Abstract
In the present study we investigated whether single nucleotide polymorphisms (SNPs) in the P2RX4, which alter the P2X4R function, are associated with the development of osteoporosis and whether an interaction between the P2X4R and P2X7R confer a synergistic effect of these two receptors on osteoporosis risk. Patients with fracture (690 females and 231 males, aged ≥50 years) were genotyped for three non-synonymous P2X4R SNPs. Bone mineral density (BMD) was measured at the total hip, lumbar spine, and femoral neck. Subject carrying the variant allele of the Tyr315Cys polymorphism showed a 2.68-fold (95 % CI, 1.20–6.02) higher risk of osteoporosis compared with wild-type subject. Furthermore, significant lower lumbar spine BMD values were observed in subjects carrying the Cys315 allele as compared with wild-type (0.85 ± 0.17 and 0.93 ± 0.17 g/cm2, respectively; p < 0.001). Assuming a recessive model, carriers of the variant allele of the Ser242Gly polymorphism showed increased BMD values at the lumbar spine compare to wild-type subject (1.11 ± 0.35 and 0.92 ± 0.17 g/cm2, respectively; p = 0.0045). This is the first study demonstrating an association of non-synonymous polymorphisms in the P2RX4 and the risk of osteoporosis, suggesting a role of the P2X4R in the regulation of bone mass.
Electronic supplementary material
The online version of this article (doi:10.1007/s11302-012-9337-0) contains supplementary material, which is available to authorized users.
Keywords: P2X4 receptor, Osteoporosis, Bone mineral density, Polymorphisms
Introduction
Bone remodeling is the process that maintains the bone quality during life. Mechanical loading is one of the factors that controls the local bone remodeling process via regulating bone formation by osteoblasts and bone resorption by osteoclasts [1]. As a result of mechanical loading, ATP is released from mechanically stimulated cells [2–4]. ATP is known as one of the main mediators of the response of bone cells to mechanical loading. ATP acts on bone cells through its binding to specific P2 purinergic receptors. The P2 receptor family includes seven members of P2X receptors (P2X1–7), and eight members of P2Y receptors (P2Y1,2,4,6,11,12,13,14) [5]. Several P2X and P2Y receptors have been shown to be expressed in bone cells (reviewed in [6]), including the P2X4 receptor (P2X4R) and P2X7 receptor (P2X7R).
Expression of the P2X4R on osteoblasts was first described by Nakamura et al. [7], who demonstrated the presence of mRNA coding for the P2X4R in human osteoblast-like MG-63 cells. This result was confirmed in human osteoblastic SaM-1 cells and in cells from human osteosarcoma HOS, MG63, and SaOs cell lines [8–10], as well as from a rat osteoblast primary cell line Orriss (abstract) [27]. In osteoclasts, mRNA transcripts for P2X4 were first identified in rabbit osteoclasts purified by micromanipulation [8]. Expression of the P2X4R was later confirmed in isolated rat osteoclasts and human osteoclastic cells [9, 10].
The fact that P2X4Rs are expressed in osteoblasts and osteoclasts would suggest a potential role in bone physiology for this receptor subtype. Binding of ATP to the P2X4R causes a conformational change of this receptor, which results in channel formation and membrane depolarization. This leads to an increase of intracellular [Ca2+] both by direct Ca2+ ion influx and by an inward Na+ current that depolarizes the cell and opens voltage-operated Ca2+ channels [11–13].
To date only a few studies have investigated the possible role of the P2X4R in bone physiology. Naemsch and colleagues [8] reported that the activation of rabbit osteoclasts by ATP resulted in a rapid inward current which showed several characteristics consistent with those of P2X4-mediated currents. The authors suggested that nucleotides released as a result of mechanical stimulation may act through binding to P2X4Rs to enhance the activity of osteoclasts and, consequently, the resorption of bone [8]. Possible involvement of the P2X4R in bone resorption was also shown by the group of Hoebertz et al. [10]. More recently further evidence for a role of the P2X4R in the activation of osteoclasts was reported by Binderman et al. [14], who showed that the expression of P2X4R in marginal gingival cells was significantly up-regulated after surgical separation of the marginal gingiva in an in vivo experiment. Moreover, these studies showed that a single local application of apyrase (an enzyme that degrades ATP) during surgery reduced alveolar bone loss, suggesting that the P2X4R was directly involved in activation of osteoclasts. In addition to its apparent effects on osteoclast function, a functional effect of the P2X4R on osteoblast-like cells has been suggested by Liu and Chen [15]. They showed that both the P2X7R and P2X4R may be involved in an ATP-induced cell proliferation in human osteosarcoma HOS cells.
Given the above observations, single nucleotide polymorphisms (SNPs) in the P2X4 receptor gene (P2RX4) that have putative effects on the function of this receptor subtype may affect bone mass and quality in humans. Therefore, the P2RX4 may be a good candidate gene to predict osteoporosis risk as well as the associated fracture risk.
The P2RX4 is located on the long arm of chromosome 12 (12q24.32), spans over 50 kbp with 12 exons. To date, four non-synonymous SNPs have been found in the P2RX4. Stokes et al. [16] showed a loss of receptor function for one of these non-synonymous SNPs (Tyr315Cys), but no major effect on receptor function could be shown for the other three non-synonymous SNPs (Ala6Ser, Ile119Val, and Ser242Gly). Because the P2RX4 lies only 23 kbp away from P2RX7 to which it is closely related, these separate receptor subtypes may have evolved by a process of gene duplication [17]. The P2RX7 is highly polymorphic with at least eight non-synonymous SNPs leading to a functional effect on the receptor. Several of these non-synonymous P2X7 SNPs were shown to be significantly associated with bone mineral density (BMD) [18–21]. It is conceivable that the significant associations found between SNPs within the P2RX7 gene and BMD arise through a genetically linked effect of the P2X7R and P2X4R on bone remodeling processes. Therefore, it is possible that an interaction of SNPs at the P2RX7/P2RX4 locus exerts a maximal effect on BMD and thus leads to high susceptibility for osteoporosis.
We here report on the association between P2X4R SNPs and osteoporosis in a Dutch cohort of fracture patients. Furthermore, we investigated whether an interaction between SNPs in the P2X4R and P2X7R confers the maximal effect on osteoporosis risk. We hypothesized that non-synonymous SNPs within the P2RX4 causing a functional defect of the P2X4R would be related with the risk of osteoporosis and that an interaction between SNPs at the P2X7/P2X4 locus would have a synergistic effect in affecting BMD. To our knowledge this is the first study focusing on the association between SNPs within the P2RX4 and osteoporosis.
Methods
Study population and design
The study population consisted of men and women aged ≥50 years, recruited at the osteoporosis outpatient clinic at the Maastricht University Medical Centre (MUMC+), The Netherlands, from patients receiving regular medical follow-up for a recent fracture. The regular medical follow-up procedure for fracture patients as well as the recruitment of the patients for the present study has been fully described previously [22].
Patients were divided in either cases [i.e., fracture patients suffering from osteoporosis (defined by T-score ≤ −2.5)] or controls [i.e., non-osteoporotic fracture patients (T-score > −2.5)]. The study was approved by the ethical committee of the University Hospital Maastricht and Maastricht University.
Bone density measurements
As part of the standard medical follow-up of fracture patients, bone mineral density (in grams/square centimeters) of the lumbar spine (L2–L4), femoral neck, and total hip (trochanter and neck) was assessed by dual X-ray absorptiometry, using the cross-calibrated Hologic QDR 4500 Elite densitometer (Waltham, MA, USA).
DNA extraction
Blood samples
DNA was extracted from blood using the Maxwell DNA purification system. EDTA anticoagulated blood (400 μL) was used, and the isolation procedure was performed according to the manufacturer’s instructions.
Saliva samples
A plain cotton swab collection device (SalivetteTM, Sarstedt AG & Co., Numbrecht, Germany) was used to collect small amount of saliva for DNA extraction. Upon return the SalivetteTM containing the saliva swab was stored in a refrigerator at 4 °C until DNA extraction. First, the swab which was kept in the collection tube was centrifuged at 4,000 rpm for 10 min, and the saliva was transferred to a 15-mL Nunc tube which was kept at 5 °C overnight. Using a pair of sterile tweezers, the swab was then transferred from the collection tube to a 50-mL Nunc tube; 4 mL sterile water was added, and the tube was kept at room temperature overnight. The next day the swab plus water was transferred back into the collection tube and again centrifuged at 4,000 rpm for 10 min; the saliva yield was again transferred to the 15-mL Nunc tube already containing the saliva yield from the day before. Next, cells were isolated from the saliva by centrifuging the saliva-containing 15 mL Nunc tube at 4,000 rpm for 10 min. Subsequently, the supernatant was carefully removed, leaving a cell pellet of 600–800 μl cells over the pellet. DNA extraction was carried out using Maxwell 16 DNA Purification Kits on the Maxwell 16 Instrument (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Genotyping
Genotyping was done by Sequenom (Sequenom, Hamburg, Germany) using the Sequenom MassARRAY® iPLEX Gold assay. The mass difference between SNP alleles was analyzed using matrix-assisted laser desorption ionization time-of-flight mass spectrometry.
Internal validation study
To assess the accuracy of the genotyping assay, an internal validation study was performed in which a randomly selected number of samples (N = 45) was genotyped for the second time, using restriction enzyme digestion of appropriate PCR products or Taqman assay. This was done according to a previously published protocol Hansen et al. [28]. When the results were compared with the original genotyping, we observed a discrepancy of ~4.2 % between the two different genotyping methods. The discrepancy appeared to be smaller (~2.7 %) if the original genotyping with the Sequenom MassARRAY® iPLEX Gold assay had failed for the maximum one SNP. Therefore, all subjects who had failed more than two SNPs in original genotyping were excluded from the statistical analysis.
Statistical analysis
Descriptive statistics were used to determine the prevalence of osteoporosis in the cohort of fracture patients, as well as to assess distributions of possible risk factors, including sex, age (in years), and body mass index (BMI, in kg/cm2), and to describe the occurrence of different fracture types. These analyses were performed using SAS, version 9.1.
Deviation of genotype frequencies from those expected under Hardy–Weinberg equilibrium was tested in the non-osteoporotic control subjects by the χ2 test. The software package PLINK [23] was used to test for association between genetic variations and BMD and osteoporosis. All analyses were adjusted for age, sex, and BMI. Furthermore, we performed analyses stratified by sex. Both single SNPs and haplotypes were tested for association and different models of inheritance—additive, recessive, and dominant—were used.
Haplotype construction and linkage disequilibrium analysis
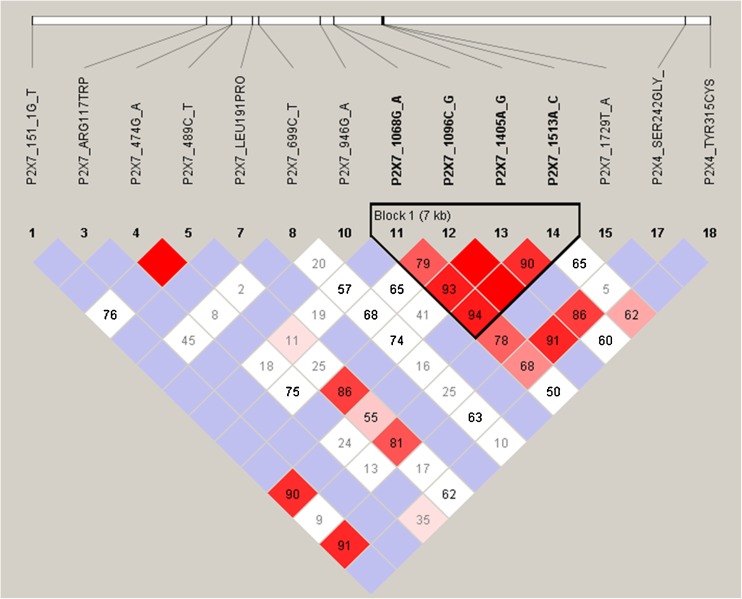
To test whether the P2X4 SNPs had an independent effect on osteoporosis risk and whether an interaction between P2X4 and P2X7 SNPs conferred a synergistic effect of these two receptors on osteoporosis risk, haplotypes were constructed by means of Haploview v4.1, using 17 marker SNPs that change receptor function (i.e., two P2X4 and 15 previously genotypes P2X7 non-synonymous SNPs [19]. The nonrandom distributions of the haplotype were assessed by calculating pairwise linkage disequilibrium (LD) coefficient, D′. The value of D′ = 1 indicates complete dependency, i.e., no evidence for recombination between the SNP pairs; while D′ = 0 indicates no LD, i.e., linkage equilibrium. Strong LD was defined as having pairwise D′ > 0.85. Patterns of LD are visualized using Haploview v4.1. Haplotype block structure was examined if the LD color scheme indicated bright red or shade of pink/red.
Results
Study population
Results of the recruitment procedures have been published previously [19]. Briefly, we collected blood from a total of 381 fracture patients and saliva from a total of 1,064 patients with recent fracture. In blood samples genotyping failed in five (1.3 %) patients. In saliva samples DNA extraction failed in 27 (2.5 %) saliva samples and genotyping in 492 (46.2 %) samples. In total 921 samples were successfully genotyped and used for subsequent analyses.
We identified 283 (31 %) cases, of which 61 (22 %) are men and 222 (78 %) are women, and 638(69 %) controls, of which 170 are men (27 %) and 468 (73 %) are women. Characteristics of the case and control groups are shown in Table 1.
Table 1.
Characteristics of the study population
| Characteristics | Total (N = 921) | Casesa (N = 283) | Controls (N = 638) |
|---|---|---|---|
| Age (years) | 65.0 (9.8) | 67.3 (9.6) | 64.0 (9.7) |
| Weight (kg) | 72.5 (13.8) | 67.7 (12.8) | 74.6 (13.7) |
| Height (cm) | 165.8 (9.1) | 163.8 (9.3) | 166.7 (8.8) |
| BMI (kg/m2) | 26.3 (4.2) | 25.2 (3.9) | 26.8 (4.3) |
| Femoral neck BMD (g/cm2) | 0.69 (0.13) | 0.60 (0.10) | 0.72 (0.12) |
| Total hip BMD (g/cm2) | 0.84 (0.15) | 0.72 (0.13) | 0.88 (0.14) |
| Lumbar spine BMD (g/cm2) | 0.93 (0.17) | 0.77 (0.12) | 0.99 (0.14) |
BMD bone mineral density
aCases of osteoporosis as defined by BMD T-score values of T ≤ −2.5
P2X4 polymorphisms and linkage disequilibrium
Subjects were genotyped for three non-synonymous SNPs within the P2RX4 (Fig. 1). We found no subjects carrying the minor allele of the Ile119Val (rs28360470) polymorphism. The Ser242Gly (rs25644) polymorphism was found to have a minor allele frequency of 0.105 (n = 182), and the loss-of-function Tyr315Cys (rs28360472) polymorphism was found to have a minor allele frequency of 0.013 (n = 24) (Table 2). The genotype distributions for the polymorphisms Ser242Gly and Tyr315Cys polymorphisms were in Hardy–Weinberg equilibrium (Table 2). Pairwise LD calculations showed that the Tyr315Cys polymorphism was in weak LD with loss-of-function polymorphisms Gly150Arg (P2X7_474G_A) (D′ = 0.35) and the Glu496Ala (P2X7_1513A_C) (D′ = 0.62) within the P2RX7 (Fig. 2). The Ser242Gly polymorphism was found to be in LD with a haplotype block within the P2RX7, containing four functional SNPs, including two loss-of-function SNPs, Thr357Ser (P2X7_1096C_G),and Glu496Ala (P2X7_1513A_C); one gain-of-function SNP, Ala348Thr (P2X7_1068G_A); and one marker gain-of-function SNP (i.e., the clinical effect of this polymorphism is caused by another polymorphism in linkage disequilibrium with this polymorphism), Gln460Arg (P2X7_1405A_G).
Fig. 1.
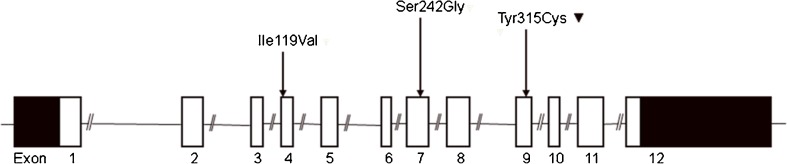
non-synonymous SNPs in the P2X4 receptor gene which were assessed in the study population. Inverted triangle indicates polymorphisms with reduced receptor function
Table 2.
HWE in the study population and genotype distribution in cases (i.e., patients suffering from osteoporosis as defined by BMD T-score ≤ −2.5) and controls (i.e., patients having a BMD T-score > 2.5) for each single P2X4 SNP
| Cases | Controls | MAF | HWE p value | |
|---|---|---|---|---|
| Ser242Gly (rs25644) | – | – | 0.105 | 1 |
| AA (%) | 224 (80) | 500 (80) | – | – |
| AG (%) | 53 (19) | 119 (19) | – | – |
| GG (%) | 3 (1) | 1 | – | – |
| χ2 | OR = 0.99 (95 % CI, 0.7165–1.37), χ2 = 0.003, p = 0.956 | – | – | |
| Tyr315Cys (rs28360472) | – | – | 0.013 | 1 |
| AA (%) | 270 (95) | 622 (98) | – | – |
| AG (%) | 13 (5) | 11 (2) | – | – |
| GG | – | – | – | – |
| χ2 | OR = 2.68 (95 % CI, 1.194–6.024), χ2 = 6.169, p = 0.013 | – | – |
OR odds ratio; CI confidence interval; MAF minor allele frequency; HWE Hardy–Weinberg equilibrium
Fig. 2.
Haploview analysis of pairwise linkage disequilibrium between P2X4 and P2X7 SNPs using 14 marker SNPs that change receptor function. Plot of relative D′/LOD scores between the P2X4 and P2X7 SNPs from Dutch Caucasian subjects produced by the Haploview program. Standard color scheme is displayed as follows: bright red (D′ = 1; LOD ≥ 2), blue (D′ = 1; LOD < 2), shade of pink/red (D′ < 1; LOD ≤ 2), and white (D′ < 1; LOD < 2). Numbers represent D′ scores for pairwise linkage disequilibrium
Associations of P2X4 polymorphisms with osteoporosis
Table 2 shows the genotype distribution in cases and controls. No difference in genotype frequency between cases and controls was found for the Ser242Gly polymorphism. The frequency of carrying the G allele of the Tyr315Cys polymorphism was significantly higher among cases (5 %) compared to controls (2 %) (p = 0.013). The additive risk model showed a 2.68-fold higher odds ratio (95 % CI, 1.20–6.02) on the risk of osteoporosis for carriers of the 315Cys mutation.
Association between P2X4 polymorphisms and BMD
The Tyr315Cys polymorphism, which showed an association with osteoporosis, was also associated with lumbar spine BMD (Table 3). Subjects carrying the Cys315 allele had lower BMD at the lumbar spine 0.85 ± 0.17 g/cm2 (mean ± SD) as compared with wild-type subjects (0.93 ± 0.17 g/cm2) (p < 0.001) (Table 3). Similar results were found in the sex-stratified analyses. Both men and women harboring the Cys315 SNP showed significantly decreased lumbar spine BMD values compared to wild-type subjects (men, 0.96 ± 0.17 g/cm2 versus 0.98 ± 0.17 g/cm2, p = 0.015; women, 0.82 ± 0.16 g/cm2 versus 0.91 ± 0.17 g/cm2, p = 0.012).
Table 3.
BMD values for the individual genotypes for each single SNP
| SNP | WT | HET | HOMO | p valuea | p valueb | p valuec |
|---|---|---|---|---|---|---|
| Tyr315Cys | AA | AG | GG | – | – | – |
| N | 885 | 24 | – | – | – | – |
| BMD TH (g/cm2) | 0.84 (0.15) | 0.79 (0.17) | – | – | – | 0.5467 |
| BMD LS (g/cm2) | 0.93 (0.17) | 0.85 (0.17) | – | – | – | <0.001 |
| BMD FN (g/cm2) | 0.69 (0.13) | 0.66 (0.15) | – | – | – | 0.9356 |
| Female | AA | AG | GG | – | – | – |
| N | 667 | 19 | – | – | – | – |
| BMD TH (g/cm2) | 0.80 (0.13) | 0.76 (0.17) | – | – | – | 0.8819 |
| BMD LS (g/cm2) | 0.91 (0.17) | 0.82 (0.16) | – | – | – | 0.0124 |
| – | BMD FN (g/cm2) | 0.63 (0.16) | – | – | – | 0.6703 |
| Male | AA | AG | GG | – | – | – |
| N | 225 | 5 | – | – | – | – |
| BMD TH (g/cm2) | 0.95 (0.15) | 0.92 (0.09) | – | – | – | 0.9546 |
| BMD LS (g/cm2) | 0.98 (0.17) | 0.96 (0.17) | – | – | – | 0.0145 |
| BMD FN (g/cm2) | 0.76 (0.13) | 0.73 (0.06) | – | – | – | 0.9404 |
| Ser242Gly | AA | AG | GG | – | – | – |
| N | 718 | 169 | 10 | – | – | – |
| BMD TH (g/cm2) | 0.84 (0.15) | 0.82 (0.15) | 0.85 (0.17) | 0.8075 | 0.5728 | 0.6127 |
| BMD LS (g/cm2) | 0.92 (0.17) | 0.92 (0.15) | 1.11 (0.35) | 0.3448 | 0.0045 | 0.4189 |
| BMD FN (g/cm2) | 0.69 (0.13) | 0.68 (0.13 | 0.70 (0.11) | 0.7087 | 0.8949 | 0.6131 |
| Female | AA | AG | GG | – | – | – |
| N | 549 | 125 | 6 | – | – | – |
| BMD TH (g/cm2) | 0.80 (0.13) | 0.79 (0.13) | 0.78 (0.17) | 0.6206 | 0.5859 | 0.6883 |
| BMD LS (g/cm2) | 0.91 (0.16) | 0.91 (0.15) | 1.09 (0.36) | 0.5507 | 0.0202 | 0.9283 |
| BMD FN (g/cm2) | 0.67 (0.12) | 0.65 (0.11) | 0.65 (0.10) | 0.5907 | 0.8804 | 0.5889 |
| Male | AA | AG | GG | – | – | – |
| N | 175 | 47 | 4 | – | – | – |
| BMD TH (g/cm2) | 0.95 (0.15) | 0.93 (0.17) | 0.97 (0.12) | 0.8178 | 0.8931 | 0.8280 |
| BMD LS (g/cm2) | 0.98 (0.16) | 0.98 (0.16) | 1.14 (0.38) | 0.6539 | 0.0510 | 0.9131 |
| BMD FN (g/cm2) | 0.76 (0.13) | 0.74 (0.14) | 0.77 (0.10) | 0.8568 | 0.9493 | 0.8231 |
p values are shown for PLINK association analysis for the bone mineral density (BMD) parameters adjusted for age, BMI and sex. Numbers are mean (SD)
WT wild-type; HET heterozygote; HOMO homozygote; TH total hip; LS lumbar spine; FN femoral neck
aAdjusted for age, BMI, and sex: additive model
bAdjusted for age, BMI, and sex: recessive model
cAdjusted for age, BMI, and sex: dominant model
.
For the Ser242Gly polymorphism, we observed increased BMD values at all sites for homozygous subjects. Using a recessive model, we found a significant increase in BMD values at the lumbar spine in subjects carrying the Gly242 mutation (1.11 ± 0.35 g/cm2 versus 0.92 ± 0.17 g/cm2, p = 0.005). Again, similar results were shown in gender-stratified analyses (Table 3).
Associations of haplotypes with osteoporosis
Although LD was shown between the P2X7 Glu496Ala (rs3751143) polymorphism and the Tyr315Cys polymorphism (Fig. 2), it was not included in the haplotype analysis because it did not affect the BMD values. Furthermore, by including the Glu496Ala, the statistical power would be reduced since it would subdivide haplotype H11 in two haplotypes.
Subjects who carries either of Gly150Arg (rs28360447) and Tyr315Cys SNPs [i.e., H12, H13, and H14 (H12, Arg150-Tyr315; H13, Gly150-Cys315; H14, Arg150-Cys315)] showed a significantly increased risk of osteoporosis compared to wild-types [OR = 2.27 (95 % CI 1.23–4.17), p = 0.008].
Haplotypes H12, H13, and H14 showed an increased risk of osteoporosis when compared to the other haplotypes [H12 versus H11, H13, and H14, OR = 2.08 (95 % CI, 0.89–4.85); H13 versus H11 and H12, H14, OR = 2.44 (95 % CI, 0.84–7.13); H14 versus H11, H12, and H13, OR = 2.60 (95 % CI, 0.64–10.50)] (Table 4). However these increased risks did not reach statistical significance (p = 0.009, p = 0.103, and p = 0.180, respectively) (Table 4).
Table 4.
Haplotype frequency in the study population in both affected and unaffected subjects
| P2X7, Gly150Arg | P2X4, Tyr315Cys | Population frequency | Affected frequency | Unaffected frequency | OR | 95 % CI | p-Value | Number | |
|---|---|---|---|---|---|---|---|---|---|
| H11a | G | A | 97.4 | 95.5 | 98.2 | 0.44 | (0.24–0.81) | 0.008 | 867 |
| H12b | A | A | 1.3 | 2.1 | 0.88 | 2.08 | (0.89–4.85) | 0.090 | 22 |
| H13c | G | G | 0.8 | 1.4 | 0.56 | 2.44 | (0.84–7.13) | 0.103 | 15 |
| H14d | A | G | 0.5 | 0.88 | 0.31 | 2.60 | (0.64–10.50) | 0.18 | 9 |
Furthermore, the table shows the osteoporosis risk for each haplotype compared to all other haplotypes and their osteoporosis risk
OR odds ratio; CI confidence interval
aWild-type
bH12: Arg150-Tyr315
cH13: Gly150-Cys315
dH14: Arg150-Cys315
Associations of haplotypes with BMD
Haplotypes containing at least one mutation of the Gly150Arg and Tyr315Cys polymorphisms (i.e., H12–H14) showed significantly decreased lumbar spine BMD values when compared to wild-types [i.e., haplotype H11) (0.87 ± 0.16 versus 0.93 ± 0.17, respectively (p = <0.001)] (data not shown).
Discussion
This is the first study investigating the association between non-synonymous SNPs in the P2RX4 with osteoporosis. Results showed that the Tyr315Cys polymorphism was significantly associated with decreased BMD values and increased risk of osteoporosis. Furthermore, the Ser242Gly polymorphism showed an association with increased lumbar spine BMD values. These results provide supportive evidence to our hypothesis that the P2X4R, together with the P2X7R, plays a role in bone physiology, as it has been shown that both receptors involve in activation of both osteoclasts and osteoblasts [8, 10, 14]. However, the overall effect of the P2X4R and P2X7R in bone metabolism is unclear.
The allele frequencies for the Tyr315Cys and Ser242Gly polymorphism in our population were almost identical to the previously published data Stokes et al. [26]. Both SNPs were shown to be in HWE, indicating a valid population for current study. However, the minor allele of Ile119Val SNP was not found in this population.
Previous in vitro studies showed two functional effects for the Tyr315Cys SNP. First, the Cys315 mutation showed a reduction in agonist (i.e., ATP) potency, suggesting that the Tyr315 locus may be involved in ATP binding [16, 24, 25]. Second, it reduced the maximum response of the P2X4R caused by disruption in ion channel function [16]. Therefore, the Tyr315Cys polymorphism is considered to be a loss-of-function polymorphism of the P2X4R. Combining these in vitro results with the results found in the present study, which shows decreased lumbar spine BMD values and increased risk of osteoporosis in subjects harboring the minor allele of the Tyr315Cys SNP, the overall effect of the P2X4R in bone appears to be mainly pro-osteogenic.
As shown by LD calculations, the Tyr315Cys mutation is in weak LD with the functional Gly150Arg SNP in the P2RX7. We previously showed that this P2X7 SNP is associated with decreased BMD values (Supplementary Table 2) [19]. Furthermore, the frequency of carrying the G allele of the Gly150Arg polymorphism was significantly higher among cases (5 %) compared to controls (2 %) (p = 0.021) (Supplementary Table 3). It could therefore be possible that the association of the Tyr315Cys polymorphism with BMD and osteoporosis found in the present study is in fact due to functional effects of the Gly150Arg polymorphism in the P2RX7. However, haplotype analyses showed that both haplotype H12, containing Arg150-Tyr315, and H13, containing Gly150-Cys315, were associated with decreased lumbar spine BMD values and increased risk of osteoporosis, suggesting independent effects of both the Tyr315Cys and Gln150Arg polymorphisms. An independent effect was confirmed by the similar values for D′ (D′ = 0.35) found in cases and controls (data not shown). No conclusions could be drawn whether these SNPs confer a synergistic effect on BMD values and on the risk of osteoporosis, since the numbers of subjects harboring both mutated alleles was too low (N = 9). Replication in larger cohort studies is therefore warranted.
Although in vitro studies showed no functional effect of the Ser242Gly polymorphism [16], we found significantly increased lumbar spine BMD values in homozygous subjects. Haploview showed strong LD for this SNP with a haplotype block containing four functional SNPs within the P2RX7, of which two (Ala348Thr and Gln460Ala) are known to be associated with increased BMD values [19–21]. However, the LD between the SNPs was shown to be caused by the co-inheritance of the Gly242 minor allele with the major alleles (i.e., wild-type) of Ala348Thr and Gln460Ala polymorphisms (Supplementary Table 1). Therefore, the protective effect on osteoporosis found for the Ser242Gly polymorphism in the present study is unlikely due to the genetic linkage with either the Ala348Thr or Gln460Ala gain-of-function SNP. However, it remains possible that other yet-to-be-identified functional polymorphisms in LD with the Ser242Gly SNP are responsible for the effect of this SNP on BMD found in the present study.
The present study has several limitations. First, the low number of subjects may have influenced the precision of the results, especially when performing stratified and haplotype analyses. Replication in large cohort studies is therefore necessary. Another limitation that could have influenced the precision of the results is the lack of access to the risk factors influencing the development of osteoporosis. However, genetic association studies are not likely to be confounded by behavioral and environmental factors, as these factors are very unlikely to show an association with the genotype (i.e., a necessary condition for confounding).
In conclusion this is the first study demonstrating an association of non-synonymous polymorphisms in the P2RX4 and the risk of osteoporosis, suggesting a role of the P2X4R in the regulation of bone mass. However, to elucidate the exact role of P2X4R SNPs in bone physiology and to evaluate the effect of the interaction between P2RX4 and P2RX7 on the risk of osteoporosis, future study in larger cohorts is necessary.
Electronic supplementary material
(DOC 32 kb)
(DOC 39 kb)
(DOC 29 kb)
Acknowledgments
The work was supported by the European Commission under the 7th Framework Programme, performed as a collaborative project “Fighting Osteoporosis by blocking nucleotides: purinergic signaling in bone formation and homeostasis” (ATPBone), with participants; Copenhagen University Hospital, University College London, Maastricht University, University of Ferrara, University of Liverpool, University of Sheffield, and Université Libre de Bruxelles.
Contributor Information
Anke Wesselius, Phone: +31-43-3882876, FAX: +31-43-3884128, Email: anke.wesselius@maastrichtuniversity.nl.
Lodewijk van Rhijn, Email: i.punt@mumc.nl.
References
- 1.Allori AC, Sailon AM, Pan JH, Warren SM. Biological basis of bone formation, remodeling, and repair-part III: biomechanical forces. Tissue Eng Part B Rev. 2008;14(3):285–293. doi: 10.1089/ten.teb.2008.0084. [DOI] [PubMed] [Google Scholar]
- 2.Genetos DC, Kephart CJ, Zhang Y, Yellowley CE, Donahue HJ. Oscillating fluid flow activation of gap junction hemichannels induces ATP release from MLO-Y4 osteocytes. J Cell Physiol. 2007;212:207–214. doi: 10.1002/jcp.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, Genetos DC, Shao Y, Geist DJ, Li J, Ke HZ, Turner CH, Duncan RL. Activation of extracellular-signal regulated kinase (ERK1/2) by fluid shear is Ca(2+)- and ATP-dependent in MC3T3-E1 osteoblasts. Bone. 2008;42(4):644–652. doi: 10.1016/j.bone.2007.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genetos DC, Geist DJ, Liu D, Donahue HJ, Duncan RL. Fluid shear-induced ATP secretion mediates prostaglandin release in MC3T3-E1 osteoblasts. J Bone Miner Res. 2005;20(1):41–49. doi: 10.1359/JBMR.041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbracchio MP, Burnstock G. Purinoceptors: are there families of P2X and P2Y purinoceptors? Pharmacol Ther. 1994;64(3):445–475. doi: 10.1016/0163-7258(94)00048-4. [DOI] [PubMed] [Google Scholar]
- 6.Wesselius A, Bours MJ, Agrawal A, Gartland A, Dagnelie PC, Schwarz P, Jorgensen NR (2011) Role of purinergic receptor polymorphisms in human bone. Front Biosci 17:2572–2585. http://dx.doi.org/10.2741/3873 [DOI] [PubMed]
- 7.Nakamura E, Uezono Y, Narusawa K, Shibuya I, Oishi Y, Tanaka M, Yanagihara N, Nakamura T, Izumi F. ATP activates DNA synthesis by acting on P2X receptors in human osteoblast-like MG-63 cells. Am J Physiol Cell Physiol. 2000;279:C510–C519. doi: 10.1152/ajpcell.2000.279.2.C510. [DOI] [PubMed] [Google Scholar]
- 8.Naemsch LN, Weidema AF, Sims SM, Underhill TM, Dixon SJ. P2X(4) purinoceptors mediate an ATP-activated, non-selective cation current in rabbit osteoclasts. J Cell Sci. 1999;112(Pt 23):4425–4435. doi: 10.1242/jcs.112.23.4425. [DOI] [PubMed] [Google Scholar]
- 9.Buckley KA, Hipskind RA, Gartland A, Bowler WB, Gallagher JA (2002) Adenosine triphosphate stimulates human osteoclast activity via upregulation of osteoblast-expressed receptor activator of nuclear factor-kappa B ligand. Bone 31(5):582–590. http://dx.doi.org/10.1016/S8756-3282(02)00877-3 [DOI] [PubMed]
- 10.Hoebertz A, Townsend-Nicholson A, Glass R, Burnstock G, Arnett TR. Expression of P2 receptors in bone and cultured bone cells. Bone. 2000;27(4):503–510. doi: 10.1016/S8756-3282(00)00351-3. [DOI] [PubMed] [Google Scholar]
- 11.Soto F, Garcia-Guzman M, Stuhmer W. Cloned ligand-gated channels activated by extracellular ATP (P2X receptors) J Membr Biol. 1997;160(2):91–100. doi: 10.1007/s002329900298. [DOI] [PubMed] [Google Scholar]
- 12.Khakh BS, North RA. P2X receptors as cell-surface ATP sensors in health and disease. Nature. 2006;442(7102):527–532. doi: 10.1038/nature04886. [DOI] [PubMed] [Google Scholar]
- 13.Surprenant A, North RA. Signaling at purinergic P2X receptors. Annu Rev Physiol. 2009;71:333–359. doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- 14.Binderman I, Bahar H, Jacob-Hirsch J, Zeligson S, Amariglio N, Rechavi G, Shoham S, Yaffe A (2007) P2X4 is up-regulated in gingival fibroblasts after periodontal surgery. J Dent Res 86(2):181–185. doi:10.1177/154405910708600214 [DOI] [PubMed]
- 15.Liu PS, Chen CY. Butyl benzyl phthalate suppresses the ATP-induced cell proliferation in human osteosarcoma HOS cells. Toxicol Appl Pharmacol. 2010;244(3):308–314. doi: 10.1016/j.taap.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Stokes L, Scurrah K, Ellis JA, Cromer BA, Skarratt KK, Gu BJ, Harrap SB, Wiley JS. A loss-of-function polymorphism in the human P2X4 receptor is associated with increased pulse pressure. Hypertension. 2011;58(6):1086–1092. doi: 10.1161/HYPERTENSIONAHA.111.176180. [DOI] [PubMed] [Google Scholar]
- 17.Williams EJ, Bowles DJ. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14(6):1060–1067. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lise B. Husted TH, Liselotte Stenkjaer, Mette Carstens, Niklas R. Jorgensen, Bente L. Langdahl (2012) Functional polymorphisms in the P2X7 receptor gene are associated with osteoporosis. Bone (in press)
- 19.Anke Wesselius MJLB, Niklas R. Jørgensen, Peter Schwarz, Zanne Henriksen, Susanne Syberg SP, Svenhjalmar van Helden, Pieter C. Dagnelie (2012) Association of P2X7 receptor polymorphisms with bone mineral density and osteoporosis risk in a cohort of Dutch fracture patients. Osteoporosis International (in press) [DOI] [PMC free article] [PubMed]
- 20.Gartland A, Skarratt KK, Hocking LJ, Parsons C, Stokes L, Jorgensen NR, Fraser WD, Reid DM, Gallagher JA, Wiley JS (2012) Polymorphisms in the P2X7 receptor gene are associated with low lumbar spine bone mineral density and accelerated bone loss in post-menopausal women. Eur J Hum Genet. doi:10.1038/ejhg.2011.245 [DOI] [PMC free article] [PubMed]
- 21.Jorgensen NR, Husted LB, Skarratt KK, Stokes L, Tofteng CL, Kvist T, Jensen JE, Eiken P, Brixen K, Fuller S, Clifton-Bligh R, Gartland A, Schwarz P, Langdahl BL, Wiley JS (2012) Single-nucleotide polymorphisms in the P2X7 receptor gene are associated with post-menopausal bone loss and vertebral fractures. Eur J Hum Genet. doi:10.1038/ejhg.2011.253 [DOI] [PMC free article] [PubMed]
- 22.van Helden S, Cauberg E, Geusens P, Winkes B, van der Weijden T, Brink P. The fracture and osteoporosis outpatient clinic: an effective strategy for improving implementation of an osteoporosis guideline. J Eval Clin Pract. 2007;13(5):801–805. doi: 10.1111/j.1365-2753.2007.00784.x. [DOI] [PubMed] [Google Scholar]
- 23.PLINK (2009) PLINK v1.07. Whole genome association analysis toolset. Available via http://pngu.mgh.harvard.edu/~purcell/plink/reference.shtml. Accessed on February 2012
- 24.Yan Z, Liang Z, Obsil T, Stojilkovic SS. Participation of the Lys313-Ile333 sequence of the purinergic P2X4 receptor in agonist binding and transduction of signals to the channel gate. J Biol Chem. 2006;281(43):32649–32659. doi: 10.1074/jbc.M512791200. [DOI] [PubMed] [Google Scholar]
- 25.Roberts VH, Webster RP, Brockman DE, Pitzer BA, Myatt L. Post-translational modifications of the P2X(4) purinergic receptor subtype in the human placenta are altered in preeclampsia. Placenta. 2007;28(4):270–277. doi: 10.1016/j.placenta.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Stokes L, Fuller SJ, Sluyter R, Skarratt KK, Gu BJ, Wiley JS (2010) Two haplotypes of the P2X(7) receptor containing the Ala-348 to Thr polymorphism exhibit a gain-of-function effect and enhanced interleukin-1beta secretion. Faseb J 24(8):2916–2927 [DOI] [PubMed]
- 27.Orriss IR, Burnstock G, Arnett TR (2009) Expression of multiple P2 receptor subtypes by osteoblasts and osteoclasts. Bone 44:S304–S304
- 28.Hansen T, Jakobsen KD, Fenger M, Nielsen J, Krane K, Fink-Jensen A, et al. (2008) Variation in the purinergic P2RX(7) receptor gene and schizophrenia. Schizophr Res 104(1-3):146–152 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 32 kb)
(DOC 39 kb)
(DOC 29 kb)