Abstract
Allosteric regulation of protein function is a critical component of metabolic control. Its importance is underpinned by the diversity of mechanisms and its presence in all three domains of life. The first enzyme of the aromatic amino acid biosynthesis, 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, shows remarkable variation in allosteric response and machinery, and both contemporary regulated and unregulated orthologs have been described. To examine the molecular events by which allostery can evolve, we have generated a chimeric protein by joining the catalytic domain of an unregulated 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with the regulatory domain of a regulated enzyme. We demonstrate that this simple gene fusion event on its own is sufficient to confer functional allostery to the unregulated enzyme. The fusion protein shares structural similarities with its regulated parent protein and undergoes an analogous major conformational change in response to the binding of allosteric effector tyrosine to the regulatory domain. These findings help delineate a remarkably facile mechanism for the evolution of modular allostery by domain recruitment.
Keywords: ACT domain, shikimate
Protein allostery, where ligand binding is coupled to a functional change at a remote site, is critical for the control of metabolism. For enzymes of key metabolic pathways, allostery allows precise control of catalysis in response to cellular demand. Although this important phenomenon was first described many years ago, it is only more recently that the molecular details that govern this precise control of enzyme function have been explored for a diverse range of protein systems (1–3). Functional change results from changes in the protein energy landscape elicited by ligand binding (4). This change in energy landscape may lead to conformational change and/or may be more subtly expressed as a change in protein molecular dynamics (5–12).
The importance of protein allostery is underpinned by the observation that it is ubiquitous. As more proteins are studied in detail, one of the striking revelations is the variety of allosteric mechanisms that are used to control protein function. These mechanisms can be broadly divided into three groups that reflect the evolutionary path taken to acquire the allosteric functionality (5). In the first group, modification of existing structural features of a protein creates an allosteric site. Second, the formation of homo- and heterooligomeric assemblies may lead to the development of allosteric sites at the interface of subunits. Third, allostery may be endowed by domain fusion to create a modular protein with a distinct domain responsible for binding of the allosteric effector. A detailed understanding of this modular allostery, in which the allosteric effector binding site is associated with a discrete protein domain, offers the potential to generate engineered proteins with tailored functionality.
The ACT domain has been identified as a modular regulatory unit associated with the control of a variety of metabolic processes (13–16). The name of this domain originates from the three proteins in which it was first identified: aspartokinase, chorismate mutase, and prephenate dehydrogenase (17). This domain, of ∼80 amino acids in length, was first identified as a ligand-binding regulatory module in a number of proteins of diverse function, leading to its description as a “conserved evolutionarily mobile ligand-binding regulatory module” (17). Most proteins in which the ACT domain has been recognized catalyze early steps in amino acid and purine metabolic pathways, and the ACT domain serves as a binding site for pathway end products. The ligand-binding sites for the allosteric effector(s) are typically associated with interfaces between two or more ACT domains. The underlying details of the allosteric mechanisms associated with the ACT domain are now starting to be revealed: generally, ligand binding in these sites elicits structural changes that alter the catalytic function of the remote active site (13, 15, 16, 18, 19).
3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase (DAH7PS) is the first enzyme of the shikimate pathway, which is responsible for the biosynthesis of the aromatic amino acids Phe, Tyr, and Trp (Fig. 1A). Although all DAH7PS enzymes share a core catalytic (β/α)8 barrel, this barrel is variably decorated with additional structural elements, creating a remarkable array of diverse examples of modular allostery (Fig. S1). The most intricate system discovered to date is demonstrated by the DAH7PS from Mycobacterium tuberculosis, where multiple subdomains and ligand-binding sites grafted onto the (β/α)8 barrel are associated with a complex synergistic allostery by Phe and Trp, largely mediated by changes in protein flexibility (20). The DAH7PS enzymes from Escherichia coli and Saccharomyces cerevisiae have N-terminal barrel extensions providing a single aromatic amino acid binding site (21, 22). Other DAH7PS enzymes are fused to a functional chorismate mutase domain for the purposes of allosteric control (23, 24). The simplest contemporary DAH7PS enzymes show no extra barrel extensions and hence are unregulated. This latter group includes the structurally characterized enzymes from Pyrococcus furiosus (25) and Aeropyrum pernix (26). We recently demonstrated that the DAH7PS from Thermotoga maritima, structurally composed of an ACT domain fused to the N terminus of a catalytic (β/α)8 barrel, undergoes a remarkable conformational change associated with ligand binding (27). The associated domain reorganization controls enzyme functionality by physically gating substrate access to the active site upon binding of Tyr (Fig. 1B).
Fig. 1.
(A) DAH7PS catalyzes the first step of the shikimate pathway, the condensation of PEP and E4P to give 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAH7P), for the biosynthesis of the aromatic amino acids. (B) A superposition of the barrels of the open Tyr-free (PDB ID code 1RZM, green) and closed Tyr-bound (PDB ID code 3PG9, purple) conformations of TmaDAH7PS showing the rearrangement of the ACT domains in the presence of Tyr.
How readily can this modular allostery demonstrated by Tyr-regulated T. maritima DAH7PS (TmaDAH7PS) be acquired by a simple gene fusion event? The DAH7PS family, with contemporary examples of both unregulated enzymes and those containing a discrete ACT regulatory domain controlling catalysis by physical gating of the active site, is an ideal family to examine the evolution of this important enzyme property. In this work, we describe a regulated DAH7PS chimera (TmaACT-PfuDAH7PS), constructed by combining the ACT domain from TmaDAH7PS with the unregulated catalytic (β/α)8 barrel domain from P. furiosus DAH7PS (PfuDAH7PS). This article demonstrates the transfer of functional allostery from one contemporary protein to the ortholog from another genetically distinct species. These findings demonstrate that the acquisition of allosteric properties by gene fusion is surprisingly facile.
Results
Design Considerations: Construction of Fusion Protein TmaACT-PfuDAH7PS.
Unregulated PfuDAH7PS, whose subunit comprises the basic (β/α)8 barrel housing the active site, was chosen as the scaffold for fusion to the regulatory ACT domain of TmaDAH7PS. Both PfuDAH7PS and TmaDAH7PS crystallize with a similar homotetrameric architecture, which has been shown to be critical for the allosteric mechanism of TmaDAH7PS (25, 28). The crystal structures of both proteins show the existence of a β-hairpin at the N-terminal end of the (β/α)8 barrel (Fig. S2). For TmaDAH7PS, the ACT regulatory domain is connected to this hairpin via a flexible linker of 19 amino acids. The ligand-bound crystal structure of TmaDAH7PS illustrates the importance of the flexibility afforded by the linker for allosteric inhibition of the enzyme by Tyr (27). A fusion construct (TmaACT-PfuDAH7PS) was designed with its ACT domain, linker sequence, and β-hairpin from TmaDAH7PS (residues 1–94) and the catalytic barrel (from residue 24) from PfuDAH7PS (Fig. S3).
The coexpression of TmaACT-PfuDAH7PS with the E. coli chaperonins resulted in soluble protein with a molecular mass identical to that inferred from the TmaACT-PfuDAH7PS ORF. Circular dichroism spectrophotometry indicated the enzyme was correctly folded (Fig. S4).
TmaACT-PfuDAH7PS Is Catalytically Active and Inhibited by Tyr.
TmaACT-PfuDAH7PS is catalytically active with kinetic parameters similar to those of WT PfuDAH7PS, the enzyme from which the catalytic barrel of TmaACT-PfuDAH7PS was derived (Table 1). The chimeric enzyme displays limited cooperativity with respect to substrate concentrations and negligible change in the Hill coefficient is observed in the absence and presence of Tyr (Fig. S5).
Table 1.
Kinetic parameters for TmaACT-PfuDAH7PS, PfuDAH7PS, and TmaDAH7PS
| Enzyme | KmPEP (µM) | KmE4P (µM) | kcat (s−1) | kcat/KmE4P (s−1⋅µM−1) |
| TmaACT-PfuDAH7PS | 46 ± 4 | 10 ± 1 | 2.41 ± 0.02 | 0.24 |
| PfuDAH7PS* | 120 ± 10 | 22 ± 2 | 9.70 ± 0.03 | 0.44 |
| TmaDAH7PS | 4.85 ± 0.04 | 13 ± 1 | 11.7 ± 0.2 | 0.9 |
| TmaACT-PfuDAH7PS (with Tyr)† | 42 ± 2 | 20 ± 2 | 1.6 ± 0.1 | 0.08 |
| TmaDAH7PS (with Tyr)b | 21 ± 1 | 118 ± 10 | 2.59 ± 0.07 | 0.02 |
*The kinetic parameters for PfuDAH7PS determined in the absence and presence of Tyr are identical.
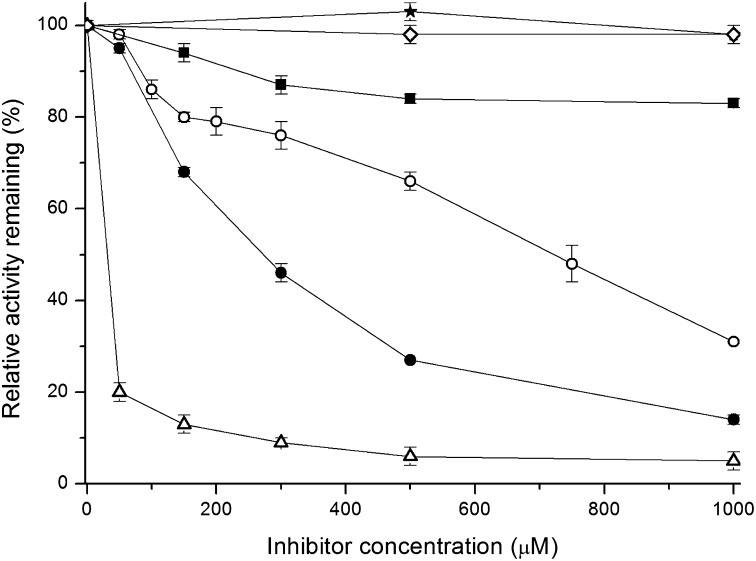
TmaDAH7PS is strongly inhibited by Tyr and to a lesser extent inhibited by Phe (27). These two amino acids were tested as inhibitors of TmaACT-PfuDAH7PS (Fig. 2). Both Tyr and Phe had an inhibitory effect on the enzyme, with Tyr being a more effective inhibitor. The inhibition by Tyr was less pronounced for TmaACT-PfuDAH7PS than for TmaDAH7PS.
Fig. 2.
TmaACT-PfuDAH7PS is inhibited by Tyr and Phe. Inhibitory response of TmaACT-PfuDAH7PS toward increasing concentrations of Tyr (open circles) and Phe (closed squares) in comparison with the inhibition of TmaDAH7PS achieved by Tyr (open triangles) and Phe (closed circles), and the effect on the activity of PfuDAH7PS in the presence of Tyr (open diamonds) and Phe (closed stars). The catalytic activity of TmaACT-PfuDAH7PS was determined in the presence of 300 µM PEP, 100 µM MnSO4, 110 µM E4P and 2 uL enzyme at 6.2 mg mL−1 in 50 mM BTP, pH 7.3 and 0–1 mM of either Tyr or Phe. The catalytic activity of PfuDAH7PS was determined in the presence of 585 µM PEP, 112 µM E4P, 100 µM MnSO4, and 2 µL enzyme at 1.2 mg⋅mL−1 in 50 mM BTP, pH 6.8, and 0–1 mM of either Tyr or Phe. Triplicate assays were performed and averaged.
Kinetic parameters were determined for TmaACT-PfuDAH7PS in the presence of Tyr (Table 1). The apparent Km for phosphoenolpyruvate (PEP) remained relatively unchanged by the presence of Tyr. On the other hand, the TmaACT-PfuDAH7PS enzyme showed a twofold reduced affinity for erythrose 4-phosphate (E4P) and a 30% lower kcat. A reduced kcat/KmE4P in the presence of Tyr is also observed for WT TmaDAH7PS, and the reduction in this parameter is also found for TmaACT-PfuDAH7PS (27).
Small-Angle X-Ray Scattering Measurements Indicate the Same Allosteric Mechanism for TmaACT-PfuDAH7PS and TmaDAH7PS.
Tyr binding to TmaDAH7PS is associated with a major conformational change as the regulatory ACT domains dimerize to occlude access to the active sites (Fig. 1B) (27). This large structural change between the open (Tyr-free) and closed (Tyr-bound) forms is readily observable by altered scattering profiles measured by small-angle X-ray scattering (SAXS). Scattering profiles of TmaACT-PfuDAH7PS were compared with those obtained for TmaDAH7PS in both the presence and absence of Tyr (Fig. 3 and Fig. S6). In the absence of Tyr, the scattering profile of TmaACT-PfuDAH7PS closely resembles the experimentally determined scattering and theoretical scattering profiles calculated from the crystal structure for the open form of TmaDAH7PS (Fig. 3A).
Fig. 3.
SAXS of TmaACT-PfuDAH7PS (open circles) measured in the absence (A) and presence (B) of Tyr. Theoretical scattering profiles were generated from crystallographic coordinates from both the open Tyr-free (PDB ID code 1RZM, solid line) and closed Tyr-bound (PDB ID code 3PG9, dashed line) TmaDAH7PS crystal structures using CRYSOL (29). Discrepancy from the fit of the calculated open and closed crystal structures to the experimentally determined data (χ) in the absence of Tyr are 0.56 and 2.2, respectively, and in the presence of Tyr are 2.5 and 0.90. P(r) function calculations and scattering parameters are provided in Table S1 and Fig. S6.
In the presence of Tyr, the scattering profile for TmaACT-PfuDAH7PS was significantly altered (Fig. 3B). This change is consistent with the changes in the scattering profile observed for TmaDAH7PS and indicates the engineered TmaACT-PfuDAH7PS enzyme behaves similarly to TmaDAH7PS in the presence of Tyr, and adopts a more compact, globular structure relative to the Tyr-free structures.
Tyr Binds Between the Interfaces of Two ACT Domains Formed From Diagonally Opposite Subunits in Tetrameric TmaACT-PfuDAH7PS.
The structure of TmaACT-PfuDAH7PS in the presence of Tyr was determined by X-ray crystallography. TmaACT-PfuDAH7PS adopts a tetrameric structure in the crystalline form, consistent with the structures of the undecorated PfuDAH7PS (Fig. S7A) and TmaDAH7PS, with or without Tyr bound. The TmaACT-PfuDAH7PS structure is found in the closed conformation where ACT domains from diagonal subunits interact (Fig. 4). This arrangement is very similar to that observed for the structure of Tyr-bound TmaDAH7PS (Fig. 1B). Likewise, Tyr is observed to bind in the same location in the chimeric protein as in TmaDAH7PS, with two Tyr molecules bound between each pair of diagonally opposite ACT domains (Fig. S7B). There is close agreement between the theoretical scattering profile generated from the crystallographic coordinates of this Tyr-bound TmaACT-PfuDAH7PS structure with the SAXS profile for TmaACT-PfuDAH7PS in the presence of Tyr (χ of 0.78, Fig. S8).
Fig. 4.

The X-ray crystal structure of TmaACT-PfuDAH7PS with Tyr bound. The (β/α)8 barrel portion, which derives from PfuDAH7PS, of each subunit in the homotetramer is colored slate, whereas the ACT domain and residues derived from TmaDAH7PS are colored magenta. The bound Tyr ligands are shown as spheres with carbon atoms colored cyan.
Comparison of the individual catalytic (β/α)8 barrel domains of TmaACT-PfuDAH7PS (residues 80–332) and WT PfuDAH7PS (residues 24–262) reveals negligible overall differences (rmsd 0.36–0.98 Å between equivalent Cα atoms for the various pairwise superpositions of the four domains). There are minor changes in the disposition of the subunits that form the tetramer as the TmaACT-PfuDAH7PS tetramer superimposes on the PfuDAH7PS tetramer with an increased rmsd of 0.65 Å (Fig. S7A).
The comparison of the tetrameric structures of Tyr-bound TmaACT-PfuDAH7PS and the Tyr-bound TmaDAH7PS tetramer suggests minor structural variation (rmsd 1.18 Å). This variation arises primarily from the slight twist of one pair of diagonally opposed subunits relative to the other for TmaACT-PfuDAH7PS compared with TmaDAH7PS, with the (β/α)8 barrel of the chimeric protein more closely resembling its parent PfuDAH7PS. Superpositions of isolated ACT domains (residues 1–66) from TmaACT-PfuDAH7PS onto those from TmaDAH7PS, and separately, of the catalytic barrel domain (residues 80–332 for TmaACT-PfuDAH7PS onto residues 95–332 for TmaDAH7PS) have very similar rmsd values of 0.36–0.45 Å and 0.80–1.31 Å, respectively, indicating close structural similarity of the two domains between the Tyr-bound structures.
Closure clearly results in contacts between the regulatory domains and Tyr, and these contacts are conserved between the Tyr-bound TmaDAH7PS and TmaACT-PfuDAH7PS structures. There are also some contacts that form between the regulatory domain and the diagonally opposed (β/α)8 barrel in the Tyr-bound closed conformation. These contacts are well preserved between the ACT and catalytic (βα)8 barrel domains in the comparison of Tyr-bound TmaDAH7PS with the Tyr-bound TmaACT-PfuDAH7PS. In general, however, for the Tyr-bound TmaACT-PfuDAH7PS and TmaDAH7PS, the residues in close proximity between the ACT domains (which share common sequence) and the catalytic barrel domains (which do not share a common sequence, Fig. S3) are not conserved. Whereas this region of the catalytic barrel has evolved to be optimally functional for TmaDAH7PS, this evolution has not occurred for TmaACT-PfuDAH7PS. In particular, the Gly310 (TmaDAH7PS) to Ser310 (TmaACT-PfuDAH7PS) exchange forces a change in contacts between Ser55 and the conserved Arg277. Likewise, the Lys311 (TmaDAH7PS) to Gln311 (TmaACT-PfuDAH7PS) and Glu261 (TmaDAH7PS) to Leu261 (TmaACT-PfuDAH7PS) substitutions weaken electrostatic interactions with Glu54 and Lys67, respectively. These changes likely account in part for the decreased sensitivity toward Tyr.
Although not making direct contact with the ACT domain, the E4P-binding loop is in close proximity, and the conformations of the E4P-binding loop (residues 130–142) are perturbed by movement of the ACT domain upon binding of Tyr for both TmaDAH7PS and TmaACT-PfuDAH7PS. For the latter structure, this loop is poorly defined in all subunits. In addition, one intrasubunit contact between the hydroxyl moiety of Tyr47 and the side chain of Glu222 is poorly defined in the chimeric protein. The ratio of the average B factor for the ACT domain to that for the barrel domain for the Tyr-bound structures is noticeably higher at 1.3 for TmaACT-PfuDAH7PS than that of 1.1 for TmaDAH7PS. This indicates that the ACT domain is less tightly locked over the active site for the Tyr-bound TmaACT-PfuDAH7PS than for the corresponding TmaDAH7PS. This difference, along with the altered twist of the tetramer, may also contribute to the reduced response to Tyr.
Discussion
It is generally understood that new protein functions have evolved by the modification of existing protein scaffolds, including “domain swapping”, whereby rearranged proteins are created by recombining gene sequences that encode for different domains or subdomains (30, 31). In this way, several functions can be linked in a single polypeptide chain and modular proteins of altered functionality can be generated. There is intense interest in the creation of proteins with engineered allosteric function for the generation of protein molecular switches (for example, for biosensors and reporting on cellular activities), in which protein function is coupled to a signal received at a remote allosteric site (9, 32, 33). As such, several previous studies have sought to artificially incorporate allosteric behavior by the addition of protein modules to enzymes (32, 34). For example, random domain insertion of the β-lactamase gene into maltose-binding protein created enzymes with activity both up- and down-regulated by maltose, and inhibited, in one chimera, by Zn2+ that binds at an interface between the two domains (30, 33, 35). The full structural details of this system have only recently been reported (36, 37). Light-dependent catalytic activity has been observed when dihydrofolate reductase was linked to a light-sensing signaling domain (32). And, complementarily, fluorescent proteins have been grafted onto other proteins, especially ones that undergo conformational changes in response to ligand binding, to endow biosensory activity (38). However, only rarely has detailed structural information been obtainable to discern the mechanism of allostery.
There are fundamental differences between these systems and our fusion protein. The ACT domain used here is a discrete protein module, which is found linked to a diverse group of enzymes, thereby conferring allosteric functionality (13, 16). Allostery is mediated by ligand binding to the ACT domain. These binding sites are usually found at the interface between two or more of these domains, and the catalytic function of the host enzyme is generally altered by a change in structure associated with changes in the interactions between the ACT domains. This change is very obvious for TmaDAH7PS and TmaACT-PfuDAH7PS, in which a major conformational change occurs on ligand binding to disrupt catalysis. Here, the regulatory mechanism depends upon not only the quaternary structure of the catalytic domains, but also on that of the regulatory unit, which in this case rotates by more than 90° about a hinge at residue 66, to form a paired association on binding Tyr.
Our system constitutes, we believe, an instance in which a known allosteric regulatory element has been transferred from one enzyme to an unregulated ortholog from a different species. This suggests a very simple mechanism of domain recruitment, especially as in this particular instance the organisms, despite being from different domains, are known to have exchanged and used genetic material (39). The readiness with which this allosteric mechanism can be transferred to another contemporary DAH7PS catalytic unit is entirely consistent with the physical gating mechanism by which it operates, simply requiring a flexible linker and the regulatory domains appended to the subunits of a conserved catalytic homotetrameric core. The key contacts that enable regulation and association of the regulatory domains are encoded within the regulatory domain itself, and the regulatory domain truly befits the description as an evolutionarily mobile module (17). In this study, we have mimicked the evolutionary pathway likely adopted by the DAH7PS family to provide the regulation manifest in contemporary enzymes. This observation of the facile acquisition of allosteric properties is entirely consistent with the importance of gene fusion events in the evolution of functional allostery.
Methods
Cloning, Enzyme Expression and Purification, and Kinetic Assays.
A synthetic gene encoding for the TmaACT-PfuDAH7PS fusion protein was designed to incorporate residues 1–94 of TmaDAH7PS and 24–262 of PfuDAH7PS (Figs. S2 and S3). TmaACT-PfuDAH7PS was expressed in the pDEST14 vector in E. coli Chaperone 3 cells. The expression and purification of the chimeric protein was performed using the same methods that have been previously described for TmaDAH7PS (27). The mass of TmaACT-PfuDAH7PS was measured by electrospray ionization MS. Kinetic parameters were measured for TmaACT-PfuDAH7PS using the procedures previously described for TmaDAH7PS and PfuDAH7PS (25, 27, 40). The kinetic analysis was performed at 60 °C, the highest temperature achievable without substrate decomposition compromising the assay. Details of the assays performed are provided in Fig. S5.
SAXS.
SAXS experiments were performed at the Australian Synchrotron SAXS/WAXS beamline using the set-up previously described (27). Scattering data were collected from TmaACT-PfuDAH7PS following elution from a size-exclusion chromatography column (Superdex 200 5/150) (applied to the column at ∼12 mg⋅mL−1), pre-equilibrated (10 mM BTP pH 7.5, 100 mM KCl, 200 µM PEP) with or without amino acid (1 mM Tyr). Data were processed as described for TmaDAH7PS (27).
X-Ray Crystallography and Structure Refinement.
TmaACT-PfuDAH7PS was crystallized by sitting-drop vapor diffusion using 0.3 µL drops at 8 °C. Protein solution (12 mg⋅mL−1 in 10 mM BTP pH 7.5, 100 mM KCl, 200 µM PEP, 0.5 mM TCEP) was mixed 1:1 (vol/vol) with the reservoir solution containing 0.2 M sodium chloride, 20% (wt/vol) polyethylene glycol 6000, 0.1 M Tris (pH 8), and 0.02% (wt/vol) sodium azide before the addition of Tyr (1 mM final concentration) to the drop. Immediately before data collection, the crystal was transferred to a cryoprotectant solution containing the reservoir solution plus 6.7% glycerol (vol/vol), 6.7% 2-methyl-2,4-pentanediol (vol/vol), and 6.7% polyethylene glycol 400 (vol/vol). X-ray diffraction data were collected at the Australian Synchrotron MX1 beamline and processed using XDS (41) and Scala (42). The phases were initially estimated using molecular replacement [Phaser (43)] with TmaDAH7PS [Protein Data Bank (PDB) ID code 1VR6] as the template. Chainsaw (44) was used to produce a corrected model and subsequent model building was accomplished with COOT (45) and the model was refined with Refmac5 (46). Simulated annealing using Phenix (47) was used to remove phase bias. The results are summarized in Table 2, along with key structure refinement details. TmaACT-PfuDAH7PS crystallized in the orthorhombic space group P212121, and diffracted to 3.00 Å, with the following unit cell dimensions: a = 76.9 Å, b = 130.9 Å, and c = 138.1 Å. The Rfree set was chosen in thin shells using Phenix (47). Validation tools of COOT and Molprobity (48) were used to check for and correct conformational infelicities. Structural alignments and root mean square deviation values were calculated using MAMMOTH-mult (49) and PyMOL (50).
Table 2.
Crystal parameters, data collection, and refinement statistics
| TmaACT-PfuDAH7PS in complex with Tyr | |
| Data collection | |
| Crystal system; space group | Orthorhombic; P212121 |
| Unit cell parameters (Å) a, b, c | 76.9, 130.9, 138.1 |
| Resolution range (Å) | 37.94–3.00 (3.08–3.00) |
| Total no. of reflections | 80,989 |
| Unique reflections | 27,079 |
| Redundancy | 3.0 (3.0) |
| Completeness (%) | 95.5 (97.6) |
| I/σ(I) | 7.0 (1.7) |
| Rmerge | 0.101 (0.581) |
| Wilson B-value (Å2) | 68.1 |
| Refinement | |
| Resolution (Å) | 37.94–3.00 |
| Rcryst | 0.192 |
| Rfree | 0.248 |
| Chain length | 333 |
| Observed number of residues (chains A, B, C, and D) | 333, 333, 332, 331 |
| Water molecules | 23 |
| Other (Tyr) | 4 |
| Mean B (Å2) | |
| Protein | 82.2 |
| Water | 30.9 |
| Other (Tyr) | 66.6 |
| rmsd from ideal values | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.1 |
| Dihedral angles (°) | 4.8 |
| Ramachandran plot | |
| Preferred (%) | 94.38 (1,243 residues) |
| Allowed (%) | 5.54 (73 residues) |
| Outliers (%) | 0.08 (1 residue) |
| PDB ID code | 4GRS |
Supplementary Material
Acknowledgments
Part of this research was undertaken on the MX and SAXS/WAXS beamlines at Australian Synchrotron, Victoria, Australia. Scholarships were provided by the University of Canterbury (to P.J.C. and T.M.A.). This work is supported by the New Zealand Marsden Fund (UOC1105).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4GRS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217923110/-/DCSupplemental.
References
- 1.Changeux J-P. Allostery and the Monod-Wyman-Changeux model after 50 years. Ann Rev Biophys. 2012;41(2):103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 2.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12(1):88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 3.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308(5727):1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 4.Tsai C-J, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: A classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5(3):207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peracchi A, Mozzarelli A. Exploring and exploiting allostery: Models, evolution, and drug targeting. Biochim Biophys Acta. 2011;1814(8):922–933. doi: 10.1016/j.bbapap.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Tsai CJ, del Sol A, Nussinov R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378(1):1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4(8):474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 8.Bahar I, Chennubhotla C, Tobi D. Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Curr Opin Struct Biol. 2007;17(6):633–640. doi: 10.1016/j.sbi.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16(1):102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Popovych N, Sun SJ, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13(9):831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Formaneck MS, Ma L, Cui Q. Reconciling the “old” and “new” views of protein allostery: A molecular simulation study of chemotaxis Y protein (CheY) Proteins. 2006;63(4):846–867. doi: 10.1002/prot.20893. [DOI] [PubMed] [Google Scholar]
- 12.Weinkam P, Pons J, Sali A. Structure-based model of allostery predicts coupling between distant sites. Proc Natl Acad Sci USA. 2012;109(13):4875–4880. doi: 10.1073/pnas.1116274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curien G, et al. Amino acid biosynthesis: New architectures in allosteric enzymes. Plant Physiol Biochem. 2008;46(3):325–339. doi: 10.1016/j.plaphy.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Grant GA. The ACT domain: A small molecule binding domain and its role as a common regulatory element. J Biol Chem. 2006;281(45):33825–33829. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- 15.Siltberg-Liberles J, Martinez A. Searching distant homologs of the regulatory ACT domain in phenylalanine hydroxylase. Amino Acids. 2009;36(2):235–249. doi: 10.1007/s00726-008-0057-2. [DOI] [PubMed] [Google Scholar]
- 16.Liberles JS, Thórólfsson M, Martínez A. Allosteric mechanisms in ACT domain containing enzymes involved in amino acid metabolism. Amino Acids. 2005;28(1):1–12. doi: 10.1007/s00726-004-0152-y. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287(5):1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, et al. Structural view of the regulatory subunit of aspartate kinase from Mycobacterium tuberculosis. Protein Cell. 2011;2(9):745–754. doi: 10.1007/s13238-011-1094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robin AY, et al. A new mode of dimerization of allosteric enzymes with ACT domains revealed by the crystal structure of the aspartate kinase from Cyanobacteria. J Mol Biol. 2010;399(2):283–293. doi: 10.1016/j.jmb.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Jiao W, Hutton RD, Cross PJ, Jameson GB, Parker EJ. Dynamic cross-talk among remote binding sites: The molecular basis for unusual synergistic allostery. J Mol Biol. 2012;415(4):716–726. doi: 10.1016/j.jmb.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 21.Shumilin IA, Zhao C, Bauerle R, Kretsinger RH. Allosteric inhibition of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase alters the coordination of both substrates. J Mol Biol. 2002;320(5):1147–1156. doi: 10.1016/s0022-2836(02)00545-4. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann M, et al. Evolution of feedback-inhibited β /α barrel isoenzymes by gene duplication and a single mutation. Proc Natl Acad Sci USA. 2003;100(3):862–867. doi: 10.1073/pnas.0337566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Woodard RW. New insights into the evolutionary links relating to the 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase subfamilies. J Biol Chem. 2006;281(7):4042–4048. doi: 10.1074/jbc.M512223200. [DOI] [PubMed] [Google Scholar]
- 24.Light SH, Halavaty AS, Minasov G, Shuvalova L, Anderson WF. Structural analysis of a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain. Protein Sci. 2012;21(6):887–895. doi: 10.1002/pro.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schofield LR, et al. Substrate ambiguity and crystal structure of Pyrococcus furiosus 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase: An ancestral 3-deoxyald-2-ulosonate-phosphate synthase? Biochemistry. 2005;44(36):11950–11962. doi: 10.1021/bi050577z. [DOI] [PubMed] [Google Scholar]
- 26.Zhou L, et al. Structure and characterization of the 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Aeropyrum pernix. Bioorg Chem. 2012;40(1):79–86. doi: 10.1016/j.bioorg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross PJ, Dobson RC, Patchett ML, Parker EJ. Tyrosine latching of a regulatory gate affords allosteric control of aromatic amino acid biosynthesis. J Biol Chem. 2011;286(12):10216–10224. doi: 10.1074/jbc.M110.209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shumilin IA, Bauerle R, Wu J, Woodard RW, Kretsinger RH. Crystal structure of the reaction complex of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Thermotoga maritima refines the catalytic mechanism and indicates a new mechanism of allosteric regulation. J Mol Biol. 2004;341(2):455–466. doi: 10.1016/j.jmb.2004.05.077. [DOI] [PubMed] [Google Scholar]
- 29.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 30.Ostermeier M, Benkovic SJ. Evolution of protein function by domain swapping. Adv Protein Chem. 2000;55:29–77. doi: 10.1016/s0065-3233(01)55002-0. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Fast W, Benkovic SJ. Structural and functional modularity of proteins in the de novo purine biosynthetic pathway. Protein Sci. 2009;18(5):881–892. doi: 10.1002/pro.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, et al. Surface sites for engineering allosteric control in proteins. Science. 2008;322(5900):438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci USA. 2005;102(32):11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park H-S, et al. Design and evolution of new catalytic activity with an existing protein scaffold. Science. 2006;311(5760):535–538. doi: 10.1126/science.1118953. [DOI] [PubMed] [Google Scholar]
- 35.Guntas G, Ostermeier M. Creation of an allosteric enzyme by domain insertion. J Mol Biol. 2004;336(1):263–273. doi: 10.1016/j.jmb.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 36.Ke W, et al. Structure of an engineered β-lactamase maltose binding protein fusion protein: Insights into heterotropic allosteric regulation. PLoS ONE. 2012;7(6):e39168. doi: 10.1371/journal.pone.0039168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright CM, Majumdar A, Tolman JR, Ostermeier M. NMR characterization of an engineered domain fusion between maltose binding protein and TEM1 beta-lactamase provides insight into its structure and allosteric mechanism. Proteins. 2010;78(6):1423–1430. doi: 10.1002/prot.22657. [DOI] [PubMed] [Google Scholar]
- 38.Doi N, Yanagawa H. Design of generic biosensors based on green fluorescent proteins with allosteric sites by directed evolution. FEBS Lett. 1999;453(3):305–307. doi: 10.1016/s0014-5793(99)00732-2. [DOI] [PubMed] [Google Scholar]
- 39.Carballeira NM, et al. Unusual fatty acid compositions of the hyperthermophilic archaeon Pyrococcus furiosus and the bacterium Thermotoga maritima. J Bacteriol. 1997;179(8):2766–2768. doi: 10.1128/jb.179.8.2766-2768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schofield LR, Patchett ML, Parker EJ. Expression, purification, and characterization of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Pyrococcus furiosus. Protein Expr Purif. 2004;34(1):17–27. doi: 10.1016/j.pep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein N. CHAINSAW: A program for mutating pdb files used as templates in molecular replacement. J Appl Cryst. 2008;41(3):641–643. [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lupyan D, Leo-Macias A, Ortiz AR. A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics. 2005;21(15):3255–3263. doi: 10.1093/bioinformatics/bti527. [DOI] [PubMed] [Google Scholar]
- 50. Schrödinger, LLC (2010) The PyMOL Molecular Graphics System, Version 1.3r1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.