Abstract
A LightCycler real-time PCR hybridization probe-based assay which detects a partial Klebsiella pneumoniae 16S rRNA gene was developed for the rapid identification of K. pneumoniae directly from growth-positive blood culture bottles (BACTEC 9240 system) within 2 h. No cross-reactivity was observed with 65 negative-control blood cultures that grew bacteria other than K. pneumoniae and 48 negative blood cultures from double-blind experiments, thus demonstrating 100% specificity when compared to results of conventional biochemical characterization. The assay also showed 100% sensitivity, as it correctly identified all 142 positive-control blood cultures and 4 from double-blind trials.
Klebsiella pneumoniae is an important hospital-acquired pathogen that causes severe morbidity and mortality among the newborn, the elderly, and immunocompromised patients (3, 4, 15). K. pneumoniae infections acquired in hospitals can be difficult to treat, as many strains are highly resistant to treatment with broad-spectrum cephalosporins and aminoglycosides (1, 7, 8, 10, 13). The isolation of bacteria from blood culture bottles is usually indicative of a serious invasive infection (bacteremia) which requires urgent antimicrobial therapy. Microorganisms isolated from patients usually have different antimicrobial susceptibilities, and successful treatment is dependent on the prompt administration of the appropriate antibiotic (6, 9, 11, 14). K. pneumoniae is identified by various automated and manual products such as Vitek (BioMérieux, Inc.) and the Analytab Products (API) system (BioMérieux, Inc.) or by traditional biochemical tests, and the whole process requires at least 24 to 48 h.
In recent years, real-time PCR has emerged as a valuable tool for the rapid testing of various biological specimens and body fluids for the presence of microorganisms (2, 5, 12, 17). The development of highly sensitive and specific PCR assays has alleviated the problems typically associated with microorganisms that are found in low numbers in tissues or body fluids, that are difficult to culture, or that are serologically similar. The LightCycler system (Roche Molecular Biochemicals, Mannheim, Germany) offers two different fluorescence detection formats. The first employs SYBR Green I, which is a dye that binds nonspecifically to double-stranded DNA (16); the second uses hybridization probes which allow sequence-specific detection by using fluorescence energy transfer (FRET) between two fluorophores. This result is achieved by attaching the two fluorophores to two oligonucleotide probes designed to hybridize to a complementary region of the target gene, leaving a 1-nucleotide-wide gap. The LightCycler instrument has been developed for fast cycling and real-time monitoring of the amplification process and thus eliminates the need to carry out gel electrophoresis of the samples after PCR.
In this study, we describe a real-time PCR assay with specific primers and hybridization probes targeting the 16S rRNA gene for the direct detection of K. pneumoniae from positive blood culture bottles. The assay also provides specific and sensitive quantification of K. pneumoniae DNA.
We studied 142 blood cultures that were known to be positive controls for K. pneumoniae, 65 that were negative controls for K. pneumoniae, and 52 double-blind samples (Table 1) that showed positive growth in BACTEC Plus aerobic medium in the BACTEC 9240 system. DNA extractions were performed in all experiments by using the QIAamp blood and tissue kit (QIAGEN, Valencia, Calif.) according to the manufacturer's instructions. DNA extracted from type strains which acted as positive and negative controls was stored at −20°C until use. Oligonucleotide primers were designed to amplify a 126-bp target sequence of the 16S rRNA gene spanning nucleotide positions 44 to 170 of the K. pneumoniae strain with EMBL databank accession no. X93214. The forward primer K16SF (15-mer; position 44 to 58) and reverse primer K16SR (16-mer; position 155 to 170) were used. Within the 16S rRNA gene, fluorescence-labeled probes were designed to hybridize to the species-specific region of K. pneumoniae. The sensor probe used was the K. pneumoniae sensor (19-mer; melting temperature [Tm] = 62.7°C) oligonucleotide labeled with LightCycler Red 640 at the 5′ end and phosphorylated at the 3′ end to block extension. The corresponding fluorescein isothiocyanate-labeled anchor probe at the 3′ end was the K. pneumoniae anchor (30-mer, Tm = 69.9°C) oligonucleotide that binds to the template strand at a distance of one base to the bound sensor probe.
TABLE 1.
Real-time PCR hybridization probe-based testing of microorganisms
| Sample type and species or groupa | No. of samples | PCR sensitivity (%) | PCR specificity (%) |
|---|---|---|---|
| Positive-control organisms | |||
| Klebsiella pneumoniae (type strain ATCC 13883) | 1 | ||
| Klebsiella pneumoniae (clinical isolates) | 141 | 100 | |
| Total | 142 | ||
| Negative-control organisms | |||
| Klebsiella oxytoca (clinical samples) | 3 | 100 | |
| Escherichia coli (ATCC 25922, ATCC 35218, NCTC 10418) | 10 | 100 | |
| Escherichia coli (O157:H7, clinical isolates) | 10 | 100 | |
| Pseudomonas aeruginosa (ATCC 27853, clinical isolates) | 10 | 100 | |
| Streptococcus pneumoniae (ATCC 49619, NCTC 7465) | 10 | 100 | |
| Streptococcus pyogenes (group A) | 1 | 100 | |
| Streptococcus agalactiae (group B) | 1 | 100 | |
| Staphylococcus aureus (ATCC 25923, ATCC 29213) | 7 | 100 | |
| Staphylococcus aureus (MRSAb; ATCC 43300) | 4 | 100 | |
| Enterococcus faecalis (ATCC 29212, clinical isolate) | 3 | 100 | |
| Enterococcus faecium (EQASc, clinical isolate) | 2 | 100 | |
| Haemophilus influenzae (ATCC 49247, clinical isolate) | 2 | 100 | |
| Neisseria meningitidis (EQAS) | 1 | 100 | |
| Neisseria gonorrhoeae (ATCC 49226) | 1 | 100 | |
| Total | 65 | 100 | |
| Double-blind samples | |||
| First group (tested positive for K. pneumoniae) | 4 | 100 | |
| Second group (no above-background fluorescence) | 48 | 100 | |
| Total | 52 |
Strain numbers and/or further descriptive information is given in parentheses.
MRSA, methicillin-resistant S. aureus.
EQAS, External Quality Assurance System.
Real-time PCR sample preparation.
The master mixture contained 2 μl of 10× LightCycler-FastStart DNA master hybridization probes (Roche Diagnostics), 3 mM MgCl2 (final concentration), 0.5 μM final concentration of K. pneumonia-specific primers, and 0.2 μM final concentration of hybridization probes. To complete the PCR mixtures, 18 μl of master mixture and 2 μl of a DNA preparation were added into the LightCycler glass capillaries. After a short centrifugation (700 × g for 10 s), the sealed capillaries were placed into the LightCycler.
Real-time PCR amplification and melting-curve analysis.
Thermocycling conditions were optimized to one cycle of denaturation at 95°C for 10 min, followed by 30 amplification cycles (temperature transition rate of 20°C/s), each comprising denaturation (95°C for 10 s), annealing (55°C for 15 s), and extension (74°C for 10 s). PCR amplification was followed by melting-curve analysis from 40 to 95°C (temperature transition rate of 0.1°C/s) with continuous fluorescence readings. Fluorescence was measured continuously during the slow temperature rise to monitor the dissociation of the LightCycler Red 640-labeled sensor probe at the F2 channel. Fluorescence signals from F2 were plotted automatically in real time versus temperature (T) to produce melting curves for amplicons. Melting curves were then converted into melting peaks by plotting the negative derivative of fluorescence versus T (−dF2/dT versus T and −dF3/dT versus T). Water was used as the negative control and was included in each set of melting peak determinations. The entire process took approximately 40 min. The presence of the 126-bp amplicon was verified by agarose gel electrophoresis.
Sensitivity of real-time PCR assay for K. pneumoniae.
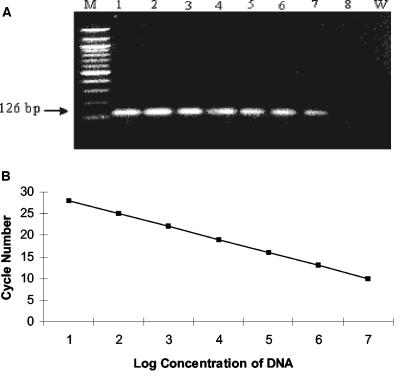
The sensitivity of the real-time PCR assay was optimized to 10 fg of purified K. pneumoniae DNA. Agarose gel electrophoresis of the amplicons showed that the band intensity correlated well with the calculated concentrations of amplicons (Fig. 1 A), and the sequence was analyzed (data not shown). Real-time PCR sensitivity optimization performed by using the K. pneumoniae type strain (ATCC 13883) resulted in a linear-regression curve across 107 to 1 genomic equivalents (10 ng to 1 fg of DNA) with an error rate of <1% and a correlation coefficient at −1 (Fig. 1B). The intraassay variation of the threshold/cycle (CT) values was evaluated by using K. pneumoniae DNA dilutions containing 107, 106, 105, 104, 103, 102, and 10 copies. CT values were established between 10 and 27. The CT values, which are inversely related to the quantity of organisms, ranged between 10 and 27 corresponded to 107 to 10 organisms (Fig. 1B).
FIG. 1.
Standard curve for K. pneumoniae DNA quantification. (A) Agarose gel electrophoresis of a 126-bp PCR product. Lane W represents the negative control (water). Lane M represents the 100-bp ladder DNA marker; lanes 1 to 8 represent the 10-fold-dilution series of genomic equivalents of 107 copies (lane 1), 106 copies (lane 2), 105 copies (lane 3), 104 copies (lane 4), 103 copies (lane 5), 102 copies (lane 6), 10 copies (lane 7), and 1 copy (lane 8) of purified K. pneumoniae DNA. (B) Plasmid containing one copy of the target sequence was used to construct the standard curve for K. pneumoniae DNA quantification. Serial 10-fold dilutions ranging from 107 copies to 1 copy of plasmid were used for determination of real-time PCR assay detection limits for in vitro sensitivity testing. Cycle number plotted against the log of calculated concentration values resulted in a standard curve with an error of 0.122 and correlation coefficient at unity. Negative-control organisms did not fluoresce above the background signal level.
Specificity of the real-time PCR assay.
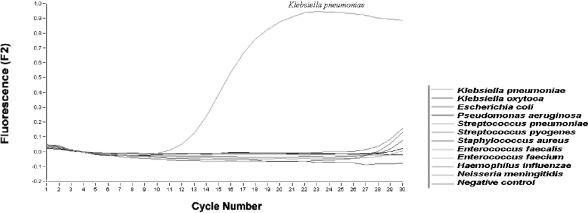
The specificity of the real-time PCR assay for K. pneumoniae with primers specific for the 16S rRNA gene was investigated by testing 1 ng of purified DNA from each of the 65 negative-control organisms (Table 1). Amplification of Pseudomonas aeruginosa, Streptococcus spp., Staphylococcus aureus, Enterococcus faecalis, Haemophilus influenzae, and Neisseria meningitidis remained negative when tested at all concentrations (10 ng to 1 fg), whereas the CT values for Klebsiella oxytoca, Escherichia coli, and Enterococcus faecium were greater than 29 at a concentration of 1 ng/μl. By comparison, a CT value of ≤13 was achieved for K. pneumoniae at the same concentration of 1 ng/μl (Fig. 2). Upon agarose gel electrophoresis of PCR mixtures derived from negative-control organisms, no amplicon was ever observed. The target sequence was shown to be highly specific for K. pneumoniae, as it failed to detect any other bacteria that are known to cause bacteremia (Fig. 2).
FIG. 2.
Specificity of the real-time PCR assay for K. pneumoniae. Different bacterial species were subjected to DNA extraction and quantitation by the real-time PCR assay. Data for fluorescence versus cycle number plots for clinical samples are shown.
Melting-curve analysis of real-time PCR assay for K. pneumoniae.
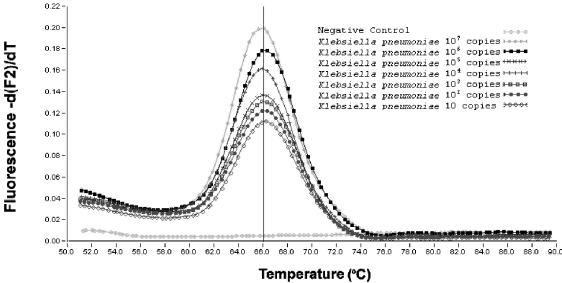
When the temperature reached the Tm of the probes, a rapid loss of fluorescence was observed as the two adjacently bound probes dissociated from their complementary targets and FRET stopped. This was plotted as the negative derivative of fluorescence versus temperature to define the template-specific melting curves (Fig. 3). Without changing the MgCl2 concentration, we could still maintain a sensitivity of 10 organisms per sample, as shown by a 10-fold dilution series from 107 to 1 genome equivalent(s) of our reference strain. With the hybridization probes, the PCR products were found to reproducibly yield a melting peak of 66°C (Fig. 3).
FIG. 3.
Melting curve for the LightCycler KPS and KPA FRET hybridization probes. The specific melting peak can be observed with the quantified number of copies of K. pneumoniae DNA.
Double-blind testing of blood culture bottles.
Four of 52 double-blind samples tested positive for K. pneumoniae, with a CT value of <20, and 48 samples displayed no detectable fluorescence above the background level. Double-blind PCR-based testing results were concordant with those of biochemical identification.
PCR-based testing of all 259 blood samples by real-time PCR demonstrated a sensitivity of 100% (146 of 146) and a specificity of 100% (113 of 113). The molecular assay was completed within 2 h upon receiving BACTEC 9240 positive blood culture bottles. No contamination was observed at any time during the study. No false-positive signal was detected when the negative control (which comprised sterile water, primers, and probes) was amplified in the reaction capillaries. In contrast to conventional PCR methods, they are less prone to contamination because postamplification handling is eliminated (12). Our real-time PCR-based approach for identification of K. pneumoniae resulted in significant time saving in comparison to traditional biochemical identification of K. pneumoniae from BACTEC 9240 positive blood cultures.
In conclusion, real-time PCR offers a fast tool with high sensitivity and specificity for the identification of K. pneumoniae from positive blood cultures. If in the future it becomes possible to detect more pathogens by multiplex real-time PCR, this method could become a suitable technology for implementation in routine diagnostic laboratories. It is also suitable for laboratories which encounter high isolation rates for K. pneumoniae or in outbreak situations.
REFERENCES
- 1.Arlet, G., M. J. Sanson-Le Pors, M. Rouveau, G. Fournier, O. Marie, B. Schlemmer, and A. Philippon. 1990. Outbreak of nosocomial infection due to Klebsiella pneumoniae producing SHV-4 β-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 9:797-803. [DOI] [PubMed] [Google Scholar]
- 2.Bellin, T., M. Pulz, A. Matussek, H. G. Hempen, and F. Gunzer. 2001. Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J. Clin. Microbiol. 39:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingen, E. H., P. Desjardins, G. Arlet, F. Bourgeois, P. Marianikurkdjian, N. Y. Lambertzechovsky, E. Denamur, A. Philippon, and J. Elion. 1993. Molecular epidemiology of plasmid spread among extended broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J. Clin. Microbiol. 31:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coovadia, Y. M., A. P. Johnson, R. H. Bhana, G. R. Hutchinson, R. C. George, and I. E. Hafferjee. 1992. Multiresistant Klebsiella pneumoniae in a neonatal nursery: the importance of maintenance of infection control policies and procedures in the prevention of outbreaks. J. Hosp. Infect. 22:197-205. [DOI] [PubMed] [Google Scholar]
- 5.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, A. J. Fox, and E. B. Kaczmarski. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunney, R. J., E. B. McNamara, N. Alansari, B. Loo, and E. G. Smyth. 1997. The impact of blood culture reporting and clinical liaison on the empiric treatment of bacteraemia. J. Clin. Pathol. 50:1010-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Champs, C., D. Sirot, C. Chanal, M. C. Poupart, M. P. Dumas, and J. Sirot. 1991. Concomitant dissemination of three extended-spectrum β-lactamases among different Enterobacteriaceae isolated in a French hospital. J. Antimicrob. Chemother. 27:441-457. [DOI] [PubMed] [Google Scholar]
- 8.Jacoby, G. A. 1996. Antimicrobial-resistant pathogens in the 1990s. Annu. Rev. Med. 47:169-179. [DOI] [PubMed] [Google Scholar]
- 9.Jamulitrat, S., U. Meknavin, and S. Thongpiyapoon. 1994. Factors affecting mortality outcome and risk of developing nosocomial bloodstream infection. Infect. Control Hosp. Epidemiol. 15:163-170. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, A. P., M. J. Weinren, B. Ayling-Smith, S. K. Du Bois, S. G. B. Amyes, and R. C. George. 1992. Outbreak of infection in two U.K. hospitals caused by a strain of Klebsiella pneumoniae resistant to cefotaxime and ceftazidime. J. Hosp. Infect. 20:97-103. [DOI] [PubMed] [Google Scholar]
- 11.Lebovici, L. S., I. Shraga, M. Drucker, H. Konigsberger, Z. Samra, and S. D. Pitlik. 1998. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med. 244:379-386. [DOI] [PubMed] [Google Scholar]
- 12.Logan, J. M. J., K. J. Edwards, N. A. Saunders, and J. Stanley. 2001. Rapid identification of Campylobacter spp. by melting peak analysis of biprobes in real-time PCR. J. Clin. Microbiol. 39:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nouvellon, M., J. L. Pons, D. Sirot, M. L. Combe, and J. F. Lemeland. 1994. Clonal outbreaks of extended-spectrum β-lactamase-producing strains of Klebsiella pneumoniae demonstrated by antibiotic susceptibility testing, β-lactamase typing, and multilocus enzyme electrophoresis. J. Clin. Microbiol. 32:2625-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen, G., H. C. Schonheyder, and H. T. Sorensen. 1997. Antibiotic therapy and outcome of monomicrobial gram-negative bacteremia: a 3-year population-based study. Scand. J. Infect. Dis. 29:601-606. [DOI] [PubMed] [Google Scholar]
- 15.Rice, L. B., S. H. Willey, G. A. Papanicolaou, A. A. Medeiros, G. M. Eliopoulos, R. C. Moellering, Jr., and G. A. Jacoby. 1990. Outbreak of ceftazidime resistance caused by extended-spectrum β-lactamases at a Massachusetts chronic-care facility. Antimicrob. Agents Chemother. 34:2193-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skeidsvoll, J., and P. M. Ueland. 1995. Analysis of double-stranded DNA by capillary electrophoresis with laser-induced fluorescence detection using the monomeric dye SYBR Green I. Anal. Biochem. 231:359-365. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]