Abstract
Although variola virus was eradicated by the World Health Organization vaccination program in the 1970s, the diagnosis of smallpox infection has attracted great interest in the context of a possible deliberate release of variola virus in bioterrorist attacks. Obviously, fast and reliable diagnostic tools are required to detect variola virus and to distinguish it from orthopoxviruses that have identical morphological characteristics, including vaccinia virus. The advent of real-time PCR for the clinical diagnosis of viral infections has facilitated the detection of minute amounts of viral nucleic acids in a fast, safe, and precise manner, including the option to quantify and to genotype the target reliably. In this study a complete set of four hybridization probe-based real-time PCR assays for the specific detection of orthopoxvirus DNA is presented. Melting analysis following PCR enables the identification of variola virus by the PCR product's characteristic melting temperature, permitting the discrimination of variola virus from other orthopoxviruses. In addition, an assay for the specific amplification of variola virus DNA is presented. All assays can be performed simultaneously in the same cycler, and results of a PCR run are obtained in less than 1 h. The application of more than one assay for the same organism significantly contributes to the diagnostic reliability, reducing the risk of false-negative results due to unknown sequence variations. In conclusion, the assays presented will improve the speed and reliability of orthopoxvirus diagnostics and variola virus identification.
Smallpox was declared eradicated in 1979 (30), and the last naturally occurring variola virus infection occurred in 1977 (2). Orthopoxviruses have recently been discussed as biological weapons (14, 29). Orthopoxviruses are characterized by large brick-shaped virus particles, containing a double-stranded DNA genome of approximately 200,000 bp (20). Among the characterized orthopoxvirus genomes, the highest sequence homology of orthopoxvirus species is found in the middle of the genome, while the terminal sections of the genome can exhibit considerable variability, probably reflecting differences in host range, tissue tropism, and virulence (12, 19, 21). While humans are the only natural host for variola virus, both vaccinia virus and cowpox virus have a much broader host spectrum, and the natural reservoir for cowpox virus is most likely the rodent. Although zoonotic infections of humans with monkeypox and other orthopoxviruses are known (5, 6, 15, 28), potential variola virus infections bear by far the most lethal risk for humans. Therefore, the rapid and sensitive identification of variola virus and its differentiation from other orthopoxviruses, such as vaccinia, monkeypox, or cowpox virus, is fundamental for reliable diagnostics. Electron microscopy is an extremely fast tool for the differentiation of herpesviruses (e.g., varicella-zoster virus) and orthopoxviruses. However, differentiation of the members of the genus Orthopoxvirus (Vaccinia virus, Cowpox virus, Camelpox virus, Monkeypox virus, and Ectromelia virus) and most importantly the unambiguous identification of variola virus are not possible, and sensitivity is limited (13). In contrast, due to its extreme sensitivity, PCR has evolved to be the diagnostic method of choice (18). Unfortunately, conventional PCR methods are time-consuming, require laborious post-PCR handling, and include a high risk of unwanted carryover contamination by processed PCR product, particularly when genotyping by restriction fragment length polymorphism analysis is used for differentiation (17). The implementation of real-time PCR has addressed these methodological drawbacks. Recently, the first real-time PCR-based LightCycler assay for the identification of variola virus was reported (9). The assay amplifies a part of the hemagglutinin gene, a heterogeneous gene region frequently used for genotyping different orthopoxvirus species. By subsequent melting analysis of the PCR product, variola virus can be distinguished from cowpox virus, monkeypox virus, and vaccinia virus in the same reaction mixture. Unfortunately, due to the appearance of new poxvirus sequences in GenBank, these hybridization probes also display identity to certain cowpox and camelpox virus strains.
However, the consequences of a false-positive or a false-negative result in smallpox diagnosis caused by unexpected sequence variation is considerable, and identification of variola virus should be based on specific detection in several independent PCRs.
Therefore, a set of four LightCycler assays was designed, encompassing different regions of the orthopoxvirus genome and enabling the specific detection of variola virus DNA, either by melting analysis subsequent to generic orthopoxvirus DNA amplification or by specific amplification of variola virus DNA. The A13L gene was previously shown to display significant diversity in different species of the genus Orthopoxvirus, permitting clear identification of variola virus. In vaccinia virus the A13L gene encodes the small membrane protein p8 of the intracellular mature virus (27). The rpo18 gene and the virus early transcription factor (VETF) gene were chosen because they encode highly conserved proteins in orthopoxvirus species. The rpo18 gene product is an 18-kDa subunit of the RNA polymerase in vaccinia virus (1), and the VETF gene product is a transcription factor involved in early transcriptional events (3, 4, 16). We have consciously refrained from using more-established genes such as crmB or hemagglutinin genes because these gene sequences were known to vary considerably among orthopoxviruses and were previously used to assess phylogenetic relationships among orthopoxvirus species (17). In contrast, our approach required highly conserved genomic regions featuring low sequence variability with orthopoxvirus species-specific nucleotides.
All assays were designed and optimized with regard to sensitivity, specificity, speed, and identical conditions to allow all PCR assays to be performed under identical reaction conditions in the same run. Therefore, reliable results can be obtained from extracted DNA in a single PCR run in less than 1 h. In addition, for clinical specimens, the PCR assays described can be combined with a published varicella-zoster virus-specific assay for differential diagnosis (10). Because authentic variola virus DNA is not available, artificial variola virus DNA produced by site-directed mutagenesis of camelpox virus DNA was used. As demonstrated by comparing DNA from 12 non-variola virus orthopoxviruses, including ectromelia, camelpox, cowpox, monkeypox, and vaccinia virus, each individual assay could clearly differentiate between these orthopoxviruses and the variola virus DNA.
MATERIALS AND METHODS
PCR templates.
Extraction of orthopoxvirus DNA from cell culture supernatants was performed with the QIAGEN (Hilden, Germany) DNA kit according to the manufacturer's recommendations. Available orthopoxviruses are shown in Table 1. As calibration standards plasmids containing the respective PCR amplicon were cloned (Table 1) using the TOPO TA cloning kit (Invitrogen, Breda, The Netherlands) as described previously (25, 26). Plasmids with variola virus-specific sequences were generated by site-directed mutagenesis of closely related orthopoxvirus sequences (GenExpress, Berlin, Germany). The plasmid pVar-rpo was constructed by site-directed mutagenesis of plasmid pCam-rpo at position 110 from T (camelpox virus) to C (variola virus). Plasmid pVar-VETF was constructed accordingly, by modification of plasmid pCam-VETF at the following four positions: 46 (T/C), 134 (g/A), 158 (T/C), and 185 (T/C). The variola virus-specific A13L plasmid was synthesized as a synthetic gene (GenExpress). All plasmids pVar-rpo, pVar-VETF, and pVar-A13L were identical to the respective regions of the GenBank entries X69198, X67119, and L22579. Plasmids were diluted in λ-DNA (1 ng/μl; MBI Fermentas, Leon-Roth, Germany) as carrier DNA.
TABLE 1.
Characteristic Tms of the various PCR products of tested orthopoxviruses
| OPV no. | Virus | ABBa |
Tm (°C) of PCR productb
|
|||
|---|---|---|---|---|---|---|
| rpo18 | VETF | A13L Var | A13L CAM | |||
| 1 | Camelpox CP-1 | CAM | 59 (1) | 58 (3) | 59 (3) | |
| 2 | Camelpox CP-19 | CAM | 59 (1) | 58 (3) | 59 (3) | |
| 3 | Cowpox Süd | CPX | 59 (1) | 44 (6) | 59 (3) | |
| 4 | Cowpox Brighton Red | CPX BR | 59 (1) | 49 (5) | 59 (3) | |
| 5 | Cowpox 81/02 | CPX | 59 (1) | 58 (3) | 54 (4) | |
| 6 | Monkeypox AP-1 | MPX | 59 (1) | 49 (5) | 54 (4) | |
| 7 | Monkeypox MP | MPX | 59 (1) | 49 (5) | 54 (4) | |
| 8 | Ectromelia MP-1 | ECT | 59 (1) | 58 (3) | —c | |
| 9 | Ectromelia MP-Nü | ECT | 59 (1) | 58 (3) | — | |
| 10 | Vaccinia VV M1 | VV | 59 (1) | 54 (5) | 59 (3) | |
| 11 | Vaccinia Elstree | VV | 59 (1) | 58 (3) | 54 (4) | |
| 12 | Vaccinia MVA | VV | 59 (1) | 54 (4) | 54 (4) | |
| pVar-A13L (77 bp) | 65 (0) | |||||
| pCam-A13L (83 bp) | 59 (3) | |||||
| pVar-rpo18 (205 bp) | 63 (0) | |||||
| pCam-rpo18 (205 bp) | 59 (1) | |||||
| pVar-VETF (230 bp) | 63 (0) | |||||
| pCam-VETF (230 bp) | 58 (3) | |||||
ABB, abbreviations used in the figures.
The number of mismatches in the binding region of the hybridization probes is given in parentheses.
—, no Tm determined (>10 mismatches).
PCR protocols.
All PCRs contained 10 pmol of each primer (TIB Molbiol, Berlin, Germany), 3 pmol of each hybridization probe (TIB Molbiol), 2 μl of LightCycler-Fast Start DNA master hybridization probes mix (Roche, Mannheim, Germany), 5 mM Mg2+, and 5 μl of template DNA. Cycling conditions included initial denaturation for 10 min at 95°C, followed by 45 cycles of 10 s at 95°C, 10 s at 55°C, and 10 s at 72°C. Subsequent melting analysis was performed with 30 s at 95°C, 20 s at 38°C, and heating to 85°C with a ramping rate of 0.2°C/s. The same conditions can be used for varicella-zoster virus detection as published previously (10).
Primer and hybridization probe sequences for all orthopoxvirus assays are given in Table 2.
TABLE 2.
Primers and hybridization probes used for orthopoxvirus detection
| Oligonucleotide name | Sequencea | X69198 | Tmb (°C) |
|---|---|---|---|
| rpo OPV F1 | CTgTAgTTATAAACgTTCCgTgTg | S 93663-93686 | 54.9 |
| rpo OPV R1 | TTATCATACgCATTACCATTTCgA | A 93867-93844 | 55.4 |
| rpo Var302 Sen | AgATTAAATCTCCgCATTgAATAgTTAC X | A 93785-93758 | 56.5 |
| rpo Var302 Anc | L-TTTgATTCATCTTCgATgTTTAATgTTCCTCT p | A 93756-93725 | 62.6 |
| VETF OPV F1 | ACCAACTATATTACCTCATCAgTTAgC | S 91577-91603 | 54.1 |
| VETF OPV R1 | TTAAACAAgTTCATAgCTACACCCA | A 91807-91783 | 55.6 |
| VETF Var181 Sen | CgCCTTgATAgCTTCCAgATTTAA X | S 91703-91726 | 58.3 |
| VETF Var181 Anc | L-AAggTTCACATTCTAgTgCCgAACATTAAC p | S 91728-91757 | 63.6 |
| A13L Var F | TgTTTCTggAggAggCAAg | S 114285-114303 | 55.3 |
| A13L OPV F | TgTTTCTgAAggAggCgAA | S 126481-126499c | 55.2 |
| A13L FL | CggACTTggATTTTgTgAgTTCTTgAT X | S 114312-114338 | 62.2 |
| A13L R iLC | TCCTATACgCgATgTATAATAAgA T L CA | A 114361-114335 | 54.6 |
Boldface type indicates variola virus-specific position. Lowercase letters are for typographical clarity. Abbreviations: X, fluorescein; L, LC Red640; p, 3′ phosphate to prevent extension.
Tm was calculated by the nearest-neighbor method.
Sequencing of the PCR products.
PCR products were purified using the QiaQuick PCR purification kit (QIAGEN). Sequencing was performed according to the manufacturer's instructions with the ABI PRISM BigDye terminator cycle sequencing kit (version 3.0; Applied Biosystems, Foster City, Calif.) and analyzed using the DNAStar software package (version 5.0).
RESULTS
In total, four different PCR assays covering three different regions of the orthopoxvirus genome were established.
rpo18 assay.
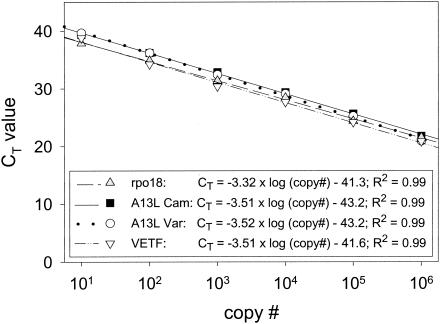
The rpo18 assay was designed as a generic assay to detect the DNA from all orthopoxviruses with subsequent specific identification of variola virus by melting analysis. The variola virus-specific base is the C at position 93772 (referring to GenBank accession no. X69198) that appears as T in all 10 non-variola virus orthopoxviruses deposited in GenBank so far. Amplification of 10-fold serial dilutions of a plasmid with the variola virus-like sequence of rpo18 (pVar-rpo18) revealed a linear detection range from 106 to 10 copies per reaction mixture, with an R2 of 0.99 and a PCR efficiency of 99.5%, indicating a PCR efficiency close to the ideal amplification factor of 2 per PCR cycle (Fig. 1). Similar results were obtained when rpo18 sequences from camelpox virus (pCam-rpo18), as a representative of non-variola virus sequences, were used. However, when amplified products were subjected to melting analysis, variola and camelpox virus sequences were clearly distinguishable, with characteristic melting temperatures (Tms) of 63°C for variola virus and 59°C for camelpox virus. Even mixtures of 10 pCam-rpo18 plasmids and 10 pVar-rpo18 plasmids could be detected and typed correctly (data not shown). When the ratio of pCam-rpo18 to pVar-rpo18 exceeded a factor of 5 to 10, representing 10 to 20% of variola virus DNA in non-variola virus orthopoxvirus DNA, the variola virus-specific peak was reduced in height and no longer clearly detectable. The same effect was visible for contrary proportions.
FIG. 1.
Calibration curves of the four LightCycler assays. PCRs for each individual assay were carried out with serial 10-fold dilutions of the corresponding quantified plasmid. CT values were plotted against initial plasmid copy number (copy #). The correlation of CT value and plasmid copy number is shown in the box.
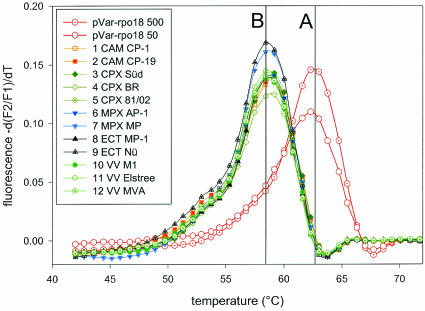
When analyzing DNA from the 12 different orthopoxvirus strains listed in Table 1, all orthopoxviruses exclusively showed the non-variola virus-specific Tm of 59°C (Fig. 2, peak B), whereas the plasmid pVar-rpo18 could be clearly identified by a Tm of 63°C (Fig. 2, peak A), suggesting that the rpo18 assay allows one to clearly distinguish variola virus from all other orthopoxviruses.
FIG. 2.
Melting analysis of assay rpo18 of variola virus. Melting curves of amplified DNA from 12 different orthopoxviruses (as listed in Table 1), covering camelpox, cowpox, monkeypox, ectromelia, and vaccinia virus, compared to 50 and 500 copies of the variola virus-resembling plasmid pVar-rpo18. Only the variola virus-resembling plasmid showed a melting point of 63°C (peak A), attributed to perfectly matching hybridization probes. Due to mismatches in the hybridization probe-binding region, DNA of all other orthopoxviruses show a decreased Tm of 59°C (peak B).
VETF assay.
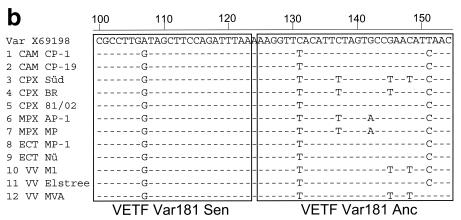
The VETF assay was designed to amplify DNA of all orthopoxviruses, including variola virus. Identification of variola virus was based on two specific nucleotides at position 91710 (A in variola virus, G in nonvariola virus), covered by the FL sensor probe, and in position 91734 (C in variola virus, T in nonvariola virus), covered by the LC anchor probe (accession no. X69198). Assay characteristics as deduced from 10-fold serial dilutions of plasmid pVar-VETF were as shown in Fig. 1, indicating a PCR efficiency of 93%. When comparing the variola virus-resembling plasmid pVar-VETF to the panel of non-variola virus orthopoxvirus DNA samples in melting analyses, the variola virus-resembling plasmid pVar-VETF showed the highest Tm (63°C) due to the perfect match of both the FL sensor probe and the LC anchor probe (Fig. 3a, peak A). All other orthopoxvirus DNA samples displayed lower Tms, and according to the number of mismatches in the probe regions they fell into four separate peaks: B, C, D, and E. The two ectromelia virus strains MP-1 (orthopoxvirus [OPV] 8) and MP-Nü (OPV 9) as well as the two camelpox virus strains CP-1 (OPV 1) and CP-19 (OPV 2) had a characteristic Tm of 58°C (peak B), pointing out mismatches in the binding regions of the two hybridization probes. Sequencing of the respective amplicons proved the presence of three mismatches covered by the hybridization probes. Interestingly, cowpox virus strain 81/02 (OPV 5) and vaccinia virus strain Elstree (OPV 11) also appeared in peak B, and sequencing of the amplicons also revealed three mismatches covered by the hybridization probes. Two other vaccinia virus strains, VVM1 (OPV 10) and MVA (OPV 12), displayed a melting peak at ∼54°C (peak C) and revealed four and five mismatches, respectively, for the sequence stretch covered by the hybridization probes. At 50°C, melting peak D represented monkeypox virus (OPV 6 and 7) and the cowpox virus strain Brighton Red (OPV 4), corresponding to five mismatches. Finally, cowpox virus strain Süd (OPV 3) had a distinct melting peak of 44°C that could be attributed to six mismatches in the hybridization probe-binding region (peak E). A sequence alignment including all the variations confirmed by sequencing is shown in Fig. 3b. Despite the higher number of mismatches, the two monkeypox virus strains (OPV 6 and 7), the cowpox virus strain Brighton Red (OPV 4, peak D), and the cowpox virus strain Süd (OPV 3, peak E) could still be detected in the melting analysis, but due to the reduced bond strength, reflected by Tms of 50 and 44°C, respectively, the amplification could not be visualized during PCR with an annealing temperature of 55°C.
FIG. 3.
Melting analysis of the VETF assay. (a) Melting curves of amplified DNA from 12 different orthopoxviruses (as listed in Table 1), covering camelpox, cowpox, monkeypox, ectromelia, and vaccinia virus, compared to 50 and 500 copies of the variola virus-resembling plasmid pVar-VETF. Besides the characteristic melting peak for the variola virus-resembling plasmid pVar-VETF at 63°C (peak A), all non-variola virus orthopoxviruses fall into the four distinct peaks—peaks B, C, D, and E—as described in the text. (b) Alignment of the VETF sequence corresponding to the two LightCycler hybridization probes VETF Var181 Sen and VETF Var181 Anc. Subsequent to LightCycler PCR, amplicons were sequenced and aligned with the variola virus-specific sequence. Only nucleotide differences are shown. Identical nucleotides are indicated as dots. Sequences for the hybridization probes are boxed.
A13L assay.
The A13L assay was performed either as a non-variola virus orthopoxvirus-specific assay using primer A13L OPV F, which detects various orthopoxviruses except variola virus, or as a variola virus-specific assay applying primer A13L Var F (Table 2). Both assays used the LC Red640 internally labeled reverse primer and the fluorescein-labeled hybridization probe. The fluorescein-labeled probe was designed to match the variola virus sequence, while the other 22 orthopoxvirus A13L sequences found in public databases showed a 6-bp deletion. Again, amplification of serial dilutions of a plasmid containing the variola virus-like A13L sequence revealed a linear correlation (R2) of 0.99 between the threshold cycle (CT) value and copy number, and PCR efficiency was 93%. Nearly identical results were obtained for the non-variola virus orthopoxvirus-specific assay when a camelpox virus-specific plasmid (pCam-A13L) was used as the target (Fig. 1). The A13L CAM assay was unable to amplify plasmid pVar-A13L, but all of the orthopoxviruses tested were amplified, except ectromelia virus, which has a large deletion in this region, preventing amplification with the primers used. In contrast, the A13L VAR assay detected exclusively plasmid pVar-A13L and none of the orthopoxviruses listed above. Melting analysis of the PCR products obtained from amplification of plasmid pVar-A13L with the A13L VAR assay and in parallel from amplification products of 12 orthopoxvirus DNAs with the A13L CAM assay allowed a clear identification of variola virus DNA (Fig. 4). As expected, plasmid pVar-A13L displayed the highest Tm of 65°C (peak A), indicating perfect matching hybridization probes. All other orthopoxviruses showed lower Tms and were divided into two groups, with either a Tm of 59°C (peak B) for camelpox (OPV 1 and 2), cowpox (OPV 3 and 4), and vaccinia VV M1 (OPV 10) virus or a Tm of 54°C (peak C) for monkeypox virus (OPV 6 and 7), vaccinia virus strains Elstree and MVA (OPV 11 and 12), and cowpox virus 81/02 (OPV 5). As expected, ectromelia virus DNA (OPV 8 and 9) showed no signal in amplification or melting analysis (Table 1).
FIG. 4.
Melting analysis of the A13L VAR and A13L CAM assays. Shown are melting curves of amplified DNA from 12 different non-variola virus orthopoxviruses (as listed in Table 1) amplified with assay A13L CAM compared to melting curves of 50 and 500 copies of the artificial variola virus A13L sequence amplified with the A13L VAR assay. Melting curve analysis shows characteristic peaks for variola virus at 65°C (peak A) and two additional peaks (peaks B and C) for the non-variola virus orthopoxviruses as described in the text.
To test the applicability of the described assays, we applied them to clinical samples of a poxvirus outbreak in a colony of New World monkeys in a German private zoo (22). DNA from several monkeys was prepared from bronchoalveolar fluid, serum, liver, spleen, lymph nodes, skin, labium, and whole blood and subjected to PCR. No amplification was obtained with the A13L Var assay, ruling out variola virus. In contrast specific amplification was obtained with the A13L OPV assay. Furthermore, the rpo18 assay and the VETF assay gave positive results for the amplification and with melting curves characteristic for non-variola virus orthopoxvirus (59°C for rpo18 and 44°C for VETF), supporting the argument for the absence of variola virus. At the same time these results confirmed the presence of a non-variola virus orthopoxvirus in these samples. The origin of this orthopoxvirus is currently under investigation.
Specificity and precision of the assays.
None of the presented assays led to amplification products or fluorescence signals with human or mouse genomic DNA (data not shown). The overall variability of the assays as determined by repeated amplification of the respective plasmids was below 30%. Interassay Tm variation was in the range of ±0.5°C but always identical in the same run for identical samples. None of the assays showed cross-reactions to human DNA.
DISCUSSION
Due to the successful eradication of naturally occurring variola virus infections by the global World Health Organization vaccination program in the 1970s (11), molecular detection of variola virus has not been a priority for more than 20 years. Although clinical criteria for the diagnosis of variola virus infections in humans are well described, the identification of zoonotic orthopoxvirus infections can be difficult. As the monkeypox virus infections of humans in the United States and in the Democratic Republic of Congo have shown, symptoms of smallpox and monkeypox are closely related in the early stages of disease (5, 24). In these cases reliable molecular tests were required to identify the infectious virus in time. With the increasing threat of bioterrorist attacks using weaponized anthrax (23), the fear of an attack using variola virus or other orthopoxviruses has risen (14). Should a suspected bioterrorist attack occur, fast and reliable differentiation is of the highest importance, since a diagnosis and risk assessment may have to be provided in the absence of a clear clinical picture. Consequently, there is an urgent need for rapid, safe, and specific detection methods to identify variola virus and to differentiate it from other members of the genus Orthopoxvirus. The evolution from end-point PCR to real-time PCR has provided new dimensions regarding speed and sensitivity of PCR detection (18). Moreover, with this technique an accurate and reproducible quantification is feasible.
In order to improve variola virus diagnostics, we established a set of four real-time PCR assays, based on the amplification of three different genomic regions that can either specifically amplify variola virus DNA or distinguish variola virus from other orthopoxviruses by melting analysis of the PCR product following orthopoxvirus-specific PCR. All PCR assays permit accurate and reproducible quantification of as few as 10 genome equivalents of poxvirus DNA in less than 1 h. However, unequivocal identification of variola virus is more important than quantification in primary diagnostics. In this context, the use of hybridization probes binding to the amplified DNA provides a gain of specificity, making positive results more reliable.
To date, there is one published real-time-based assay for the detection of variola virus (9). Unfortunately, new poxvirus GenBank entries appearing recently contain sequence variation at the previous publication's oligoprobe hybridization sites. This variation occurs not only with variola virus but also with cowpox virus strains and camelpox virus strains. Therefore, it has to be considered that positive results may demonstrate variola virus as well as cowpox virus, and a clear differentiation might not be possible. However, as demonstrated for other infectious agents (8), the targeting of different genomic regions within the same pathogen enormously improves the dependability of positive as well as negative results. In orthopoxviruses sequence variation is frequently found between different isolates. If unexpected sequence variation appears in the primer-binding region, reduced primer binding can lead to false-negative results. Furthermore, it is obvious that the number of mismatches covered by both of the hybridization probes reflects the binding stability of the probes and thus the characteristic Tm for a given pathogen. Unfortunately, the presence of variola virus DNA containing just one unknown mutation in the probe-binding region would lead to a reduced Tm and consequently to a misinterpretation of results, indicating the presence of a non-variola virus orthopoxvirus. Exceeding a critical number of mismatches would finally completely eliminate the binding of the hybridization probe, suggesting the absence of any orthopoxvirus at all. Although the genomic regions chosen are well conserved among the variola virus isolates known to date, this risk can be minimized by the use of more than one independent assay system.
Therefore, we established a set of independent assays. All of the assays can be run under identical reaction conditions, allowing simultaneous processing and reliable analysis in less than 1 h after template preparation. The rpo18 assay melting analysis could distinguish clearly between an artificial variola virus sequence stretch and 12 different non-variola virus orthopoxviruses. The assay design had to be based on only the 14 sequences deposited in GenBank at that time. Practically, the rpo18 gene was found to be highly conserved among all orthopoxviruses we tested.
With the VETF assay, variola virus was clearly distinguishable from all other tested orthopoxviruses. While camelpox, ectromelia, and monkeypox virus strains showed individual melting peaks, vaccinia and cowpox virus split up into several peaks. This result is not surprising, because vaccinia virus and cowpox virus are closely related, exhibit a broad host range, and appear frequently in adjacent clusters in phylogenetic tree analyses. Cowpox virus isolates are usually classified according to the species from which they have been isolated; therefore, a genuine vaccinia virus isolated from a cow might be classified as cowpox virus (7). Our results underline the sequence heterogeneity of vaccinia and of cowpox viruses. The VETF assay was designed based on 14 sequences deposited in GenBank. Since none of the 12 orthopoxviruses investigated showed the variola virus-specific Tm, it can be assumed that the polymorphism used is representative of and conserved among variola virus strains; however, experiments with further orthopoxvirus strains are ongoing to prove the assay's validity.
While rpo18 and the VETF assay were used to identify variola virus by melting analysis, the A13L assay provided the possibility to identify variola virus by specific amplification. This approach is important in cases where mixtures of orthopoxviruses have to be analyzed, because melting analyses are vulnerable to heterogeneous target populations with ratios of >10:1, although the exact ratio is assay specific. A low-abundance variant will often be masked by a highly abundant variant, and false-negative results can be obtained for the rare target. The specific amplification of variola virus DNA is not susceptible to mixtures of orthopoxviruses. It could be shown that amplification occurs exclusively with variola virus sequences, and moreover, the variola virus-specific hybridization probe allowed subsequent melting analysis to verify the results. Even in the case of a potential non-variola virus-specific amplification or due to unknown sequence variation in the primer-binding region of a non-variola virus orthopoxvirus, the amplified DNA could still be characterized by melting analysis and variola virus could be excluded. The established primers and probes for the A13L assay were based on 27 sequences deposited in GenBank. Sequencing of the 12 orthopoxvirus amplicons used in this study confirmed mismatches for all orthopoxviruses in the probe-binding region, reflected by the decreased Tms (Table 1).
In conclusion, although PCR cannot replace cell culture experiments to detect infectious virus particles, the use of more than one diagnostic PCR assay, each directed against a different target distributed across the viral genome, is superior to the use of single assays. As shown for a poxvirus outbreak in New World monkeys in a German zoo (22), the four assays presented here provide an extremely rapid, safe, sensitive, and reliable tool for the detection of orthopoxvirus genomes and the identification or exclusion of variola virus. Each assay can be used separately and leads to reliable results, but, with respect to the consequences deriving from a false-positive or false-negative result in variola virus PCR, we propose using a set of detection systems for distinct regions of the orthopoxvirus genome for the PCR diagnostics of clinical and especially environmental samples. The reaction pattern obtained with different test systems allows us to perform a risk assessment for the presence or absence of variola virus sequences in a suspected sample, also with respect to genome variation within the genus Orthopoxvirus. How many independent assays should be run to confirm or exclude variola virus from a suspected sample is more a political than a scientific question.
Acknowledgments
We are grateful to Sabrina Wendt and Jung-Won Sim-Brandenburg for excellent technical assistance, to Horst Emmel and Siegfried Pociuli for excellent sequencing, and to Ian M. Mackay for critical reading of the manuscript.
REFERENCES
- 1.Ahn, B. Y., E. V. Jones, and B. Moss. 1990. Identification of the vaccinia virus gene encoding an 18-kilodalton subunit of RNA polymerase and demonstration of a 5′ poly(A) leader on its early transcript. J. Virol. 64:3019-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behbehani, A. M. 1983. The smallpox story: life and death of an old disease. Microbiol. Rev. 47:455-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broyles, S. S., and B. S. Fesler. 1990. Vaccinia virus gene encoding a component of the viral early transcription factor. J. Virol. 64:1523-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broyles, S. S., and J. Li. 1993. The small subunit of the vaccinia virus early transcription factor contacts the transcription promoter DNA. J. Virol. 67:5677-5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2003. Multistate outbreak of monkeypox—Illinois, Indiana, and Wisconsin, 2003. Morb. Mortal. Wkly. Rep. 52:537-540. [PubMed] [Google Scholar]
- 6.Czerny, C. P., A. M. Eis-Hubinger, A. Mayr, K. E. Schneweis, and B. Pfeiff. 1991. Animal poxviruses transmitted from cat to man: current event with lethal end. Zentbl. Veterinarmed. B 38:421-431. [DOI] [PubMed] [Google Scholar]
- 7.Damaso, C. R., J. J. Esposito, R. C. Condit, and N. Moussatche. 2000. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology 277:439-449. [DOI] [PubMed] [Google Scholar]
- 8.Ellerbrok, H., H. Nattermann, M. Ozel, L. Beutin, B. Appel, and G. Pauli. 2002. Rapid and sensitive identification of pathogenic and apathogenic Bacillus anthracis by real-time PCR. FEMS Microbiol. Lett. 214:51-59. [DOI] [PubMed] [Google Scholar]
- 9.Espy, M. J., F. R. Cockerill III, R. F. Meyer, M. D. Bowen, G. A. Poland, T. L. Hadfield, and T. F. Smith. 2002. Detection of smallpox virus DNA by LightCycler PCR. J. Clin. Microbiol. 40:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espy, M. J., R. Teo, T. K. Ross, K. A. Svien, A. D. Wold, J. R. Uhl, and T. F. Smith. 2000. Diagnosis of varicella-zoster virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:3187-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenner, F. 1977. The eradication of smallpox. Prog. Med. Virol. 23:1-21. [PubMed] [Google Scholar]
- 12.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-263. [DOI] [PubMed] [Google Scholar]
- 13.Hazelton, P. R., and H. R. Gelderblom. 2003. Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg. Infect. Dis. 9:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, D. A., T. V. Inglesby, J. G. Bartlett, M. S. Ascher, E. Eitzen, P. B. Jahrling, J. Hauer, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. Perl, P. K. Russell, K. Tonat, et al. 1999. Smallpox as a biological weapon: medical and public health management. JAMA 281:2127-2137. [DOI] [PubMed] [Google Scholar]
- 15.Lewis-Jones, S. 2002. The zoonotic poxviruses. Dermatol. Nurs. 14:79-82, 85-86. [PubMed] [Google Scholar]
- 16.Li, J., and S. S. Broyles. 1993. Recruitment of vaccinia virus RNA polymerase to an early gene promoter by the viral early transcription factor. J. Biol. Chem. 268:2773-2780. [PubMed] [Google Scholar]
- 17.Loparev, V. N., R. F. Massung, J. J. Esposito, and H. Meyer. 2001. Detection and differentiation of old world orthopoxviruses: restriction fragment length polymorphism of the crmB gene region. J. Clin. Microbiol. 39:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massung, R. F., J. J. Esposito, L. I. Liu, J. Qi, T. R. Utterback, J. C. Knight, L. Aubin, T. E. Yuran, J. M. Parsons, and V. N. Loparev. 1993. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature 366:748-751. [DOI] [PubMed] [Google Scholar]
- 20.Massung, R. F., L. I. Liu, J. Qi, J. C. Knight, T. E. Yuran, A. R. Kerlavage, J. M. Parsons, J. C. Venter, and J. J. Esposito. 1994. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology 201:215-240. [DOI] [PubMed] [Google Scholar]
- 21.Massung, R. F., V. N. Loparev, J. C. Knight, A. V. Totmenin, V. E. Chizhikov, J. M. Parsons, P. F. Safronov, V. V. Gutorov, S. N. Shchelkunov, and J. J. Esposito. 1996. Terminal region sequence variations in variola virus DNA. Virology 221:291-300. [DOI] [PubMed] [Google Scholar]
- 22.Mätz-Rensing, K., H. Ellerbrok, B. Ehlers, G. Pauli, A. Floto, M. Alex, C. P. Czerny, and F. J. Kaup. 2003. Fatal pox virus outbreak in a colony of New World monkeys. Verh. ber. Erkrg. Zootiere 41:135-139. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy, M. 2001. Anthrax in USA—attacks “deadly but treatable.” Lancet 358:1520. [DOI] [PubMed] [Google Scholar]
- 24.Meyer, H., M. Perrichot, M. Stemmler, P. Emmerich, H. Schmitz, F. Varaine, R. Shungu, F. Tshioko, and P. Formenty. 2002. Outbreaks of disease suspected of being due to human monkeypox virus infection in the Democratic Republic of Congo in 2001. J. Clin. Microbiol. 40:2919-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, H. Ellerbrok, G. Pauli, and W. Siegert. 2000. Detection of human cytomegalovirus DNA by real-time quantitative PCR. J. Clin. Microbiol. 38:2734-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, and W. Siegert. 1999. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin. Chem. 45:1932-1937. [PubMed] [Google Scholar]
- 27.Pulford, D. J., H. Meyer, and D. Ulaeto. 2002. Orthologs of the vaccinia A13L and A36R virion membrane protein genes display diversity in species of the genus Orthopoxvirus. Arch. Virol. 147:995-1015. [DOI] [PubMed] [Google Scholar]
- 28.Stephenson, J. 2003. Monkeypox outbreak a reminder of emerging infections vulnerabilities. JAMA 290:23-24. [DOI] [PubMed] [Google Scholar]
- 29.Whitley, R. J. 2003. Smallpox: a potential agent of bioterrorism. Antivir. Res. 57:7-12. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. 1980. Declaration of global eradication of smallpox. Wkly. Epidemiol. Rec. 55:145-152. [Google Scholar]