Abstract
DNA methylation is an epigenetic mark that silences transposable elements (TEs) and repeats. Whereas the establishment and maintenance of DNA methylation are relatively well understood, little is known about their dynamics and biological relevance in plant and animal innate immunity. Here, we show that some TEs are demethylated and transcriptionally reactivated during antibacterial defense in Arabidopsis. This effect is correlated with the down-regulation of key transcriptional gene silencing factors and is partly dependent on an active demethylation process. DNA demethylation restricts multiplication and vascular propagation of the bacterial pathogen Pseudomonas syringae in leaves and, accordingly, some immune-response genes, containing repeats in their promoter regions, are negatively regulated by DNA methylation. This study provides evidence that DNA demethylation is part of a plant-induced immune response, potentially acting to prime transcriptional activation of some defense genes linked to TEs/repeats.
Keywords: epigenetics, RNA silencing, MAMP
The innate immune response is the first line of defense against pathogens and plays a critical role in antimicrobial defense. This response is initiated by host-encoded pattern-recognition receptors (PRRs) that recognize evolutionarily conserved pathogen-derived signatures, known as Microbe-Associated Molecular Patterns (MAMPs), and activate MAMP-Triggered Immunity (MTI) (1). In plants, the few well-characterized PRRs encode transmembrane receptor-like kinases with extracellular leucine-rich repeat (LRR) domains and intracellular kinase domains (1). Furthermore, plants have evolved another strategy to perceive microbial pathogens through disease resistance (R) proteins. Most R proteins belong to the nucleotide-binding site leucine-rich repeat (NB-LRR) class, which shares structural homologies with mammalian innate immune receptors, such as Nucleotide-binding oligomerization domain-containing protein 1 (NOD1) and NOD2, and are thus often referred to as NOD-like receptors (NLRs). They recognize, directly or indirectly, divergent pathogen virulence determinants known as effector proteins, and establish effector-triggered immunity (ETI) (2). In most cases, plant NLRs recognize indirectly pathogen effectors by sensing their effects on plant target proteins, as postulated by the “guard hypothesis” (3). Upon detection of MAMPs or pathogen effectors, plant cells rapidly induce a series of signaling events that involve, for instance, activation of mitogen-activated protein kinase (MAPK) cascades, production of reactive oxygen species (ROS), and differential expression of hundreds of immune-response genes, including short interfering RNAs (siRNAs) and microRNAs (miRNAs) (2, 4). Recently, several siRNAs and miRNAs were found to orchestrate MTI and ETI responses (4, 5), implying a key role of RNA silencing in the regulation of the plant immune system.
Small RNA-dependent DNA methylation is an RNA silencing phenomenon that prevents overexpression and proliferation of transposable elements (TEs) in different organisms. Despite their parasitic nature, TEs have been domesticated by host genomes, notably to modulate the expression of nearby genes during biotic and abiotic stress responses (6–8). In plants, this regulatory mechanism is referred to as RNA-directed DNA methylation (RdDM) and involves the biosynthesis of siRNAs that guide the DNA methylation of TEs and repeats (9). In the Arabidopsis RdDM pathway, RNA polymerase IV (PolIV) transcribes single-stranded RNAs, subsequently used as substrates by RNA-dependent RNA polymerase 2 (RDR2) to produce double-stranded RNAs (dsRNAs) (10). These dsRNAs are processed by DICER-Like 3 (DCL3) into ∼23- to 24-nt siRNAs, which are loaded onto complexes composed in part of argonaute (AGO) proteins including AGO4 and AGO6 (10). AGO4 is recruited to target loci through base pairing between siRNAs and intergenic RNAs produced by RNA polymerase V (PolV) (11). Through a currently unidentified process, the protein Domains Rearranged Methyltransferase 2 (DRM2) is recruited onto the chromatin to direct both symmetric (CG, CHG) and asymmetric (CHH) methylation (in these methylation types H refers to any nucleotide base other than a G) (10). During DNA replication, CHH methylation is actively perpetuated by the combined action of siRNAs and DRM2, whereas CG and CHG methylation is maintained by Methyltransferase 1 (MET1) and Chromomethylase 3 (CMT3), respectively (10). Furthermore, Arabidopsis encodes DNA glycosylases/lyases that can actively erase DNA methylation, among which Repressor Of Silencing 1 (ROS1) negatively regulates RdDM (12).
Whereas DNA methylation has been mostly functionally characterized in plant and animal developmental processes (13), recent findings also indicate a role for this silencing pathway in repressing plant defense toward biotrophic pathogens (8, 14–16), which take up nutrients from living plant cells. Nevertheless, the mechanistic interplay between the dynamic regulation of the RNA silencing machinery and to the transcriptional activation of TEs and pathogen-responsive genes remains ill defined. In addition, little is known about the detailed physiological relevance of RNA-dependent DNA methylation in host–pathogen interactions. This study addresses those issues in the context of Arabidopsis antibacterial defense.
Results
Flagellin-Derived Peptide flg22 Derepresses RdDM Targets in Arabidopsis Leaves.
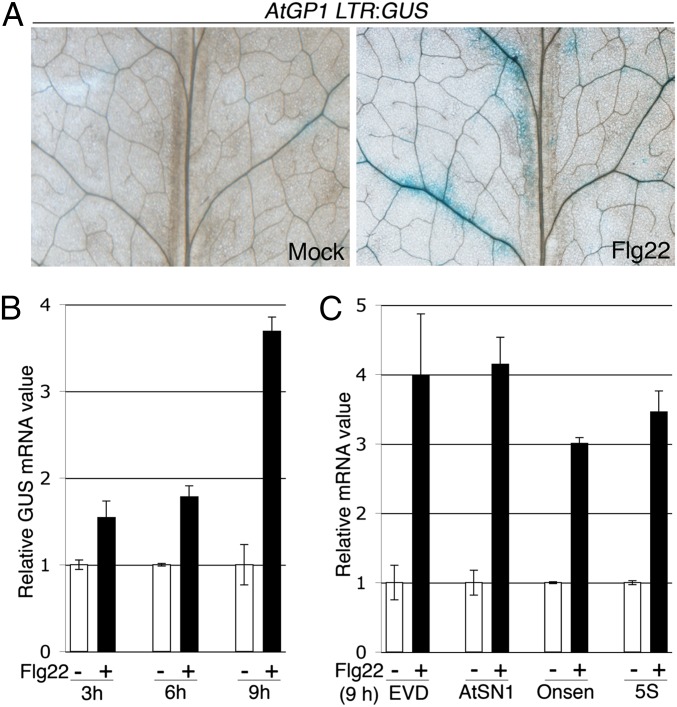
Flagellin Sensing 2 (FLS2) is a well-characterized plant PRR, which senses the bacterial flagellin-derived peptide flg22, resulting in the differential regulation of hundreds of genes (1, 17, 18). We investigated the potential impact of flg22 on transcriptional gene silencing (TGS) by monitoring the transcriptional status of AtGP1, a gypsy long terminal repeat (LTR)-retrotransposon strongly targeted by siRNA-directed DNA methylation (Fig. S1), and synergistically controlled by CG methylation (Fig. S2). An AtGP1 LTR region was fused with the β-glucuronidase GUS reporter gene and transformed into Arabidopsis. From these stable transformants we selected a reference line in which GUS expression was restored upon DNA methyltransferase inhibitor application (Fig. S3). When this line was treated with flg22, GUS expression was reactivated mostly within and around the leaf vasculature, notably at secondary veins and at the base of midveins (Fig. 1A). These effects correlated with a progressive increase in GUS and endogenous AtGP1 mRNA levels during flg22 elicitation (Fig. 1B and Fig. S4), and a higher expression of other well-characterized RdDM targets normally derepressed in DNA methylation-defective mutants (Fig. 1C and Fig. S2), suggesting that flg22 has an inhibitory effect on TGS. It is noteworthy that transcripts of Onsen, an LTR-retrotransposon strongly targeted by RdDM (Fig. S1), were induced upon flg22 treatment, but not in DNA methylation-defective mutants (Fig. 1C and Fig. S2). Thus, as recently reported (6), loss of DNA methylation is not sufficient to reactivate this TE.
Fig. 1.
(A) Five- to 6-wk-old AtGP1 LTR:GUS leaves were treated with water (mock) or 1 μM of flg22 for 24 h and stained with GUS. (B) As in A but over a 9-h time course and GUS mRNA levels were analyzed by RT-qPCR. (C) As in B at 9 h posttreatment and EVD (Evadé, AtCOPIA93), AtSN1 (A. thaliana short interspersed element 1), Onsen (AtCOPIA78), and 5S (5S rDNA) transcript levels were analyzed by RT-qPCR. Error bars: SD from three independent PCR results. Similar results were obtained in four independent experiments.
Flg22-Triggered Derepression of RdDM Targets Is Associated with the Down-Regulation of a Subset of Coregulated TGS Factors.
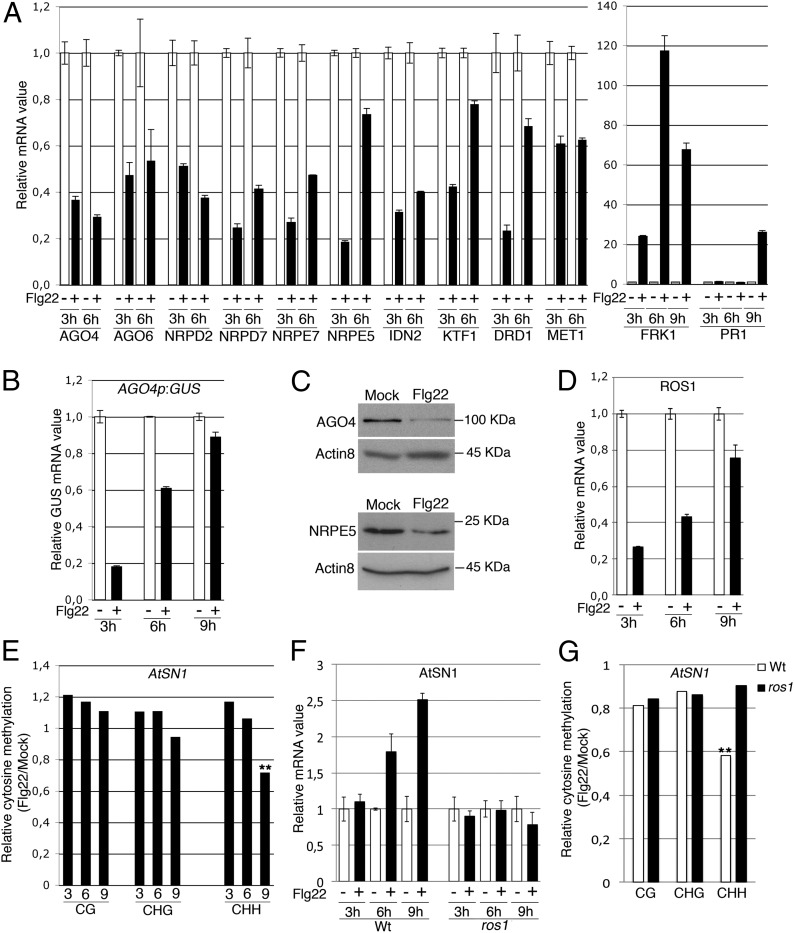
To get insights into the regulatory mechanisms by which flg22 triggers TGS release, we first monitored mRNA levels of TGS factors over a 9-h time course experiment. We found a significant down-regulation of the key components AGO4, AGO6, Nuclear RNA Polymerase D2 (NRPD2), Nuclear RNA Polymerase D7 (NRPD7), Nuclear RNA Polymerase E7 (NRPE7), Nuclear RNA Polymerase E5 (NRPE5), Involed in De Novo 2 (IDN2), KOW Domain-containing Transcription Factor 1 (KTF1), Defective in RNA-directed DNA methylation 1 (DRD1), and MET1 at 3 h and 6 h after flg22 treatment, which correlated with the up-regulation of the early defense-marker gene Flg22-induced Receptor-Kinase 1 (FRK1) (Fig. 2A). The majority of these TGS factor mRNAs, which mostly encode components of RdDM activity, regained normal levels at 9 h posttreatment (Fig. S5), when induction of the late defense-marker gene Pathogenesis-related gene 1 (PR1) typically sets in (Fig. 2A). A transgenic reporter of AGO4 transcription was also transiently decreased during flg22 elicitation (Fig. 2B), suggesting that repression of AGO4, and perhaps other coregulated TGS factors, occurs at the transcriptional level, which may be linked to an overrepresentation of three motifs within their promoters, including the pathogen-responsive W-box element (Fig. S6) (19). Importantly, flg22-triggered down-regulation of TGS factor mRNAs was also associated with a decrease in AGO4 and NRPE5 protein levels (Fig. 2C) and a reduction in the expression of ROS1 (Fig. 2D), a gene known to be robustly down-regulated in DNA methylation-defective mutants (20–22). Collectively, these results suggest that flg22 inhibits TGS, at least in part, by repressing RdDM activity.
Fig. 2.
(A) Five- to 6-wk-old WT leaves were syringe infiltrated with water (−) or flg22 (+) and mRNA levels of known TGS factors (NRPD2, NRPD7, AGO4, AGO6, NRPE7, NRPE5, IDN2, KTF1, DRD1, and MET1) were monitored at 3 and 6 h post-flg22 treatment with SD as in Fig. 1, Left. FRK1 and PR1 transcript levels were also analyzed at 3, 6, and 9 h posttreatment with SD as in Fig. 1, Right. Similar results were obtained in five independent experiments. (B) GUS mRNA levels were monitored as in A from AGO4p:GUS-elicited plants with SD as in Fig. 1. Similar results were obtained in two independent experiments. (C) WT plants were treated as in A for 9 h and AGO4, NRPE5, and ACTIN8 protein levels monitored by Western blot analyses. Similar results were obtained in two independent experiments. (D) WT plants were treated as in A and ROS1 transcript levels were analyzed at 3, 6, and 9 h posttreatment with SD as in Fig. 1. Similar results were obtained in three independent experiments. (E) Methylation levels at AtSN1 analyzed by bisulfite sequencing in plants treated as in A. The region analyzed contains four CG, seven CHG, and 33 CHH. Asterisks represent significant differences (**P < 0.01). Similar results were obtained in five independent experiments, with kinetics of DNA demethylation starting at 6 or 9 h after flg22 treatment from one experiment to the other. (F) mRNA levels of AtSN1 treated as in A in WT and ros1–4 leaves with SD as in Fig. 1. Similar results were obtained in two independent experiments. Of note, the kinetics of DNA demethylation depicted in E are not directly comparable with the kinetics of AtSN1 induction presented in F. These experiments are independent. (G) Methylation levels at AtSN1 in plants treated as in A for 6 h and analyzed as in E. Asterisks represent significant differences (**P < 0.01). Similar results were obtained in two independent experiments, with a kinetics of DNA demethylation occurring at 6 h in one experiment and 9 h in the other.
Flg22 Triggers DNA Demethylation at Well-Characterized RdDM Loci.
We next monitored DNA methylation levels at well-characterized RdDM targets during flg22 elicitation, using bisulfite sequencing (which identifies the positions of methylated and unmethylated cytosines). We found a progressive flg22-triggered demethylation at the retroelement AtSN1, which primarily occurred in the CHH context (Fig. 2E). Demethylation at Onsen’s LTR regions, which are almost exclusively composed of cytosines in the CHH context, was also detected, although a more transient effect occurred during flg22 elicitation (Fig. S7). Notably, DNA demethylation at both AtSN1 and Onsen preceded the up-regulation of their cognate transcripts in multiple independent experiments (Fig. S7), suggesting that demethylation may contribute to the transcriptional activation of these TEs, although other chromatin modifications are also likely to be involved. Noteworthy, the mild effects observed on DNA demethylation levels also suggest that these epigenetic changes may occur in specific immune-response cells such as the ones that surround leaf vasculature and where AtGP1 was transcriptionally reactivated (Fig. 1A).
ROS1 Facilitates Induction and Demethylation of AtSN1 During flg22 Elicitation.
The relatively rapid decrease in DNA methylation upon flg22 exposure suggested the possible implication of an active DNA demethylation process. Given that ROS1 is expressed in vegetative tissues and that it contributes to abiotic stress responses (12), we investigated its contribution in the above regulatory process. We first exposed a loss-of-function mutation in ROS1 plants to flg22 and monitored the levels of some TEs that are controlled by RdDM. Flg22-mediated induction of AtSN1 and AtGP1, which are known ROS1 targets (23), was altered in ros1-elicited plants (Fig. 2F and Fig. S8), whereas induction of the retrotransposon Onsen was unaffected in the same elicited mutants (Fig S8). These results indicate that transcriptional reactivation of a subset of TEs requires ROS1, which is consistent with a compromised CHH demethylation of AtSN1 observed in flg22-treated ros1 mutants (Fig. 2G). Therefore, ROS1 presumably contributes to the transcriptional activation of some TEs by constitutively pruning DNA methylation at these loci, thereby potentially accelerating the reduction in DNA methylation caused by the repression of TGS factors during the elicitation (Fig. 2 A–C). Nevertheless, it remains to be determined whether flg22-triggered repression of ROS1 mRNAs (Fig. 2D) also contributes to an eventual remethylation and resilencing of these TEs in a later phase of the elicitation.
DNA Demethylation Restricts Bacterial Multiplication in Arabidopsis Leaves and Is Associated with an Activation of the Salicylic Acid-Dependent Defense Response.
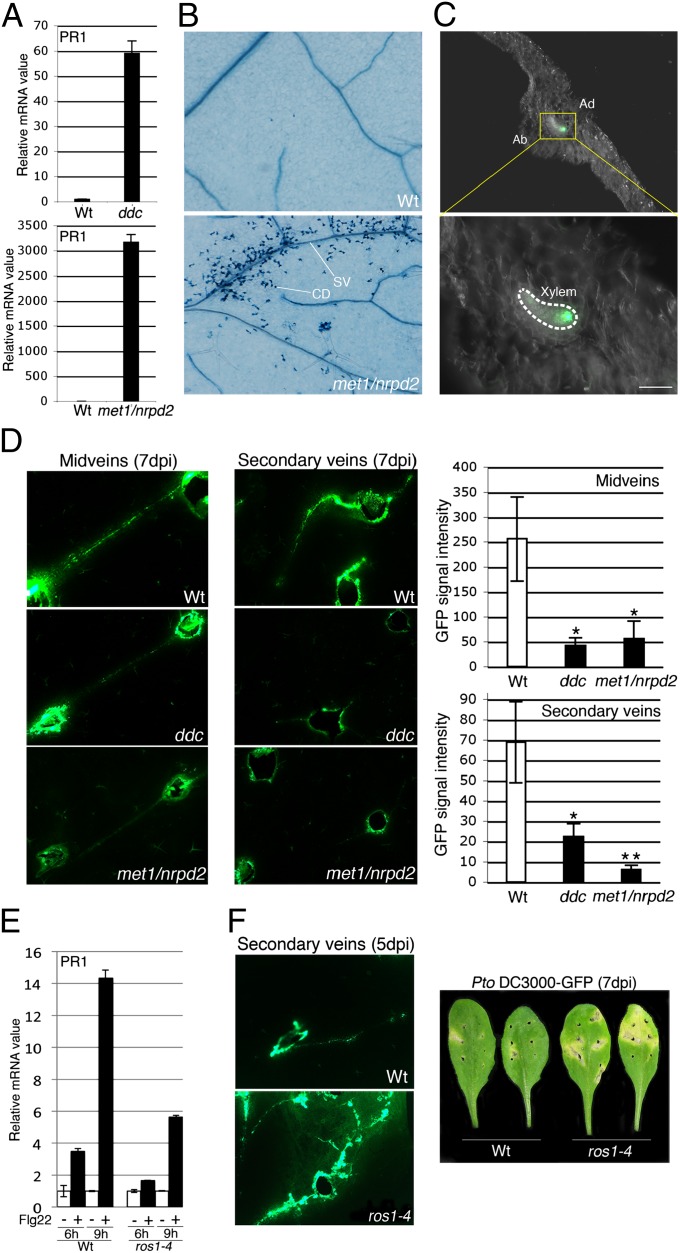
To explore the potential link between DNA demethylation and antibacterial defense, we first tested the resistance of DNA methylation mutants to the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 (Pto DC3000) by syringe inoculation assay. Using this inoculation method, a mild enhanced bacterial growth was observed in ros1-infected plants, but not in the demeter-like 2 (dml2) and dml3-infected DNA glycosylase/lyase loss-of-function mutants (Fig. S9), supporting a role for ROS1-dependent DNA demethylation in antibacterial resistance. This phenotype was also associated with an altered salicylic acid (SA)-dependent defense response as revealed by an attenuated flg22-triggered induction of PR1 in ros1- versus WT-elicited plants (Fig. 3E). On the contrary, bacterial titers were lower in mutants defective in RdDM activity (i.e., nrpd2, drm1/drm2, and Fig. S9) (8, 16), with stronger resistance achieved in mutants impaired in both the RdDM pathway and maintenance of CG or CHG methylation (i.e., drm1/drm2/cmt3 referred to as the ddc mutant, met1/nrpd2, and Fig. S9). This is consistent with the constitutive expression of PR1 observed in the ddc and met1/nrpd2 mutants (Fig. 3A), and thus mimics, to some extent, the flg22-induced response observed in wild-type plants (Fig. 2A). Enhanced PR1 expression in nontreated met1/nrpd2 was also associated with constitutive cell death resembling the hypersensitive response (HR), a physiological response that often accompanies plant antimicrobial resistance and that is typically observed in mutants exhibiting autoimmune phenotypes (Fig. 3B) (24). Interestingly, this cell death phenotype was confined around secondary veins (Fig. 3B), in the same location as the tissues in which AtGP1 was reactivated upon flg22 treatment (Fig. 1A). On the basis of these results we hypothesized that DNA demethylation may restrict bacterial propagation within and around leaf vasculature.
Fig. 3.
(A) PR1 mRNA levels with SD as in Fig. 1. Similar results were obtained in two independent experiments. (B) Trypan blue-stained nontreated WT and met1-3(−/+)/nrpd2-2 leaves. SV and CD stand for secondary vein and cell death, respectively. Cell death is observed around secondary veins but also at the base of some trichomes in the met1/nrpd2 background. Similar results were obtained in two independent experiments. (C) Five- to 6-wk-old leaves of WT were wound inoculated with a toothpick in midveins with Pto DC3000–GFP at 5 × 107 cfu/mL and GFP signal monitored under UV light from transversal sections of leaf blades performed in between two inoculation sites. Pictures were taken at 7-d postinfection (dpi). Ad and Ab stand for adaxial part of the leaf and abaxial part of the leaf, respectively. (Scale bar, 0.05 mm.) Similar results were obtained in three independent experiments. (D) Five- to 6-wk-old leaves of WT, ddc, and met1-3 (−/+)/nrpd2-2 were wound inoculated in midveins or secondary veins as in C and pictures were taken at 7 dpi (Left). GFP fluorescence intensity in midveins or in secondary veins (Right). Asterisks represent significant differences in GFP fluorescence intensity (*P < 0.05; **P < 0.01). Similar results were obtained in three independent experiments. (E) WT and ros1–4 leaves were treated with water (mock) or 1 μM of flg22 for 6 and 9 h and PR1 transcript levels analyzed by RT-qPCR. Error bars: SD from three independent PCR results. Similar results were obtained in four independent experiments. (F) WT and ros1–4 leaves were wound inoculated as in C and GFP signal monitored at 5 dpi as in C (Left). Pictures of bacterial disease symptoms on WT and ros1–4 wound-inoculated leaves (Right). Similar results were obtained in four independent experiments.
DNA Demethylation Restricts Bacterial Propagation Within Xylem Vessels.
To assess the role of DNA demethylation in vascular propagation of Pto DC3000, we first determined whether this bacterium could propagate within Arabidopsis leaf vasculature. Using wound-inoculation assay of a GFP-expressing Pto DC3000 strain in wild-type leaf midveins, we observed bacterial propagation within xylem vessels and restricted to a few vascular bundles, as recently described in Nicotiana benthamiana leaves (Fig. 3C) (25). We next wound inoculated Pto DC3000–GFP in midveins and secondary veins of ddc and met1/nrpd2 mutants and found that bacterial propagation was significantly impaired in these mutants with especially strong effects in met1/nrpd2 secondary veins (Fig. 3D). Conversely, wound-inoculated ros1 leaves displayed a significant increased bacterial spread within secondary veins, with chlorosis and necrosis developing in tissues adjacent to the inoculation sites (Fig. 3F), which is consistent with the intense transcriptional expression of ROS1 in the vasculature (Fig. S10). Collectively, these results indicate that DNA demethylation restricts bacterial multiplication and propagation in Arabidopsis leaves, suggesting that some immune-response genes are likely to be directly controlled by siRNA-directed DNA methylation and ROS1-dependent DNA demethylation.
RMG1 is a Disease Resistance Gene That Is Targeted by RdDM and ROS1-Dependent DNA Demethylation.
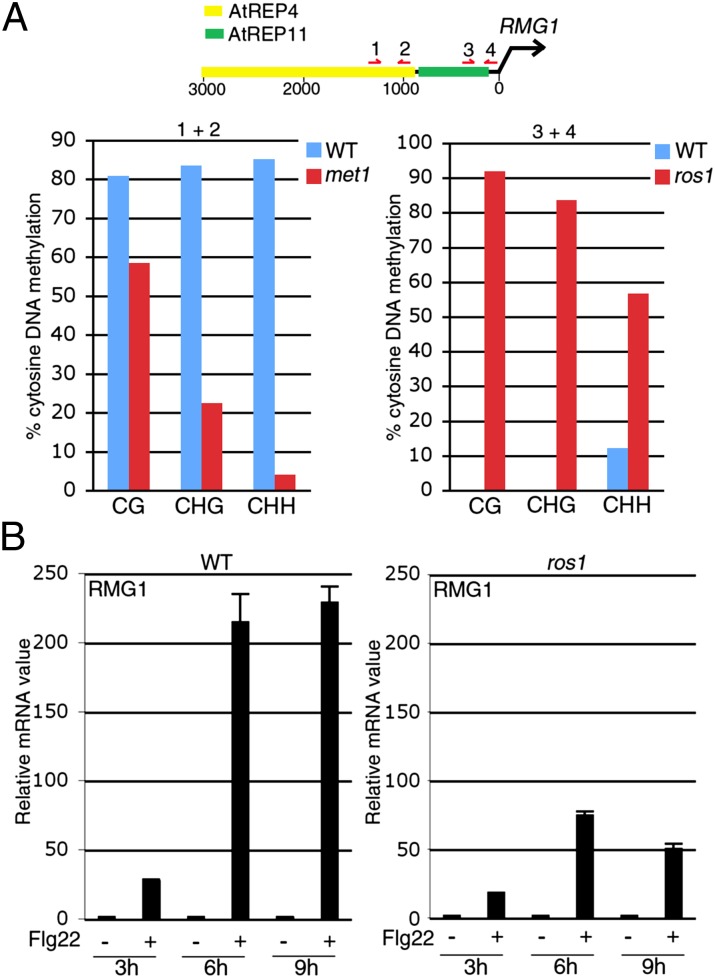
Plant NLRs encode key immune receptors whose overexpression was shown to trigger constitutive HR and/or PR1 induction in some instances (26–28). The HR-like phenotype and enhanced PR1 expression observed in met1/nrpd2 (Fig. 3 A and B) therefore suggested that some NLRs might be more expressed in this mutant background, and perhaps, directly controlled by siRNA-directed DNA methylation. Sequencing of RNA extracted from flg22-elicited wild-type leaves uncovered 55 up-regulated NLR transcripts (induced more than twofold; Dataset S1). Among those, 15 carried repeats/siRNA clusters in their vicinity and six of these NLRs were expressed at higher levels in met1/nrpd2 compared with wild-type plants (Fig. S11). One gene, At4g11170, referred to here as Resistance Methylated Gene 1 (RMG1), was expressed at high levels in response to flg22 and in naïve met1/nrpd2 relative to wild-type plants (Fig. S11) and displayed an earlier and sustained induction in flg22-treated RdDM-defective mutants compared with wild-type-elicited seedlings (Fig. S12). RMG1 encodes a NB-LRR protein with a Toll/interleukin-1 receptor (TIR) domain at its N terminus. Interestingly, this disease resistance gene contains two helitron-related repeats in its promoter region referred to as AtREP4 and AtREP11 (Fig. 4A). AtREP4, which is the most distal repeat from RMG1’s transcription start, was strongly targeted by siRNAs and heavily methylated in all cytosine sequence contexts (Fig. 4A and Fig. S13), whereas DNA methylation at a region overlapping the 3′ end of AtREP11 and the proximity of RMG1’s transcription start was weak in wild-type plants but drastically enhanced in all cytosine sequence contexts in the ros1 mutant background (Fig. 4A). Furthermore, both basal expression and flg22-triggered induction of RMG1 was compromised in ros1 mutant plants (Fig. 4B and Fig. S14). Collectively, these results indicate that the disease resistance gene RMG1 is a primary RdDM target and that both its basal expression and flg22-triggered transcriptional induction require ROS1 activity.
Fig. 4.
(A) DNA methylation at RMG1 promoter, a schematic representation of the RMG1 promoter (Upper) is presented where red arrows represent the position of primers used for bisulfite sequencing on WT and met1(−/−) leaves (Left graph) or WT and ros1–4 leaves (Right). The regions analyzed with primers (1+2) and (3+4) contain 12 CG, six CHG, 56 CHH, and two CG, three CHG, and 27 CHH, respectively. Similar results were obtained in three independent experiments. (B) RMG1 mRNA levels in 5- to 6-wk-old WT or ros1–4 leaves treated with either water (−) or flg22 (+) for 3, 6, and 9 h with SD as in Fig. 1. Similar results were obtained in four independent experiments.
Discussion
On the basis of these data and on previous findings (8, 29), we conclude that induction of some TEs/defense genes involves a DNA demethylation process during antibacterial defense. In human cells, DNA demethylation of the Interleukin-6 promoter facilitates the recruitment of specific transcriptional regulators during antiviral defense (30). Our data suggest that DNA demethylation in plants occurs in part through a combination of repression of a subset of coregulated TGS factors and constitutive ROS1-dependent active removal of DNA methylation at some TEs/defense genes. We hypothesize that such DNA demethylation may facilitate the recruitment of PolII and/or transactivators onto their promoters that contain pathogen-responsive elements (Figs. S13 and S15). If such transactivators are constitutively active, derepression will occur in unchallenged DNA methylation-defective mutants (e.g., AtGP1 or RMG1). Alternatively, derepression may require induced and/or activated transactivators and would, therefore, occur solely during antibacterial defense, as observed for the RdDM targets Onsen and WRKY22 (Fig. S2), a defense transcription factor whose flg22-triggered induction is enhanced in RdDM-defective mutants relative to wild-type seedlings (SI Text and Figs. S15–S19). We thus propose that DNA demethylation likely prime TE-, as well as defense gene-induction through the concomitant activation of their transactivators and/or the interference with other chromatin marks. Accordingly, higher levels of the active marks H3K4me3/H3K9ac, and lower levels of the repressive marks H3K9me2/H3K27me3, were detected at SA-responsive gene promoters in PolV-defective mutants (16). However, none of these promoters was directly targeted by RdDM, arguing for an indirect effect of those mutations on the chromatin-based status of SA-responsive genes. Transcriptional activation of primary RdDM targets may additionally require the constitutive presence of active chromatin marks at these loci as noticed in the body of WRKY22 and at the 5′ part of the ORF of RMG1 (http://epigara.biologie.ens.fr/cgi-bin/gbrowse/a2e/). If these epigenetic modifications are indeed present within the same cells, they would ensure a dual and antagonistic epigenetic control of these defense genes by maintaining, through DNA methylation in their promoters, a low basal expression level in normal growth conditions, while having a chromatin environment poised for a rapid and pervasive transcription under pathogen constraints when TGS is released. Such a chromatin-based regulatory mechanism would be well adapted to tightly control the basal- and pathogen-responsive- transcriptional status of immune-response genes such as NLRs, whose overexpression is often associated with severe fitness cost (26–28). Consistent with this idea, the disease resistance gene Lazarus 5 (LAZ5), whose transcriptional expression requires the active chromatin mark H3K36me3 (31), is also targeted by siRNA/DNA methylation at its 5′ and 3′ ends (http://epigenomics.mcdb.ucla.edu).
We have shown that the basal- and/or flg22-induced expression of several MAMP-responsive NLRs was enhanced in DNA methylation defective mutants (Figs. S11 and S12). Although flg22-triggered induction of many Arabidopsis NLRs was previously reported (17, 18), very little is known on the underlying mechanisms involved in this regulatory process. We propose that flg22-triggered inactivation of TGS represents one of those mechanisms because it presumably contributes to the transcriptional activation of RMG1 and perhaps other NLRs. This part of our study therefore sheds light on an as-yet unsuspected molecular link between MTI and NLR-dependent defense responses. This link may also have a posttranslational component, because flg22 triggered a decrease in the accumulation of protein levels of TGS factors (Fig. 2C) that might be sensed by NLR proteins, thereby activating an SA-dependent defense response (Fig. 2A). This scenario, although still speculative, would thus extend the classical “guard hypothesis” to the indirect detection of MAMPs by plant NLRs that would monitor the differential protein levels of “guarded” TGS factors during MAMP-triggered signaling events. Whether these hypothesized regulatory mechanisms contribute to the autoimmune phenotypes observed in the met1/nrpd2 double mutant remains to be determined.
We have also demonstrated that DNA demethylation restricts leaf vascular propagation of Pto DC3000, which possibly gains access to the vasculature from wound inoculation sites or hydathodes (Fig. S20). This plant-induced vascular defense appears to be particularly effective at the base of midveins and proximal regions of secondary veins (Fig. 1A), which represent the only tissues where cells are still dividing at late stages of leaf development (32) and where maintenance of DNA methylation is likely needed to silence RdDM targets such as AtGP1 retrotransposons. MAMP-triggered inactivation of TGS in these actively dividing cells may thus favor a potent derepression of a subset of immune-response genes in cis, including the WRKY22 and RMG1 described in this study. In a similar way, it might contribute to the strong antimicrobial defense response that is often observed in plant meristematic tissues (33). MAMP-triggered release of TGS may also lead to the production of additional TE-based substrates for DCL proteins and therefore favors the biosynthesis of trans-active siRNAs that would have the potential to silence modulators of plant defense in cells that surround sites of TE reactivation. Such a scenario would be consistent with the enhanced accumulation of TE-derived 21-nt siRNAs recently described upon SA treatment (8), and might contribute to the formation of an immune cell layer around the vasculature that would prevent bacterial propagation from xylem vessels to mesophyll cells and vice versa. Such a noncell autonomous regulatory mechanism has been initially described in the context of pollen development, where the derepression of some Athila retrotransposons in pollen vegetative cells was shown to trigger the production of 21-nt mobile TE-derived siRNAs that were trans-active in sperm cells (34). Intriguingly, the reactivation of TEs in pollen vegetative cells was associated with the down-regulation of a subset of TGS factors (34), therefore mimicking the flg22-triggered effects described in the present study. It is thus tempting to speculate that an endogenous regulatory mechanism might ensure a constitutive antimicrobial immune response in pollen vegetative cells that would protect them from pathogen invasion, thereby preserving the integrity of male gametes and limiting pollen transmission of pathogens, a common plant-to-plant spreading strategy used by many viruses (35).
Systemic acquired resistance (SAR) is an inducible broad-spectrum immune response in plants that restricts the spread of pathogens and prevents infection in distal tissues (36). The SAR signal SA, whose production is increased in response to pathogens or flg22 (37, 38), is known to trigger massive changes in gene expression and to induce DNA demethylation at SA-induced TEs (8, 39). Furthermore, bacterial-induced SAR was recently shown to confer transgenerational resistance toward unrelated pathogens including Pto DC3000 (14, 15). Based on these findings and on the present work, we speculate that pathogen- or MAMP-induced production of SA might trigger DNA demethylation of TEs/defense genes both locally but also in systemic unchallenged tissues including reproductive organs, thereby orchestrating transgenerational immune priming. Additionally, siRNA pools that are produced from pathogen- or MAMP-challenged tissues, including TE-derived siRNAs mentioned above, might trigger long-distance mobile silencing that could modulate the transcriptional response to pathogens in the offspring. Investigating the contribution of DNA demethylation and pathogen-responsive siRNAs in transgenerational immune priming will therefore be essential to unravel the mechanisms by which pathogens drive the selection of new phenotypes through epigenetic and epigenetic-directed genetic changes.
Materials and Methods
Plant Growth Conditions and Treatments.
Most of the plants used in this study were grown at 23 °C with an 8-h photoperiod. Five- to 6-wk-old leaves from different genotypes were syringe infiltrated with either water or flg22 synthetic peptide (Genscript), at 1 μM concentration. The treatments of Arabidopsis seedlings with flg22 or DNA methyltransferase inhibitor are described in SI Materials and Methods.
Transgenic Plant Materials and DNA Contructs.
AtGP1 LTR:GUS and ROS1p:ROS1-GUS constructs were generated as described in SI Materials and Methods. These constructs were transformed in the Col-0 accession. The AtGP1 LTR:GUS #16 reference line was selected based on its sensitivity to a DNA methyltransferase inhibitor (SI Materials and Methods).
Histochemical GUS Staining.
Five- to 6-wk-old leaves of AtGP1 LTR:GUS #16 were syringe-infiltrated with either water or flg22 peptide at 1 μM concentration and collected at 24 h posttreatment. They were GUS stained as described in SI Materials and Methods. Five- to 6-wk-old unchallenged ROS1p:ROS1-GUS leaves were GUS stained similarly.
Bacterial Infections.
Bacterial infections were performed by syringe infiltration or wound inoculation on 5- to 6-wk-old Arabidopsis leaves from different genotypes. Pto DC3000 and a GFP-tagged Pto DC3000 were used for this study. For syringe-inoculation assay, Pto DC3000 was used at a concentration of 105 colony-forming units per milliliter (cfu/mL) and bacterial titers were monitored by serial dilution assays. For wound inoculation, Pto DC3000–GFP was used at a concentration of 5 × 107 cfu/mL and inoculated in either midveins or secondary veins with a toothpick. Bacterial propagation was then analyzed as described in SI Materials and Methods. To determine the presence of Pto DC3000–GFP in xylem vessels, transversal sections of leaves were conducted by cutting polystyrene rod containing leaf transversally with a razor blade.
Real-Time RT-PCR Analyses.
Total RNA was extracted using RNeasy Plant Mini kit (Qiagen). RNA samples were reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Invitrogen) with a mix of random hexamers and oligodT. The cDNA was quantified using a SYBR Green qPCR kit (Roche LightCycler 480 SYBR Green I Master) and gene specific primers. PCR was performed in 384-well plates heated at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 10 s and annealing at 60 °C for 40 s. A melting curve was performed at the end of the amplification. Transcript levels were normalized to that of Actin2. All primer sequences are listed in Dataset S2.
Bisulfite Conversion and Sequencing.
Total genomic DNA was extracted using DNeasy Plant Mini kit (Qiagen) and bisulfite treated using EpiTect Bisulfite kit (Qiagen). The PCR products were purified and cloned as described in SI Materials and Methods. Height to 23 clones were sequenced from naïve leaf samples, whereas 15–25 clones were sequences from mock-treated and flg22-treated leaf samples. The bisulfite conversion efficiency was tested by confirming the absence of DNA methylation at a nonmethylated region (see SI Materials and Methods for details). All primer sequences are listed in Dataset S2.
Cell Death Observations.
Five- to 6-wk-old leaves from different genotypes were stained with Trypan Blue as described in SI Materials and Methods.
Western Blot Analyses.
Total protein extracts from 5- to 6-wk-old Arabidopsis leaves, treated with either water or flg22 synthetic peptide at 1 μM concentration, were obtained using the Tanaka method and resolved on SDS/PAGE. Protein blot analysis was performed using antibodies raised against an AGO4 and NRPE5 peptides (gifts from T. Lagrange, Laboratoire Génome et Développement des Plantes, Perpignan, France), ACTIN8 (monoclonal antiactin plant; Sigma).
Small RNA Library/Sequencing and Data Mining.
A small RNA library was made from 5-wk-old Col-0 leaf samples and deep sequenced by Fasteris (Geneva) on the Illumina HiSEq. 2000. Details of data processing are described in SI Materials and Methods.
RNA Library/Sequencing and Data Mining.
RNA libraries were made from 5- to 6-wk-old Col-0 leaf samples (treated with either water or flg22 at 1 μM concentration for 6 h) and deep sequenced by Fasteris (Geneva) on the Illumina HiSEq. 2000. Details of data processing are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Baulcombe, S.-H. He, S. Jacobsen, O. Mathieu, and T. Lagrange for materials and Peter Brodersen and members of the Navarro laboratory for critical reading of the manuscript. This work was supported by an Action Thématique et Incitative sur Programme (ATIP)/Avenir Grant from the Fondation Bettencourt Schueller (FBS) (to A.Y., G. L., and L.N.), an Emergence Grant from the Mairie de Paris (to A.Y., G. L., and L.N.), a European Research Council (ERC) grant titled “Silencing and Immunity” (to L.N.), a grant from the Fondation Pierre-Gilles de Gennes (to A.-L.A.), a prize for life science from the FBS (to O.V.), and a National Institutes of Health Grant R01GM069415 (to J.P. and R.L.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE40044).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211757110/-/DCSupplemental.
References
- 1.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23(12):454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393( *)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell. 2011;42(3):356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472(7341):115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 7.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461(7267):1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 8.Dowen RH, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA. 2012;109(32):E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA. 2007;104(11):4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21(3):367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12(8):483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 12.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111(6):803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 13.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slaughter A, et al. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2012;158(2):835–843. doi: 10.1104/pp.111.191593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luna E, Bruce TJ, Roberts MR, Flors V, Ton J. Next-generation systemic acquired resistance. Plant Physiol. 2012;158(2):844–853. doi: 10.1104/pp.111.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López A, Ramírez V, García-Andrade J, Flors V, Vera P. The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet. 2011;7(12):e1002434. doi: 10.1371/journal.pgen.1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro L, et al. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135(2):1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428(6984):764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 19.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Huettel B, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: A versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769(5-6):358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130(5):851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-Macías MI, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell. 2012;45(3):357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17(1):54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, et al. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe. 2012;11(3):253–263. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Misas-Villamil JC, Kolodziejek I, van der Hoorn RA. Pseudomonas syringae colonizes distant tissues in Nicotiana benthamiana through xylem vessels. Plant J. 2011;67(5):774–782. doi: 10.1111/j.1365-313X.2011.04632.x. [DOI] [PubMed] [Google Scholar]
- 26.Oldroyd GE, Staskawicz BJ. Genetically engineered broad-spectrum disease resistance in tomato. Proc Natl Acad Sci USA. 1998;95(17):10300–10305. doi: 10.1073/pnas.95.17.10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell. 2000;12(12):2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Dorey S, Swiderski M, Jones JD. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 2004;40(2):213–224. doi: 10.1111/j.1365-313X.2004.02201.x. [DOI] [PubMed] [Google Scholar]
- 29.Pavet V, Quintero C, Cecchini NM, Rosa AL, Alvarez ME. Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by pseudomonas syringae. Mol Plant Microbe Interact. 2006;19(6):577–587. doi: 10.1094/MPMI-19-0577. [DOI] [PubMed] [Google Scholar]
- 30.Tang B, et al. Interleukin-6 expression was regulated by epigenetic mechanisms in response to influenza virus infection or dsRNA treatment. Mol Immunol. 2011;48(8):1001–1008. doi: 10.1016/j.molimm.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Palma K, et al. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 2010;6(10):e1001137. doi: 10.1371/journal.ppat.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang J, Dengler N. Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta. 2002;216(2):212–219. doi: 10.1007/s00425-002-0847-9. [DOI] [PubMed] [Google Scholar]
- 33.Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138(4):1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mink GI. Pollen and seed-transmitted viruses and viroids. Annu Rev Phytopathol. 1993;31:375–402. doi: 10.1146/annurev.py.31.090193.002111. [DOI] [PubMed] [Google Scholar]
- 36.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 37.Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53(5):763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2(11):e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.