diversity (22, 23) with higher latitudes being less diverse than lower latitudes (Fig. 3 and Dataset S1) reinforce the concept that bacteria display biogeographical patterns (24). Interestingly, the Southern hemisphere exhibited a steeper diversity gradient than did the Northern hemisphere (Fig. 3). Recently, Ghiglione et al. (25), found a nonsignificant latitudinal gradient in bacterial diversity; however, our more comprehensive dataset supported a significant latitudinal gradient. Variability in the slope of latitudinal diversity gradients (26) in the Southern vs. the Northern hemisphere also supported hemisphere-asymmetrical distributions, suggesting that the tropics may serve as a barrier to bacterial dispersal.
Fig. 3.
A latitudinal gradient in marine epipelagic bacterial diversity. We calculated bacterial richness by using parametric methods implemented in the CatchAll program (44). Pearson correlation r values were between natural log-transformed estimated richness values and absolute latitudes. A similar correlation was obtained with richness estimation with normalized datasets (subsampling to minimum numbers of reads). Our correlation values between latitude and estimated richness were also similar to those from previous amplified ribosomal intergenic spacer analysis results (23) with 100 data points at 57 locations (Pearson r = −0.422).
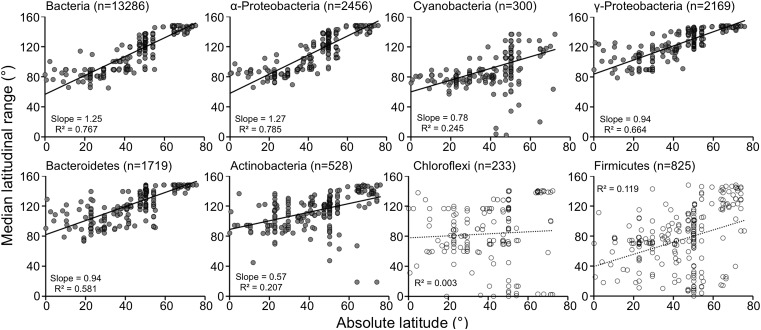
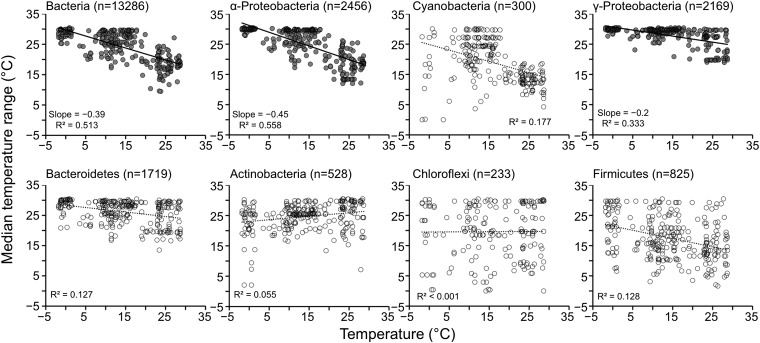
We looked over latitudinal range sizes to see if the Rapoport rule (27) explained the observed bacterial latitudinal diversity gradient. By measuring each OTU's latitudinal range size (i.e., the ranges between the northernmost latitude and southernmost latitude where an OTU occurred), we found that overall bacterial latitudinal ranges conformed to the Rapoport rule (i.e., narrower median latitudinal taxon range sizes occurred at lower absolute latitudes than at higher absolute latitudes; Fig. 4 and Fig. S2). Closer inspection of latitudinal range sizes of individual OTUs by phylum (or, in the case of Proteobacteria, by class), revealed that bacterial OTUs belonging to the phylum Bacteroidetes, Cyanobacteria, and the α-, β-, and γ-classes of Proteobacteria showed that OTUs in these groups have broader habitat ranges at higher latitudes than those compared with lower latitudes (Fig. 4, Fig. S2, and Table 1). OTUs assigned to Firmicutes, Chlamydiae, Chloroflexi, Planctomycetes, and ɛ-Proteobacteria, however, did not conform to the Rapoport rule, showing less significant or nonsignificant correlations between latitudinal range and latitude (Fig. 4, Fig. S2, and Table 1). Some genera of Chloroflexi and Firmicutes are capable of forming spores, and therefore it is possible that these taxa may passively (or stochastically) disperse as propagules. The oceans are fully connected through thermohaline circulation on the time scale of ∼1,000 to 2,700 y (28), and, therefore, if these propagules can remain viable during that time period, the few spore-forming taxa may truly disperse globally, even through environments hostile to their reproductive success.
Fig. 4.
Limited bacterial latitudinal ranges by taxonomic group. Relationship between bacterial latitudinal range sizes and latitude to test conformation to the Rapoport rule. Each point represents a median latitudinal range for each sample with taxa of a given taxonomy (n = numbers of taxa used in the calculation; “slope” indicates slope of linear regression).
Table 1.
Linear regression statistics on median latitudinal ranges and latitude, and median SST ranges and SST
| Latitude and median latitudinal ranges |
SST and median SST ranges |
|||||
| Taxonomic group | Slope ± SEM | Adjusted R2 | P value | Slope ± SEM | Adjusted R2 | P value |
| Bacteria | 0.61 ± 0.02 | 0.77 | <0.001 | −1.30 ± 0.08 | 0.51 | <0.001 |
| Acidobacteria | −0.08 ± 0.03 | 0.04 | 0.008 | 0.04 ± 0.07 | 0 | 0.596 |
| Actinobacteria | 0.36 ± 0.04 | 0.20 | <0.001 | −0.47 ± 0.12 | 0.05 | <0.001 |
| Bacteroidetes | 0.62 ± 0.03 | 0.58 | <0.001 | −0.85 ± 0.13 | 0.12 | <0.001 |
| Chlamydia | 0.09 ± 0.03 | 0.04 | 0.008 | −0.28 ± 0.08 | 0.06 | 0.001 |
| Chloroflexi | 0.02 ± 0.03 | 0 | 0.457 | 0.01 ± 0.08 | 0 | 0.915 |
| Cyanobacteria | 0.16 ± 0.03 | 0.08 | <0.001 | −0.47 ± 0.07 | 0.17 | <0.001 |
| Deferribacter | 0.31 ± 0.03 | 0.25 | <0.001 | −0.48 ± 0.08 | 0.10 | <0.001 |
| Firmicutes | 0.15 ± 0.02 | 0.12 | <0.001 | −0.47 ± 0.07 | 0.13 | <0.001 |
| Gemmatimonadetes | 0.06 ± 0.03 | 0.02 | <0.001 | −0.10 ± 0.09 | 0 | 0.310 |
| Planctomycetes | 0.10 ± 0.03 | 0.03 | 0.001 | −0.38 ± 0.08 | 0.07 | <0.001 |
| α-Proteobacteria | 0.62 ± 0.02 | 0.79 | <0.001 | −1.23 ± 0.07 | 0.56 | <0.001 |
| β-Proteobacteria | 0.42 ± 0.04 | 0.33 | <0.001 | −0.83 ± 0.10 | 0.20 | <0.001 |
| γ-Proteobacteria | 0.71 ± 0.03 | 0.66 | <0.001 | −1.66 ± 0.14 | 0.33 | <0.001 |
| δ-Proteobacteria | 0.30 ± 0.03 | 0.34 | <0.001 | −0.92 ± 0.07 | 0.38 | <0.001 |
| ɛ-Proteobacteria | 0.14 ± 0.03 | 0.08 | <0.001 | −0.20 ± 0.10 | 0.01 | 0.044 |
| Verrucomicrobia | 0.31 ± 0.03 | 0.23 | <0.001 | −0.23 ± 0.10 | 0.02 | 0.017 |
Boldface indicates Adjusted R2 value was equal or higher than 0.2.
These phylum- or class-level differences in the latitudinal range sizes of OTUs implicate characteristics such as morphology, size, and fitness to new environments, as drivers for new colonization via dispersal, extinction (i.e., narrow-range taxa possess higher extinction rates), and, ultimately, biogeographical patterns. Obviously, phylum and class taxonomic levels were coarse groupings to examine; however, it is noteworthy that trends of bacterial latitudinal range sizes deviated by even these taxonomic groupings and thus finer taxonomic levels such as species-specific groupings may yield further insights.
OTUs in abundant taxa—for instance, α- and γ-Proteobacteria in the epipelagic zone—tended to distribute more widely across datasets and had broader latitudinal range sizes; however, not all abundant OTUs exhibited broad latitudinal range sizes (Fig. S3). We hypothesize that very abundant bacteria may migrate to or emigrate from adjacent regions through passive transport (19, 29). Thus, latitudinal gradients in diversity and latitudinal range size differences of OTUs by phylum (or class) or by abundance suggest nonuniform bacterial distributions and dispersal capability (30) driven by passive processes or by fine-scale sensitivity to different environmental factors.
We also measured temperature range sizes of OTUs along with temperature to test how contemporary environmental factors related to bacterial distribution patterns. Temperature range sizes often mirrored latitudinal range sizes as a result of strong correlations between temperature and absolute latitude (Fig. S1), with some exceptions; i.e., bipolar-distributed taxa had broad latitudinal and narrow temperature range sizes. Bacteria at higher temperatures (likely at lower absolute latitudes) exhibited narrower median temperature ranges than those at lower temperature (likely at higher absolute latitudes; Fig. 5 and Fig. S4). Temperature range sizes of OTUs in Actinobacteria, Bacteroidetes, Deferribacteres, and Verrucomicrobia did not correlate with temperature gradients even though their latitudinal range sizes conformed to the Rapoport rule (Table 1). This discrepancy, conformation to the Rapoport rule with no correlation between temperature range sizes and temperature (i.e., Bacteroidetes, Cyanobacteria; Figs. 4 and 5), suggested that latitudinal distances (dispersal limitation as a component of deterministic processes) and current environmental conditions (species sorting) are bona fide factors that affect contemporary bacterial biogeography.
Fig. 5.
Limited bacterial temperature ranges by taxonomic group. Relationship between bacterial temperature range sizes and temperature to see if broader mean temperature range sizes were measured in colder locations than in moderate or warmer locations. Each point represents the median temperature range for each dataset with taxa of a given taxonomy (n = numbers of taxa used in the calculation; “slope” indicates slope of linear regression).
We suggest that contemporary environmental factors and historical events, including dispersal limitation, drive the biogeography of epipelagic marine bacteria. The driving forces for current epipelagic marine bacterial biogeography likely include environmental differences, e.g., in temperature and water mass composition (31), but also likely include differential dispersal, e.g., connectivity among communities as a result of ocean currents and physical barriers such as the landlocked nature of the Arctic Ocean. A recent study (25) suggested that environmental selection is the primary driver in differentiating epipelagic bacterial community composition between polar regions, that is, the environmental differences between the Arctic and Southern Ocean. However, here we demonstrate that stochastic processes likely play a role in global bacterial biogeography (32, 33). Our data show that observations become increasingly divergent from expectations with increasingly selectively similar but increasingly distant environments; this strongly suggests that dispersal limitation is playing an important role before environmental selection is making a difference. Our data also have important implications for microbial allopatric speciation (34) among bipolar species separated by a tropical barrier even though they exist in the high connectivity of the epipelagic zone. Understanding microbial distributions has important implications for climate change, as microbes drive major biogeochemical cycles (35) in the oceans. A growing concern is that increases in water temperature in temperate and polar provinces could lead to the decline of microbial diversity restricted to colder temperatures, including so-called bipolar species. Therefore, perturbations in microbial biogeography may serve as sentinels to irreversible change in the environment, particularly the global ocean.
Materials and Methods
Meta-Analysis of Epipelagic Bacterial Community V6 Hypervariable Region of SSU rRNA Gene.
We collected V6 hypervariable region SSU rRNA gene pyrotag data from 277 epipelagic bacterial samples from the Arctic, Atlantic, Pacific, and Southern Oceans spanning latitudes 72.4°N to 75.6°S (Fig. 1 and Dataset S1) as part of the International Census of Marine Microbes (ICoMM) effort and Palmer Station in Antarctica as part of the Microbial Inventory Research Across Diverse Aquatic Long Term Ecological Research (MIRADA-LTER) Sites project. Coordinates of latitude and longitude, depth, temperature, and numbers of reads for each sample are reported in Dataset S1. Palmer Station samples were collected and sequenced as previously described (36, 37) in a manner consistent with that used for the ICoMM samples. We sequenced the hypervariable V6 region (Escherichia coli positions 967–1046) using pyrosequencing on the GS-FLX system and trimmed sequences of adapter and primer sequences, removed low-quality reads as described previously (38), and assigned OTUs (referred to throughout the text as taxa) using the 2% single linkage preclustering and pairwise alignment with average linkage clustering method (39). This approach minimizes OTU inflation (40), and is considered comparable to sequencing error noise-reduction methods such as PyroNoise (41). All bacterial V6-pyrotag SSU rRNA gene sequences were clustered into 65,545 OTUs at the 97% similarity level. Taxonomic identifications for OTUs based on representative sequences were assigned by using Global Assignment of Sequence Taxonomy (39). National Center for Biotechnology Information Sequence Read Archive (SRA) accession numbers, including newly submitted Palmer Station bacterial SRAs (SRA059385), are located in Dataset S1. To ensure compliance with community standards (42), the open-source Investigation/Study/Assay (43) metadata-tracking framework was used to curate the datasets and format them for submission to the SRA database.
Iterative Matrix Randomization Process.
We built an observed taxon-by-sample matrix summarizing the absolute abundance (sequenced counts) of each of the 97% identical OTUs in each sample. To compare the observed results vs. null expectation patterns given the same sampling scheme and absolute OTU abundances, we permuted our observed taxon-by-sample matrix, and then analyzed the resulting permuted dataset.
Permutation.
We randomly permuted counts across our taxon-by-sample matrix, maintaining row and column sums, (i.e., the total number of counts per sample and the global absolute abundance of each OTU). This allowed us to generate matrices of populations with neither dispersal limitation nor sample or location specific selection, while controlling for sampling design and global taxon abundance. We permuted the matrix 1,000 times and stored the resulting matrices. We performed all permutations by using custom scripts in the R program (R Development Core Team).
Null distributions.
From these permuted matrices, we built a null expectation of the likelihood of three particular latitudinal distributions: bipolar, paired, and hemisphere-asymmetrical. Here, bipolarity is defined as a taxon occurring only poleward of a specified boundary latitude in both the northern and southern hemispheres, but not at any lower latitude (Fig. 2A). A paired distribution is defined as a taxon occurring poleward of specified boundary latitudes in both the northern and southern hemispheres, without regard to whether it occurs at any lower latitude (Fig. 2B). Finally, a hemisphere-asymmetrical distribution is defined as a taxon occurring poleward of specified boundary latitudes in a single hemisphere, and not occurring at any other latitude (Fig. 2C). For each permuted matrix we counted the number of taxa exhibiting each of the three distributions of interest (bipolar, paired, hemisphere-asymmetrical), at varying boundary latitudes as follows. For bipolarity, we considered boundary latitudes from 55° to 70° in steps of 5°; for paired-taxon, we considered boundary latitudes from 20° to 70° in steps of 5°; and for hemisphere-asymmetrical we considered boundary latitudes from 0° to 70° in steps of 5°. At each boundary latitude for each permuted matrix, we counted the number of taxa exhibiting the distribution in question and thereby built a null expectation distribution across all 1,000 matrices.
By comparing the permutation distribution to the number of taxa that exhibited the distribution in the nonpermuted, observed dataset, we can calculate a P value, estimating the nonparametric likelihood of the observed data under null expectations. In each case, the P value equals the number of permuted values more extreme than the observed value divided by the number of permutations minus one [P = #Extreme/(N − 1)].
Richness Estimation of Epipelagic Bacterial Communities.
We calculated bacterial richness by using parametric richness estimation methods implemented in the CatchAll program (44). We used the best parametric model estimation with frequency count data selected by CatchAll. We ran our analyses using full datasets and datasets sampled down to the lowest sampling effort by using in-house scripts that randomly resampled our original dataset matrix.
Latitudinal and Temperature Range Size Measurements.
Latitudinal range size was defined as the range between the northernmost latitude and southernmost latitude where taxa for a given taxonomic unit occurred. Temperature range size was defined as the range between the highest and lowest-temperature where taxa occurred for a given taxonomic unit. In this analysis, we used measured temperature of seawater at the time of sampling or the mean estimated SST corresponding to the week the sample was taken (Dataset S1).
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation (NSF) MIRADA-LTER Sites Grant DEB-0717390 (to L.A.A.-Z.) and NSF Grant OPP-0823101 (Palmer Long Term Ecological Research) (to H.W.D.); and by the Sloan Foundation and W. M. Keck Foundation (M.L.S.). This is an International Census of Marine Microbes contribution.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive; for the list of accession numbers, see Dataset S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212424110/-/DCSupplemental.
References
- 1.Hooker JD. The Botany of the Antarctic Voyage of H. M. Discovery Ships Erebus and Terror, in the Years 1839-1843. London: Reeve Brothers; 1847. [Google Scholar]
- 2.Ross JC. A Voyage of Discovery and Research in the Southern and Antarctic Regions, during the Years 1839-43. London: John Murray; 1847. [Google Scholar]
- 3.Darling KF, et al. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature. 2000;405(6782):43–47. doi: 10.1038/35011002. [DOI] [PubMed] [Google Scholar]
- 4.Darling KF, Kucera M, Pudsey CJ, Wade CM. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc Natl Acad Sci USA. 2004;101(20):7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montresor M, Lovejoy C, Orsini L, Procaccini G, Roy S. Bipolar distribution of the cyst-forming dinoflagellate Polarella glacialis. Polar Biol. 2003;26:186–194. [Google Scholar]
- 6.Cermeño P, Falkowski PG. Controls on diatom biogeography in the ocean. Science. 2009;325(5947):1539–1541. doi: 10.1126/science.1174159. [DOI] [PubMed] [Google Scholar]
- 7.Baas-Becking LGM. Geobiologie of Inleiding tot de Milieukunde. The Hague: W.P. Van Stockum and Zoon; 1934. Dutch. [Google Scholar]
- 8.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296(5570):1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 9.Leibold MA, et al. The metacommunity concept: A framework for multi-scale community ecology. Ecol Lett. 2004;7:601–613. [Google Scholar]
- 10.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGillicuddy DJ, Jr, et al. Eddy/wind interactions stimulate extraordinary mid-ocean plankton blooms. Science. 2007;316(5827):1021–1026. doi: 10.1126/science.1136256. [DOI] [PubMed] [Google Scholar]
- 12.Grossart HP, Dziallas C, Leunert F, Tang KW. Bacteria dispersal by hitchhiking on zooplankton. Proc Natl Acad Sci USA. 2010;107(26):11959–11964. doi: 10.1073/pnas.1000668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staley JT. Biodiversity: Are microbial species threatened? Curr Opin Biotechnol. 1997;8(3):340–345. doi: 10.1016/s0958-1669(97)80014-6. [DOI] [PubMed] [Google Scholar]
- 14.Martiny JB, et al. Microbial biogeography: Putting microorganisms on the map. Nat Rev Microbiol. 2006;4(2):102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 15.Casteleyn G, et al. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc Natl Acad Sci USA. 2010;107(29):12952–12957. doi: 10.1073/pnas.1001380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amaral-Zettler L, et al. A global census of marine microbes. In: McIntyre AD, editor. Life in the World’s Oceans: Diversity, Distribution and Abundance. New York: Wiley; 2010. pp. 223–245. [Google Scholar]
- 17.Zinger L, et al. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE. 2011;6(9):e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10(7):497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 19.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton: Princeton Univ Press; 2001. [DOI] [PubMed] [Google Scholar]
- 20.Rutherford S, D’Hondt S, Prell W. Environmental controls on the geographic distribution of zooplankton diversity. Nature. 1999;400:749–753. [Google Scholar]
- 21.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103(3):626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pommier T, et al. Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol. 2007;16(4):867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- 23.Fuhrman JA, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105(22):7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amend AS, et al. Macroecological patterns of marine bacteria on a global scale. J Biogeogr. 2012 10.1111/jbi.12034. [Google Scholar]
- 25.Ghiglione JF, et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci USA. 2012;109(43):17633–17638. doi: 10.1073/pnas.1208160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barton AD, Dutkiewicz S, Flierl G, Bragg J, Follows MJ. Patterns of diversity in marine phytoplankton. Science. 2010;327(5972):1509–1511. doi: 10.1126/science.1184961. [DOI] [PubMed] [Google Scholar]
- 27.Stevens GC. The latitudinal gradients in geographical range: How so many species co-exist in the tropics? Am Nat. 1989;133:240–256. [Google Scholar]
- 28.DeVries T, Primeau F. Dynamically and observationally constrained estimates of water-mass distributions and ages in the global ocean. J Phys Oceanogr. 2011;41:2381–2401. [Google Scholar]
- 29.Clark JS, et al. Seed dispersal near and far: Patterns across temperate and tropical forests. Ecology. 1999;80:1475–1494. [Google Scholar]
- 30.Yooseph S, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468(7320):60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 31.Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA. 2009;106(52):22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chase JM, Myers JA. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Lond B Biol Sci. 2011;366(1576):2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stegen JC, Lin X, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012;6(9):1653–1664. doi: 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitaker RJ. Allopatric origins of microbial species. Philos Trans R Soc Lond B Biol Sci. 2006;361(1475):1975–1984. doi: 10.1098/rstb.2006.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber TS, Deutsch C. Ocean nutrient ratios governed by plankton biogeography. Nature. 2010;467(7315):550–554. doi: 10.1038/nature09403. [DOI] [PubMed] [Google Scholar]
- 36.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE. 2009;4(7):e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318(5847):97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 38.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8(7):R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huse SM, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4(11):e1000255. doi: 10.1371/journal.pgen.1000255. and erratum (2008) 4(12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12(7):1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yilmaz P, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29(5):415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sansone SA, et al. Toward interoperable bioscience data. Nat Genet. 2012;44(2):121–126. doi: 10.1038/ng.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bunge JA. Estimating the number of species with CatchAll. In: Altman RB, Dunker AK, Hunter L, Murray T, Klein TE, editors. Biocomputing 2011: Proceedings of the Pacific Symposium, Kohala Coast, Hawaii, USA, 3-7 January 2011. Vol. 16. Singapore: World Scientific Publishing; 2011. pp. 121–130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.