Abstract
It is likely that many small ORFs (sORFs; 30–100 amino acids) are missed when genomes are annotated. To overcome this limitation, we identified ∼8,000 sORFs with high coding potential in intergenic regions of the Arabidopsis thaliana genome. However, the question remains as to whether these coding sORFs play functional roles. Using a designed array, we generated an expression atlas for 16 organs and 17 environmental conditions among 7,901 identified coding sORFs. A total of 2,099 coding sORFs were highly expressed under at least one experimental condition, and 571 were significantly conserved in other land plants. A total of 473 coding sORFs were overexpressed; ∼10% (49/473) induced visible phenotypic effects, a proportion that is approximately seven times higher than that of randomly chosen known genes. These results indicate that many coding sORFs hidden in plant genomes are associated with morphogenesis. We believe that the expression atlas will contribute to further study of the roles of sORFs in plants.
Keywords: transcriptome, phenome, Agilent custom microarray, transgenic plant, peptide hormone
It has been revealed that small ORFs (sORFs; 30–100 amino acids) are translated into peptides that play essential roles in eukaryotes. For example, in yeast, 21 of 247 peptides encoded by sORFs are essential for viability, as identified by KO analyses (1). In Drosophila, several peptides encoded by sORFs are involved in activating transcription factors related to development (2). In plants, a number of peptides encoded by known small genes (<150 codons) play significant roles in various aspects of plant growth and development. Specific receptors for various peptides have been identified as receptor kinases (3–18). Although peptides translated from sORFs have important roles, a high rate of false-positive prediction affects the identification of coding sORFs in genome sequences (19, 20). Therefore, in a representative plant species, Arabidopsis thaliana, many small genes had been manually identified using a restricted Markov model and similarity searching (21). To further explore the field of small genes, we developed a computational method to identify coding sORFs using the hexamer composition bias between coding sequences (CDSs) and noncoding sequences (NCDSs) (22, 23). Among available gene finders, this program package has the best performance for identifying true small genes (24).
The model plant species A. thaliana has a high-quality genome, and more than 7,000 coding sORFs were identified in the intergenic regions that lacked annotated genes (22). The coding sORFs do not have any sequence similarities to annotated genes. In the present study, to examine the functional roles of these newly identified coding sORFs, we designed an array to generate an expression atlas under 16 developmental stages and 17 environmental conditions, with three replicates. Then, we looked for evidence of expression of coding sORFs. We also examined the signatures of selective constraints on the CDSs among the coding sORFs in 16 land plant species by comparing synonymous substitutions with nonsynonymous ones. This is because most genes have undergone stronger selective constraints on nonsynonymous substitutions than on synonymous ones (25, 26). After identifying either expressed coding sORFs or those undergoing selective constraint, we generated transgenic plants that individually overexpressed 473 manually selected coding sORFs, and revealed the functional importance of coding sORFs hidden in the A. thaliana genome.
Results and Discussion
Expression Atlas of Coding sORFs.
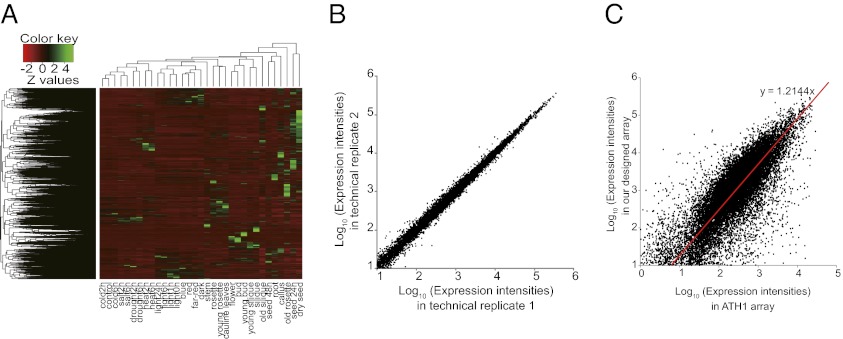
Although some coding sORFs have been annotated as coding genes by The Arabidopsis Information Resource (TAIR; www.arabidopsis.org), we focused on 7,901 coding sORFs that were identified in the A. thaliana genome by our pipelines (Fig. S1; Dataset S1). There are no functional annotations for these 7,901 coding sORFs in TAIR. To examine the expression profile of the coding sORFs, we designed custom arrays by spotting specific 60-mer sequences from each of the coding sORFs and from 26,254 annotated genes (SI Text). The expression atlases were generated under 16 developmental stages and 17 environmental conditions, with three replicates (Fig. 1A). To examine whether our arrays represent a reasonable expression atlas, we compared the expression intensities among three technical replicates in each sample. The expression intensities were significantly positively correlated among the three technical replicates in all of the samples [mean, r = 0.98 ± 0.01 (SD); Fig. 1B], indicating that our array included few errors. The expression intensities were compared with those of the ATH1 array, which is the gold standard platform, using the same sample (seedling; labeled as the control in Fig. 1A). The expression intensities in our array were significantly positively correlated with those in the ATH1 array for annotated genes (r = 0.85, P = 2.2 × 10−16; Fig. 1C), indicating that our array produced comparable results to a comprehensive, standard array. Furthermore, we used an alternative method, real-time quantitative RT-PCR (Q-RT-PCR) to analyze expressions of five selected sORFs. For each of the five sORFs, we focused on two organs with more than twofold differential expression. Differential expressions were validated for these sORFs by Q-RT-PCR (Fig. S2), indicating that our array produced comparable results to Q-RT-PCR.
Fig. 1.
Expression atlas of sORFs and annotated genes. (A) Heat map of all coding sORFs and annotated genes. Color key shows relationship between color and z-score of expression intensities in each tissue or condition. Red and green indicate low and high expression, respectively. These data can be viewed at our website (HANADB-AT: http://evolver.psc.riken.jp/seiken). (B) Correlation of expression intensities between technical replicates in our designed array and in seedling, as an example. x and y axes represent log10 values of expression intensities in technical replicate 1 and technical replicate 2 in seedlings (control in A), respectively. (C) Correlation of expression intensities between our designed array and the ATH1 array for annotated genes. x and y axes represent log10 values of expression intensities in our designed array and ATH1 array in seedlings (control in A), respectively.
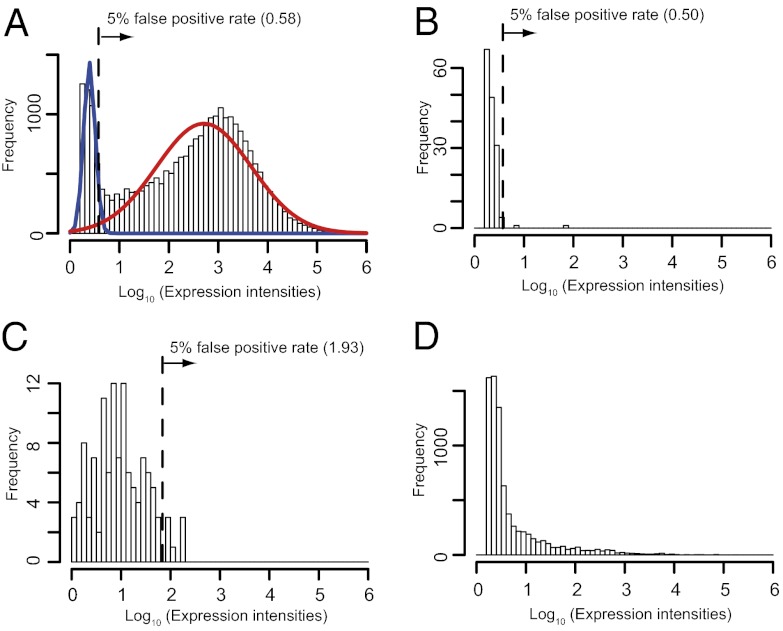
High expression intensity can be an indicator of real genes. Here, we used three criteria to identify coding sORFs with high expression intensity. First, assuming that expression intensities of genes are composed of two Gaussian distributions, expression intensities of annotated genes were fitted to the lower and higher Gaussian distribution in each of 33 conditions (Fig. 2A; Fig. S3). We focused on the lower distribution of expression intensities. At a 5% false-positive rate (FPR), which is the top 5% in the lower distribution of expression intensities in each of 33 conditions, most of the coding sORFs (7,021/7,901 = 84%) had higher expression intensities than the threshold under at least one experimental condition (Figs. S3 and S4). Second, we focused on the expression intensities of the negative controls in each of 33 conditions (Fig. 2B; Fig. S5). At a 5% FPR, which is the top 5% in expression intensities of negative controls in each of 33 conditions, we found that 96% of the coding sORFs (7,581/7,901) had higher expression intensities than the threshold under at least one experimental condition (Figs. S4 and S5). Third, we focused on the expression intensities of pseudogenes in the ATH1 array but not our designed array in one condition (seedling), because our designed array does not have any probes for pseudogenes annotated by TAIR (Fig. 2C). Expression intensities in the ATH1 array were transformed to match those in our designed array using an equation representing the relationship between our designed array and the ATH1 array (Fig. 1C). At a 5% FPR, which is the top 5% in transformed expression intensities in pseudogenes, we found that 27% of the coding sORFs (2,099/7,901) had higher expression intensities than the threshold (Fig. 2C; Fig. S4), assuming that the distribution of expression intensities in pseudogenes is the same among the 33 conditions. These observations indicate that a substantial number of coding sORFs are expressed in A. thaliana.
Fig. 2.
Expression intensities in annotated genes, negative controls, pseudogenes, and coding sORFs. x axis represents log10 values of (A) annotated genes, (B) negative controls, (C) pseudogenes, and (D) coding sORFs in the same sample (control in A). x axis indicates log10 values of expression intensities. y axis indicates frequency of probes in each bin size. In A, expression intensities of annotated genes were fitted to lower (blue solid line) and higher (red solid line) Gaussian distribution. The 5% false-positive rate, which is the top 5% in the lower distribution of expression intensities, was defined as the threshold for high expression. In B, the 5% false-positive rate, which is the top 5% in expression intensities of negative controls, was defined as the threshold for high expression. In C, the 5% false-positive rate, which is the top 5% in expression intensities of pseudogenes, was defined as the threshold for high expression.
Using 77 coding sORFs with evidence of translation from mass spectrometry analysis (27), we examined whether transcribed coding sORFs tend to have more evidence of translation than coding sORFs without evidence of transcription (Tables S1–S3). Coding sORFs with higher expression intensities than the lower group of expression intensities in annotated genes and expression intensities in negative controls tend to have more evidence of translation than coding sORFs without evidence of transcription, but this relationship is not significant (P > 0.05; χ2 test; Tables S1 and S2). Coding sORFs with higher expression intensities than pseudogenes have significantly more evidence of translation than coding sORFs without evidence of transcription (P = 0.01; χ2 test; Table S3). When we used the lower group of expression intensities in annotated genes and expression intensities in negative controls as the threshold for high expression intensities, most of the coding sORFs (84% in annotated genes and 96% in negative controls) are defined as having higher expression intensities. However, the threshold based on the expression intensities of pseudogenes produced fewer coding sORFs with higher expression intensities (27%). These results indicate that high expression intensity based on stringent criteria is a good indicator of real genes. Therefore, coding sORFs representing higher expression intensities than pseudogenes are defined as transcribed coding sORFs. However, mass spectrometry analysis tends to identify peptides translated from highly expressed genes. It is still unclear whether coding sORFs whose expression intensities are lower than pseudogenes have some functionality or not. Furthermore, the approach based on the pseudogenes threshold failed to identify coding sORFs with translational evidence, with a high false-negative rate of 61% (47/77; Table S3). Therefore, other independent criteria are required to identify functional coding sORFs.
Conservation of Coding sORFs Across Species.
The second independent criterion for assessing the functionality of coding sORFs is conservation across other plant species (Fig. S1). To assess the conservation of coding sORFs, we searched for sequences homologous to coding sORFs between A. thaliana and each of 16 other plants using a similarity search (e-value = 0.05). Among 7,901 coding sORFs, 4,844 showed at least one match to other plant genomes. For coding genes, a significantly lower nonsynonymous substitution rate than synonymous substitution rate indicates that the sequences have experienced functional constraint or purifying selection. However, the nonsynonymous substitution rate is likely to be underestimated in the alignments of amino acid sequences generated by our procedure, because the given alignment is the consequence of producing an alignment of amino acids with a maximum score. To determine the null distribution of the Ka/Ks ratio in our procedure, we generated random sequences with similar nucleotide composition to that of the coding sORFs. We identified 4,265 similar sequences of coding sORFs against these random sequences. The median of Ka/Ks ratio (0.32) in the null distribution was defined to be the Ka/Ks ratio, which represents neutrality in this procedure (Fig. S6). Applying a likelihood ratio test to these 4,844 coding sORFs, we found that 571 of them were significantly lower than the biased Ka/Ks ratio in at least 1 of the 16 plant species at the 0.05 false discovery rate (FDR; Dataset S1). The 572 coding sORFs were defined as constrained coding sORFs (Fig. S1).
We examined whether coding sORFs with evidence of translation tend to be conserved in land plants compared with those without evidence of translation (Table S4). Coding sORFs with purifying selection represent a significantly higher proportion of coding sORFs with evidence of translation than coding sORFs without purifying selection (P = 3.3 × 10−5; χ2 test). However, of 77 coding sORFs with evidence of translation, 62 were not subject to purifying selection in any of the 16 plant species, indicating that this procedure had a high false-negative rate of 81% (62/77).
Taken together, we found that 29% of the coding sORFs (2302/7901) have either evidence of transcription or functional constraint (Fig. S1; Dataset S1). Similarity searching for coding sORFs showed that 4,844/7,901 of these coding sORFs match similar sequences in other species, indicating that more than 60% of coding sORFs have potential homologs. To determine how many of these coding sORFs belong to gene families in A. thaliana, we generated similarity clusters of the sORFs and found 998 clusters (Dataset S1). Thus, these sORFs are likely to belong to novel gene families.
Overexpression Mutants of Coding sORFs.
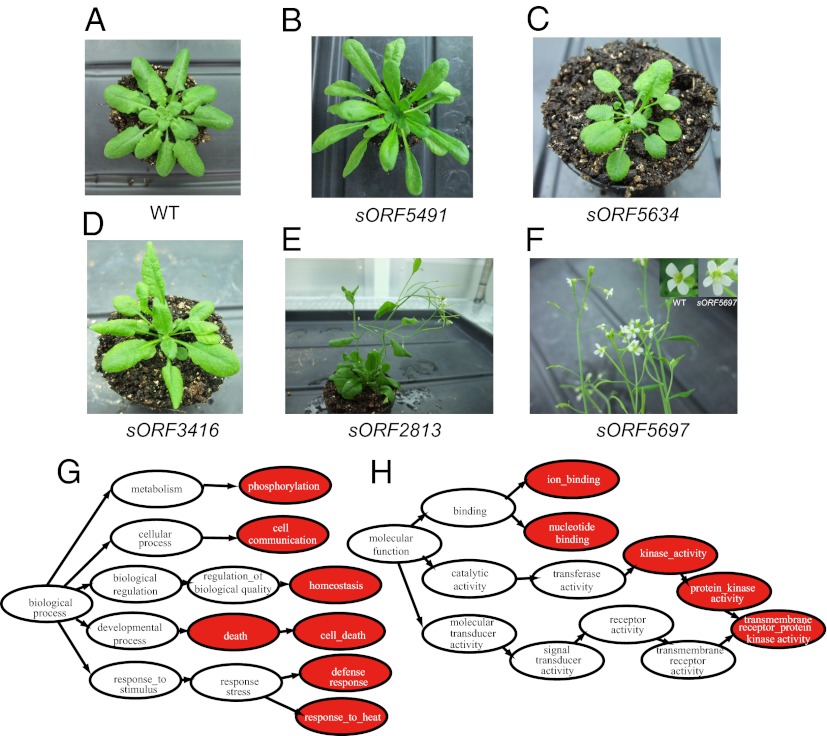
The question remains as to whether these coding sORFs play functional roles in plants. It has been reported that many types of peptides encoded by known small genes play essential roles in morphological traits of plants (5, 28–30). These functions of the small genes were discovered by overexpression analysis. Therefore, we examined the phenotypes of transgenic plants overexpressing selected coding sORFs. Of 7,901 coding sORFs, we selected 473 coding sORFs that showed high numbers of homologs in other plant species and unbiased expression intensities compared with coding sORFs that were not examined in the present study (Dataset S2). This is because sequence conservation in other species tends to be associated with evidence of translation (Tables S3 and S4). Of 473 coding sORFs, 10% (49/473) induced various morphological changes on overexpression (Fig. 3 A–F; Table 1; Dataset S2). In Arabidopsis FOX (full-length cDNA overexpressing) lines, which are derived from known genes, 1,487 of 15,547 overexpression mutants showed phenotypic changes (31). However, each mutant line includes an average of 2.6 genes. Assuming that only one gene induces a phenotypic effect by overexpression analysis, 572 (1,487/2.6) genes induced phenotypic effects of 40,422 (15,547 × 2.6) genes, indicating that 1.4% (572/40,422) of the annotated genes induced phenotypic effects on overexpression. Taken together, the phenotypic effect of coding sORFs is approximately seven times higher than that of randomly chosen known genes.
Fig. 3.
Phenotypes of transgenic plants overexpressing sORFs. (A) WT plant. (B) Mutant overexpressing sORF5491 showing a large adult plant phenotype. (C) Mutant overexpressing sORF5634 showing a small adult plant phenotype. (D) Overexpression of sORF3416 resulted in a pale green leaf phenotype. (E) Overexpression of sORF2813 resulted in a bent stem phenotype. (F) Overexpression of sORF5697 resulted in a penta-petal phenotype, whereas most WTs have tetra-petals. GO categories of biological process (G) and molecular function (H) with overrepresented numbers of coexpressed genes for 49 coding sORFs with phenotypic effects. Arrowheads point to subcategories. Red circles indicate categories with significantly (χ2 test, FDR < 0.05) more genes among the coexpressed genes.
Table 1.
sORFs with phenotypic effects
| Phenotype | sORF IDs* | Proportion in sORF | Proportion in FOX lines† | |
| Adult plant | Size (large) | sORF0484, sORF1411, sORF1819, sORF2666 | 21 (42.8%) | 1,033 (13.4%) |
| sORF2964, sORF2989, sORF3208, sORF5491 | ||||
| sORF7019, sORF6673 | ||||
| Size (small) | sORF2849, sORF4756, sORF5537, sORF5634 | |||
| sORF7728 | ||||
| Late flowering | sORF1067, sORF2686, sORF3656, sORF5884 | |||
| sORF6756, sORF7449 | ||||
| Rosette leaves | Shape | sORF0727, sORF1465, sORF3553, sORF4259 | 16 (32.6%) | 3,674 (47.6%) |
| sORF5238, sORF6780 | ||||
| Color (dark) | sORF0335, sORF2813, sORF3556, sORF5527 | |||
| Color (pale) | sORF1492, sORF1542 sORF1626, sORF3416 | |||
| Number | sORF2146, sORF2874 | |||
| Cauline leaves | Shape | — | 0 (0%) | 860 (11.1%) |
| Color | — | |||
| Number | — | |||
| Stem | Shape | sORF2743, sORF4248, sORF6982 | 3 (6.1%) | 1,594 (20.6%) |
| Color | — | |||
| Flower | Shape | sORF5697 | 1 (2%) | 155 (2%) |
| Silique | Shape | sORF3905 | 8 (16.3%) | 407 (5.3%) |
| Sterile | sORF2737, sORF5621 | |||
| Lethal | sORF0291, sORF0359, sORF1408, sORF1615 | |||
| sORF6599 | ||||
| Total | 49 | 7,723 | ||
*The sORF ID information is presented in Datasets S1 and S2.
†These values were obtained from ref. 32.
However, there is no significant difference between sORFs with and without a phenotypic effect with respect to the signatures of expression and/or purifying selection. We compared the appearance of any morphological effects with those that appeared in FOX lines (32). Lines overexpressing coding sORFs were associated with a higher proportion of growth rate or flowering timing differences compared with the FOX lines (Table 1). Peptides are known to play significant roles in various aspects of plant growth; therefore, our identified coding sORFs may encode such peptides.
Functional Categories of Coding sORFs.
Although we found 49 coding sORFs associated with phenotypic effects, the functional categories to which they belong at the molecular level remained unclear. Therefore, we examined the functional categories of genes that were coexpressed with each of the 49 coding sORFs by Gene Ontology (GO). Coexpression analysis is widely used to infer either associated genes or functional categories (33–35). We calculated R values (Pearson's coefficient of correlation) of expression intensities in 99 arrays (33 conditions × 3 replicates) in pairs between a sORF and all annotated genes and defined those annotated genes with the top 1% of R values as coexpressed genes. The overrepresented functions of coexpressed genes were related to metal-ion binding, nucleotide binding, kinase activities, phosphorylation, cell communication, cell death, homeostasis, defense response, and heat response, among the GO categories of molecular functions and biological processes (Fig. 3 G and H; Dataset S3). In particular, functional categories related to phosphorylation tend to be significantly overrepresented at lower FDR values in Dataset S3. Most of the peptides that play essential roles in signaling in plants are excreted from the cell as secreted peptides. The secreted peptides bind membrane-localized receptor kinases of target cells as ligands and activate signaling in the targeted cells (3, 5–7, 11, 12, 16). The relationship between a peptide ligand and the receptor (phosphorylated protein) has been identified by coexpression analyses (16); therefore, the overrepresentation of phosphorylation in GO categories strongly suggests that our 49 identified coding sORFs with phenotypic effects include many such peptides.
However, only 9 of 49 coding sORFs have secretion signals identified by in silico software. The proportion of secretion signals in coding sORFs with phenotypic effects is similar to the proportion of secretion signals in coding sORFs without phenotypic effects (P = 0.3; χ2 test). Thus, the existence of a secretion signal is unlikely to be essential for identifying phenotypic effects. Four known nonsecreted peptides play various essential roles in growth and development at low concentrations in plants in association with kinase proteins (29, 36–38). It is thought that these peptides might function in the cytoplasm. Therefore, some of the identified 49 coding sORFs with phenotypic effects that lack secretion signals may have a similar function or location.
Expression Atlas of Coding sORFs in Comparison with Those of Annotated Genes.
We compared the expression atlas of coding sORFs to that of negative controls, annotated coding genes, and pseudogenes. In the 33 conditions, 7,901 coding sORFs tended to show higher expression intensities than negative controls (0.8 ± 1.7 in coding sORFs, 0.35 ± 0.1 in negative controls, P = 1.0 × 10−59–1.1 × 10−11, Wilcoxon test; Fig. 2 B and D; Figs. S4 and S5). However, annotated coding genes have much higher expression intensities than these coding sORFs in 33 conditions (2.3 ± 1.2 in annotated coding genes, 0.8 ± 1.7 in coding sORFs, P < 1.0 × 10−60 in 33 conditions, Wilcoxon test; Fig. 2 A and D; Figs. S3 and S4). Pseudogenes also tend to have higher expression intensities than coding sORFs in a given condition (0.7 ± 1.7 in coding sORFs, 1.0 ± 0.5 in pseudogenes, P = 2.2 × 10−12; Fig. 2 C and D). Although it was reported that substantial numbers of pseudogenes were transcribed (39, 40), most coding sORFs are likely to be expressed at lower levels than pseudogenes. To examine whether coding sORFs are expressed or not, we used RT-PCR with specific primers to analyze transcript levels of 49 coding sORFs whose transgenic plants showed phenotypic effects. Of 49 coding sORFs, all (100%) showed targeted signals in a mixed RNA sample from 33 conditions (Fig. S7), indicating that most of the coding sORFs are indeed transcribed at levels detectable by RT-PCR.
Recently, mRNA-seq data from next generation sequencers have accumulated for A. thaliana. It is believed that mRNA-seq is one of the most sensitive analyses to identify mRNA transcription. To examine what proportion of sORFs is identified by mRNA-seq, we obtained mRNA-seq data collected under six conditions (control, drought, dry seed, flower, leaf, root) and mapped mRNA-seq tags to the A. thaliana genome. Of 1,359 coding sORFs identified by mRNA-seq, half of them (682/1,359 = 50%) were identified as transcribed coding sORFs based on the thresholds of expression intensities of pseudogenes (Fig. 2C). When we used transcriptional thresholds based on lower expression distribution in annotated genes or negative controls (Fig. 2 A and B), most sORFs (1,312/1,359 = 97% in the lower distribution of annotated genes, 1,343/1,359 = 99% in negative controls) could be identified as transcribed coding sORFs in our array data. Thus, mRNA-seq can identify sORFs with even lower expressional thresholds. However, many of the coding sORFs (671/1,353 = 50% in pseudogenes, 4,698/6,010 = 78% in lower distribution of annotated genes, 5,213/6,556 = 80% in negative controls) were not identified by mRNA-seq but were identified by our array. We think that this higher calling rate of transcribed coding sORFs in our array reflects higher variation of RNA sampling in organs and conditions (total 33 conditions) compared with that in the mRNA-seq analysis (6 conditions).
Recently, upstream ORFs (uORFs) were reported in eukaryotic genomes (41). It is thought that uORFs are located in the 5′ UTR of an mRNA that includes a long ORF. Most uORFs play a role in regulating proteins encoded by the long ORF. Thus, uORFs are cotranscribed together as a single polycistronic unit of the long ORF encoding a protein. Coding sORFs may share similar features with uORFs with respect to cotranscription. To examine whether each coding sORF represents an independent transcription unit or not, we identified neighboring annotated genes within <1 Kb distance from each coding sORF and calculated the R value (Pearson's coefficient of correlation) of expression intensities in 99 arrays (33 conditions × 3 replicates) for the pair between an sORF and its neighbor. The threshold of a neighboring gene was defined as <1 Kbp because 95% of introns are ≤850 bp in A. thaliana. R values for the pairs were compared with those for all of the neighboring pairs of annotated genes within <1 Kb. The R values for all neighboring pairs of annotated genes were not significantly different from those for the pairs of coding sORFs and their neighboring annotated genes (0.07 ± 0.24 in coding sORFs, 0.08 ± 0.28 in neighboring annotated genes, P = 0.24, Wilcoxon test; Fig. S8). This finding indicates that coding sORFs have the same degree of transcriptional independence as annotated genes. Thus, most of these coding sORFs are unlikely to be either uORFs or missing exons of known genes.
Concluding Remarks.
Using transcriptome analysis with 99 expression atlases and a comparative genomics approach in 16 other plant species, we identified a large number of coding sORFs with evidence of either expression or functional constraint. More interestingly, ∼10% of the selected coding sORFs induced visible phenotypic effects by overexpression analysis compared with 1.4% among overexpressed known genes. Our study strongly indicates that a large number of coding sORFs that are hidden in plant genomes play functional roles. To infer the functional roles of coding sORFs with phenotypic effects, we carefully examined overrepresented phenotypic effects and functional categories of genes coexpressed with sORFs. Consequently, we speculate that coding sORFs with phenotypic effects are likely to have similar features to small genes encoding ligand peptides, such as plant hormones, which play significant roles in various aspects of plant growth and development (3–18). However, it is unclear from the results of the present study whether our identified sORFs encode peptides. We strongly emphasize that future research should determine whether these coding sORFs inducing changes in morphological traits encode peptides or not. Proteome analysis has shown translational evidence for sORF5537 (27), whose overexpression mutant shows a small adult plant phenotype. This result strongly suggests that overexpression of peptides encoded by sORF5537 induces a morphological change.
There is an open question as to why coding sORFs have not been identified as annotated genes in A. thaliana, which has a highly accurate genome. One reason is that coding sORFs tend to be expressed at a significantly lower level than known genes (Fig. 2). In addition, the identification of transcripts by full-length cDNA, EST, and RNA-seq studies tends to be examined in only a few conditions. Therefore, minor transcripts will probably have been missed. Recently, it has been emphasized that most identified transcripts are expressed from known genes in model organisms (42–44). Even if some novel transcripts were identified, such transcripts tend to be expressed at a low level. Therefore, the functionality of such the novel transcripts is doubtful or is suggested to be negligible, because, in general, transcription level is highly correlated with essentiality. However, this may not be the case for some small genes, because small genes encoding functional ligand peptides are expressed at low levels (3–18).
This report describes previously uncharacterized functional analyses of coding sORFs hidden in plant genomes. However, further analyses are necessary to identify true coding genes among the coding sORFs. Two criteria for defining coding genes are that they are an independent transcription unit and that there is evidence of their translation. Although we examined the functionality of sORFs by overexpression analyses, it is still questionable whether the overexpression of a targeted sORF is really associated with its true function. This is because transgenic plants express target sORFs at high levels in most organs or conditions, unlike the expression pattern in WT. Therefore, our results represent the beginning of phenotypic analyses to find coding sORFs with functionality. In future research, knock-down or KO analyses are required to validate the functionality of coding sORFs. For such future analyses, the array data of coding sORFs shown in the present study will make a vital contribution to the functional analyses of coding sORFs. However, not only plants but also other organisms are likely to contain many novel coding sORFs in their genomes. Indeed, it was reported that more than two-thirds of the human genome can be associated with biochemical functions (45). Our results strongly indicate that a large number of coding sORFs hidden in eukaryote genomes play essential roles, and might be vital to the understanding of certain hitherto unexplained mechanisms.
Materials and Methods
Update of Coding sORFs.
There were 7,159 newly identified sORFs (30–100 codons) with high coding potential in the intergenic regions that did not have matches to annotated genes in TAIR6 (22). Of these, 692 were reannotated as small genes in TAIR8 (www.arabidopsis.org), and a further 60 sORFs were reannotated as small genes in TAIR10. The annotated small genes were used as coding sORFs in the present study. We disregarded 49 coding sORFs that were annotated as parts of neighboring annotated genes. We examined whether coding sORFs are verified or not using the available 96,358 Arabidopsis full-length cDNAs and confirmed the transcription of 71 coding sORFs. We also found 1,346 mRNA-like transcripts in the intergenic regions of the available Arabidopsis full-length cDNAs. Also, 172 coding sORFs were truncated Arabidopsis full-length cDNAs; therefore, these coding sORFs were disregarded. sORFs with high coding potential were reexamined among the transcript sequences, using the hexamer composition bias between CDSs and NCDSs. In some transcripts, multiple coding sORFs were identified in a transcriptional unit, as has been observed in eukaryotes. The analytical procedures and findings are summarized in Fig. S1. For additional details, see SI Text.
RNA Samples and Hybridization to Arrays.
In A. thaliana accession Col-0, we used 16 different organs to collect samples (dry seeds, 24-h-imbibed seeds, 48-h-imbibed seeds, callus, juvenile rosette, adult rosette, senescence leaves, cauline leaves, stems, root, young buds, mature flower buds, flowers, young siliques, mature siliques, and old siliques). Eight samples represented different light irradiation conditions (white 0 h, white 1 h, white 6 h, white 24 h, continuous dark, blue, far-red, and red light), and nine samples represented different abiotic stress conditions (control, drought 2 h, drought 6 h, heat 2 h, heat 6 h, salt 2 h, salt 6 h, cold 2 h and cold 6 h). All sampling was performed in triplicate. After extraction of RNA from each sample, hybridization and scanning were conducted. For additional details, see SI Text.
Sequence Analyses of Coding sORFs.
The following genomes were used to assess conservation of coding sORFs: Physcomitrella patens, Selaginella moellendorffii, Zea mays, Sorghum bicolor, Brachypodium distachyon, Oryza sativa, Mimulus guttatus, Vitis vinifera, Ricinus communis, Manihot esculenta, Populus trichocarpa, Cucumis sativus, Glycine max, Medicago truncatula, Carica papaya, and Arabidopsis lyrata. After aligning conserved pairs by CLUSTALW (46), the synonymous and nonsynonymous substitution rates (Ka and Ks) were calculated using PAML (47). To determine if the Ka/Ks values were significantly less than the biased Ka/Ks ratio inferred by random sequences, a likelihood ratio–based procedure was applied to sequence pairs. For each pair, two maximum likelihood values were calculated with the Ka/Ks ratio fixed at 0.32 (biased Ka/Ks ratio) and with the Ka/Ks ratio as a free parameter. The ratio of the maximum likelihood values was then compared with the χ2 distribution (SI Text). We also collected mRNA-seq datasets for 16 libraries from six conditions (48). We mapped mRNA-seq tags to the A. thaliana genome (TAIR8) with TopHat (49).
Construction of Overexpression Mutants.
Each sORF was introduced into pMDC32, which includes a double 35S promoter. The recombinant binary vector was then introduced into Agrobacterium tumefaciens (strain GV3101). Agrobacterium was infected into Arabidopsis using the floral-dip method (50). We monitored visible phenotypes, such as plant color, flowering time, and fertility, in all constructed overexpression lines. When more than three overexpression lines showed the same phenotype(s), the transformed sORF was considered responsible for the morphologies (SI Text).
Statistical Tests for Determining Overrepresented GO Categories.
GO assignments for Arabidopsis genes were obtained from The Arabidopsis Information Resource (www.arabidopsis.org). Three top GO categories, cellular components, molecular functions, and biological processes, were analyzed. Among these GO categories, we obtained the numbers of Arabidopsis genes that were coexpressed and not coexpressed with 49 coding sORFs resulting in phenotypic effects. In each GO category, the expected values were compared with the observed values using a χ2 test to determine whether the ratio of observed gene numbers in coexpressed genes to those in noncoexpressed genes was significantly higher than the expected ratio. To correct for multiple testing, the FDR was estimated by Q-VALUE software. The null hypothesis was rejected if FDR values were <0.05.
Supplementary Material
Acknowledgments
We thank S.-H. Shiu for discussions concerning the manuscript. This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN; K.H. and M.H.-T.); Grants-in-Aid for Scientific Research (to K.H. and M.H.-T.); Core Research for Evolutional Science and Technology (CREST) Program “Creation of essential technologies to utilize carbon dioxide as a resource through the enhancement of plant productivity and the exploitation of plant products” of the Japan Science and Technology Agency (JST) (K.H.); RIKEN Plant Science Center, Special Postdoctoral Researcher’s Program from RIKEN (M.O.); and a research fellowship from the Japan Society for the Promotion of Science for Young Scientists (to M.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw unfiltered microarray results are deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (subseries entry GSE34188).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213958110/-/DCSupplemental.
References
- 1.Kastenmayer JP, et al. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16(3):365–373. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondo T, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329(5989):336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, Matsubayashi Y, Nakamura K, Sakagami Y. Diversity of Arabidopsis genes encoding precursors for phytosulfokine, a peptide growth factor. Plant Physiol. 2001;127(3):842–851. [PMC free article] [PubMed] [Google Scholar]
- 4.Yang SL, et al. Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell. 2003;15(12):2792–2804. doi: 10.1105/tpc.016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(46):18333–18338. doi: 10.1073/pnas.0706403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jun JH, Fiume E, Fletcher JC. The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci. 2008;65(5):743–755. doi: 10.1007/s00018-007-7411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takayama S, et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci USA. 2000;97(4):1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gleason CA, Liu QL, Williamson VM. Silencing a candidate nematode effector gene corresponding to the tomato resistance gene Mi-1 leads to acquisition of virulence. Mol Plant Microbe Interact. 2008;21(5):576–585. doi: 10.1094/MPMI-21-5-0576. [DOI] [PubMed] [Google Scholar]
- 9.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327(5969):1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 10.Pearce G, Moura DS, Stratmann J, Ryan CA., Jr RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA. 2001;98(22):12843–12847. doi: 10.1073/pnas.201416998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohyama K, Ogawa M, Matsubayashi Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008;55(1):152–160. doi: 10.1111/j.1365-313X.2008.03464.x. [DOI] [PubMed] [Google Scholar]
- 12.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329(5995):1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 13.Butenko MA, et al. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell. 2003;15(10):2296–2307. doi: 10.1105/tpc.014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuda S, et al. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458(7236):357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- 15.Hara K, et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50(6):1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 16.Sugano SS, et al. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463(7278):241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 17.Constabel CP, Bergey DR, Ryan CA. Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc Natl Acad Sci USA. 1995;92(2):407–411. doi: 10.1073/pnas.92.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA. 2006;103(26):10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, et al. Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome Res. 2011;21(4):634–641. doi: 10.1101/gr.109280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeys Y, Rouzé P, Van de Peer Y. In search of the small ones: Improved prediction of short exons in vertebrates, plants, fungi and protists. Bioinformatics. 2007;23(4):414–420. doi: 10.1093/bioinformatics/btl639. [DOI] [PubMed] [Google Scholar]
- 21.Silverstein KA, Graham MA, Paape TD, VandenBosch KA. Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol. 2005;138(2):600–610. doi: 10.1104/pp.105.060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007;17(5):632–640. doi: 10.1101/gr.5836207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanada K, et al. sORF finder: A program package to identify small open reading frames with high coding potential. Bioinformatics. 2010;26(3):399–400. doi: 10.1093/bioinformatics/btp688. [DOI] [PubMed] [Google Scholar]
- 24.Cheng H, et al. Small open reading frames: Current prediction techniques and future prospect. Curr Protein Pept Sci. 2011;12(6):503–507. doi: 10.2174/138920311796957667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makalowski W, Boguski MS. Synonymous and nonsynonymous substitution distances are correlated in mouse and rat genes. J Mol Evol. 1998;47(2):119–121. doi: 10.1007/pl00006367. [DOI] [PubMed] [Google Scholar]
- 26.Hanada K, Shiu SH, Li WH. The nonsynonymous/synonymous substitution rate ratio versus the radical/conservative replacement rate ratio in the evolution of mammalian genes. Mol Biol Evol. 2007;24(10):2235–2241. doi: 10.1093/molbev/msm152. [DOI] [PubMed] [Google Scholar]
- 27.Castellana NE, et al. Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci USA. 2008;105(52):21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21(14):1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narita NN, et al. Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 2004;38(4):699–713. doi: 10.1111/j.1365-313X.2004.02078.x. [DOI] [PubMed] [Google Scholar]
- 30.Wen J, Lease KA, Walker JC. DVL, a novel class of small polypeptides: Overexpression alters Arabidopsis development. Plant J. 2004;37(5):668–677. doi: 10.1111/j.1365-313x.2003.01994.x. [DOI] [PubMed] [Google Scholar]
- 31.Ichikawa T, et al. The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. 2006;48(6):974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- 32.Sakurai T, et al. RiceFOX: A database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol. 2011;52(2):265–273. doi: 10.1093/pcp/pcq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22(13):1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 34.Grossmann S, Bauer S, Robinson PN, Vingron M. Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. Bioinformatics. 2007;23(22):3024–3031. doi: 10.1093/bioinformatics/btm440. [DOI] [PubMed] [Google Scholar]
- 35.Iida K, et al. ARTADE2DB: improved statistical inferences for Arabidopsis gene functions and structure predictions by dynamic structure-based dynamic expression (DSDE) analyses. Plant Cell Physiol. 2011;52(2):254–264. doi: 10.1093/pcp/pcq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djakovic S, Dyachok J, Burke M, Frank MJ, Smith LG. BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development. 2006;133(6):1091–1100. doi: 10.1242/dev.02280. [DOI] [PubMed] [Google Scholar]
- 37.Rohrig H, Schmidt J, Miklashevichs E, Schell J, John M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci USA. 2002;99(4):1915–1920. doi: 10.1073/pnas.022664799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topping JF, Lindsey K. Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell. 1997;9(10):1713–1725. doi: 10.1105/tpc.9.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou C, et al. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 2009;151(1):3–15. doi: 10.1104/pp.109.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, Takuno S, Waters ER, Gaut BS. Lowly expressed genes in Arabidopsis thaliana bear the signature of possible pseudogenization by promoter degradation. Mol Biol Evol. 2011;28(3):1193–1203. doi: 10.1093/molbev/msq298. [DOI] [PubMed] [Google Scholar]
- 41.Vilela C, McCarthy JE. Regulation of fungal gene expression via short open reading frames in the mRNA 5’untranslated region. Mol Microbiol. 2003;49(4):859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 42.Filichkin SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20(1):45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 44.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8(5):e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunham I, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi S, et al. Positional correlation analysis improves reconstruction of full-length transcripts and alternative isoforms from noisy array signals or short reads. Bioinformatics. 2012;28(7):929–937. doi: 10.1093/bioinformatics/bts065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.