Abstract
The detection of clinical isolates with decreased fluoroquinolone susceptibilities and a resistance mechanism is of epidemiological and clinical interest. We studied the susceptibilities of 62 clinical isolates and 2 American Type Culture Collection reference strains of Haemophilus influenzae to ciprofloxacin, levofloxacin, moxifloxacin, and nalidixic acid by the microdilution and disk diffusion methods. The ciprofloxacin MICs for 34 of the isolates were ≥0.12 μg/ml (range, 0.12 to 32 μg/ml), and the ciprofloxacin MICs for 28 matched control isolates were ≤0.06 μg/ml. In addition, we sequenced the quinolone resistance-determining regions (QRDRs) of gyrA and parC of all strains. The log2 MICs of all quinolones were plotted against the inhibition zone diameters. The MICs and inhibition zone diameters selected to screen for the resistance mechanism were based on the susceptibility distribution data and the presence or absence of amino acid changes in the QRDRs of GyrA and ParC. Strains for which ciprofloxacin MICs were ≤0.06 μg/ml, levofloxacin and moxifloxacin MICs were ≤0.03 μg/ml, and nalidixic acid MICs were ≤2.0 μg/ml lacked modifications in the QRDR of GyrA. In contrast, all strains for which ciprofloxacin, levofloxacin, and moxifloxacin MICs were ≥0.5 μg/ml and the vast majority of those for which nalidixic acid MICs were ≥32 μg/ml exhibited amino acid changes in GyrA and ParC. Nalidixic acid and the other three fluoroquinolones studied could be used to screen H. influenzae isolates for the detection of decreased susceptibilities to quinolones due to the acquisition of two amino acid changes in the QRDRs of GyrA and ParC (sensitivity, >95%; specificity, >80%).
Haemophilus influenzae is a common cause of pneumonia and other acute and chronic respiratory infections in children and adults. Although fluoroquinolones remain among the antimicrobial agents that are the most powerful against H. influenzae in vitro and are also highly effective as oral treatments for respiratory tract infections (15), resistance has been recognized (1, 7, 16, 25). The emergence of Streptococcus pneumoniae and H. influenzae isolates with reduced susceptibilities to ciprofloxacin and other quinolones appears to be a growing problem worldwide for clinical and public health. Recently, therapeutic failures in patients with community-acquired pneumonia associated with levofloxacin resistance in S. pneumoniae (10) and H. influenzae (3) have been described.
Cross-resistance to a large number of quinolones has been observed in H. influenzae (25), suggesting that testing for susceptibility to one quinolone would be sufficient to ascribe a loss of susceptibility to the entire antibiotic family in the majority of cases. On the basis of results of studies with a small number of isolates, we and others have suggested that subunit A of topoisomerase II (GyrA) and subunit A of topoisomerase IV (ParC) might be the first and second targets of quinolone action in H. influenzae, respectively (7, 16, 30).
Nalidixic acid has received special attention for use in a screening test for determination of the susceptibilities of gram-negative bacteria to quinolones (6, 9, 18) and the detection of strains with unique phenotypes of quinolone resistance (21, 26, 27).
We studied a collection of clinical H. influenzae isolates with decreased susceptibilities to ciprofloxacin (MIC range, 0.007 to 32 μg/ml) and other quinolones in order to (i) study the association of amino acid changes in the quinolone resistance-determining regions (QRDRs) of GyrA and ParC and decreased susceptibilities to fluoroquinolones; (ii) propose MICs and inhibition zone diameters for ciprofloxacin, levofloxacin, moxifloxacin, and nalidixic acid that might separate strains with reduced quinolone susceptibilities as a result of the acquisition of mutations in the QRDRs of gyrA and parC as a resistance mechanism; and (iii) compare the abilities of nalidixic acid and other oral fluoroquinolones to be used for screening for reduced fluoroquinolone susceptibility.
MATERIALS AND METHODS
Test isolates.
Our collection has been described elsewhere (25). Sixty-two clinical H. influenzae isolates and two reference strains from the American Type Culture Collection (ATCC) were included in this study. Three clinical isolates from the United States (1) and 31 Spanish clinical strains from our Haemophilus Reference Laboratory collection were selected on the basis of their reduced susceptibilities to ciprofloxacin (MICs, ≥0.12 μg/ml). The latter group was essentially obtained from respiratory specimens from patients with chronic respiratory infections as a result of surveillance for antibiotic resistance in Spanish clinical isolates. Strain ATCC 49247 was included as a susceptible control, as recommended by the NCCLS (24); ATCC 51907 is a strain whose whole genome has been sequenced (13). Both ATCC strains were included for quality control and comparative purposes.
Twenty-eight strains for which ciprofloxacin MICs were ≤0.06 μg/ml were selected as a susceptible control group and were matched to the clinical strains with reduced susceptibilities according to the following criteria: similar dates of isolation, isolation from similar geographical areas, isolation from patients with similar clinical diagnoses and from similar anatomical sources, and similar encapsulation statuses and biotypes. All clinical strains were nonencapsulated and belonged to different biotypes (16 biotype I, 26 biotype II, 10 biotype III, 1 biotype IV, 2 biotype V, 2 biotype VI, and 3 biotype VII strains). Nineteen of the 62 clinical strains were β-lactamase producers.
Susceptibility testing.
All H. influenzae strains were studied by the broth microdilution and disk diffusion susceptibility testing methods. The reference broth microdilution method was performed according to the NCCLS guidelines (23, 24). Haemophilus test medium (HTM) was prepared with Mueller-Hinton broth (Oxoid Ltd., Basingstoke, United Kingdom) supplemented with HTM supplement (Oxoid) and yeast extract (5‰; Difco, Detroit, Mich.). Microtiter plates were inoculated to produce a final inoculum density of approximately 5 × 105 CFU/ml, which was regularly controlled by counting the colonies. The inoculated plates were incubated at 35°C for 20 to 24 h before interpretation of the results. The MIC was defined as the lowest concentration of antibiotic that inhibited growth. The manufacturers of each of the quinolones supplied the drugs as powders of known potencies. The quinolones evaluated were ciprofloxacin, levofloxacin, moxifloxacin, and nalidixic acid.
The agar disk diffusion method was performed by following the NCCLS guidelines (23, 24) by using HTM base (Oxoid) supplemented with HTM supplement (Oxoid). Plates were inoculated with each bacterial suspension, which was adjusted to a McFarland 0.5 standard, and were incubated for 16 to 18 h at 35°C. Standard disks of ciprofloxacin (5 μg), moxifloxacin (5 μg), levofloxacin (5 μg), and nalidixic acid (30 μg) were purchased from Oxoid.
Amplification and sequence analysis of the QRDRs of gyrA and parC.
Amplification was performed in a 50-μl final volume containing 5 μl of DNA template, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1 μM each primer (Pharmacia, Piscataway, N.J.), 200 μM each deoxynucleoside triphosphate, and 2.5 U of Taq polymerase (Roche, Barcelona, Spain). The PCR program was denaturation at 94°C for 5 min and 30 amplification cycles of 94°C for 1 min, annealing at 54°C for 1 min, and polymerization at 72°C for 1 min; a final cycle of 72°C for 10 min was used to extend the amplicons fully. A 400-bp fragment including the QRDR of gyrA was amplified. The specific primers used were GYRA-F (5′-CCGCCGCGTACTATTCTCAAT-3′) and GYRA-R (5′-GTTGCCATCCCCACCGCAATACCA-3′). A 565-bp fragment including the QRDR of parC was amplified by using the following specific primers: PARC-F (5′-TCTGAACTTGGCTTAATTGCC-3′) and PARC-R (5′-GCCACGACCTTGCTCATAAAT-3′). The PCR products were purified with a PCR purification kit (Qiagen, Hilden, Germany). Both strands of the DNA of the fragments were sequenced with a Big Dye Terminator Cycle Sequencing kit (Perkin-Elmer, Warrington, United Kingdom) according to the instructions of the manufacturer. The products were resolved and analyzed with an ABI PRISM 377 DNA sequencer. Nucleotide sequences were analyzed with DNAstar (Madison, Wis.) software.
Amplification and sequence analysis of the QRDRs of gyrB and parE.
One fragment containing gyrB and parE, including the QRDRs, was sequenced from each of four nalidixic acid-resistant strains (MICs, 32 to 64 μg/ml) in which no amino acid modifications in the QRDRs of GyrA and ParC were detected. The specific primers used were GYRB-F (5′-CCTGCTCTTTCTGAAACTTTAC-3′), GYRB-R (5′CCATCTAACGCAAGGGTTAATC-3′), PARE-F (5′-TCGTTAGTGGCCCTGCATTAC-3′), and PARE-R (5′-GAACAGGGCACAGAGTAGGGT-3′). Sequencing and analysis of the sequences were done by the same method described above for GyrA and ParC. Determination of an eventual active efflux mechanism of fluoroquinolone resistance in these strains was carried out by determination of the ciprofloxacin and norfloxacin MICs in the presence and absence of carbonyl cyanide 3-chlorophenylhydrazone (CCCP; 0.01 μg/ml) and reserpine (8.0 μg/ml) (28).
Analysis of results.
The log2 MICs of all quinolones were plotted against the zone diameters obtained by the disk diffusion method. The correlation coefficients were determined by the least-squares method. The criteria used to determine the absence of a resistance mechanism (susceptibility) were the highest MICs and the smallest zone diameters for the group of strains that did not present mutations in the QRDRs of GyrA and ParC; the criteria used to determine the presence of a resistance mechanism (reduced susceptibility) were the lowest MICs and the largest zone diameters for which more than 70% of the strains presented two mutations in the QRDR of either GyrA or ParC. Along with these criteria there was an intermediate category that included strains without mutations in the QRDRs of GyrA and ParC and strains with only one amino acid change in the QRDR of GyrA. Very major errors were defined as results that indicated susceptibility by disk diffusion and reduced susceptibility by microdilution, major errors were classified as results that indicated reduced susceptibility by disk diffusion and susceptibility by microdilution, and minor errors were defined as results that indicted susceptibility in the intermediate range by one method and susceptibility or reduced susceptibility by the other method. The data were managed and the statistics were calculated by using the Whonet (WHO/CSR/DRS/99.1; World Health Organization) and GraphPad Prism (GraphPad Software, Inc.) computer programs.
RESULTS
Quinolone susceptibility and amino acid changes in QRDRs of GyrA and ParC.
2The QRDRs of GyrA and ParC of 64 isolates (62 clinical isolates and 2 ATCC reference strains) were sequenced. Thirty-four strains had no amino acid modifications (Table 1). Among the 28 strains with amino acid substitutions, 7 had one amino acid change in the QRDR of GyrA (position 84 or 88), 13 had one amino acid substitution in the QRDR of GyrA linked with one amino acid substitution in the QRDR of ParC, and 8 had two modifications in the QRDR of GyrA (affecting different combinations of positions 83, 84, and 88) linked with one amino acid change in the QRDR of ParC (Table 1).
TABLE 1.
Quinolone susceptibilities and amino acid changes in GyrA and ParC QRDR fragments of the H. influenzae isolates
| Strain or no. of strains | MIC or MIC range (μg/ml)a
|
Amino acid at the following positions in QRDR of GyrA:
|
Amino acid the following positions in QRDR of ParC:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | MOX | LEV | NAL | 83 | 84 | 88 | 82 | 83 | 84 | 88 | |
| ATCC 51907 | 0.007 | 0.03 | 0.03 | 0.5 | Asp | Ser | Asp | Gly | Asp | Ser | Glu |
| ATCC 49247 | 0.01 | 0.03 | 0.03 | 2.0 | |||||||
| 34b | 0.007-0.12 | 0.01-0.12 | 0.01-0.12 | 1.0-64 | |||||||
| 1 | 0.12 | 0.06 | 0.06 | 16 | Ala | ||||||
| 1 | 0.12 | 0.06 | 0.12 | 2.0 | Tyr | ||||||
| 1 | 0.12 | 0.12 | 0.12 | 64 | Tyr | ||||||
| 1 | 0.25 | 0.12 | 0.12 | 64 | Asn | ||||||
| 1 | 1.0 | 1.0 | 1.0 | 4.0 | Phe | ||||||
| 1 | 2.0 | 2.0 | 1.0 | 128 | Leu | ||||||
| 1 | 4.0 | 4.0 | 2.0 | 2.0 | Phe | ||||||
| 1 | 0.5 | 0.5 | 0.5 | 32 | Asn | Lys | |||||
| 1 | 0.5 | 0.5 | 0.25 | 128 | Asn | Arg | |||||
| 1 | 2.0 | 2.0 | 1.0 | 64 | Tyr | Asp | |||||
| 1 | 2.0 | 1.0 | 2.0 | 64 | Leu | Ile | |||||
| 1 | 2.0 | 1.0 | 1.0 | 128 | Phe | Arg | |||||
| 1 | 4.0 | 1.0 | 2.0 | 64 | Asn | Ile | |||||
| 1 | 4.0 | 2.0 | 2.0 | 64 | Asn | Ile | |||||
| 1 | 4.0 | 2.0 | 2.0 | 64 | Asn | Ile | |||||
| 1 | 4.0 | 2.0 | 2.0 | 128 | Asn | Ile | |||||
| 1 | 4.0 | 4.0 | 2.0 | 128 | Asn | Ile | |||||
| 1 | 4.0 | 4.0 | 2.0 | 128 | Leu | Ile | |||||
| 1 | 4.0 | 4.0 | 4.0 | 128 | Leu | Ile | |||||
| 1 | 4.0 | 2.0 | 2.0 | 64 | Phe | Lys | |||||
| 1 | 1.0 | 1.0 | 0.5 | 64 | Gly | Asp | Arg | ||||
| 1 | 4.0 | 2.0 | 2.0 | 64 | Leu | Asn | Ile | ||||
| 1 | 4.0 | 2.0 | 2.0 | 128 | Leu | Asn | Ile | ||||
| 1 | 4.0 | 2.0 | 2.0 | 128 | Leu | Asn | Ile | ||||
| 1 | 8.0 | 4.0 | 4.0 | 128 | Ile | Ala | Gly | ||||
| 1 | 16 | 8.0 | 16 | 64 | Leu | Asn | Ile | ||||
| 1 | 16 | 2.0 | 1.0 | 64 | Leu | Asn | Asn | ||||
| 1 | 32 | 32 | 32 | 2.0 | Leu | Tyr | Asp | ||||
CIP, ciprofloxacin; LEV, levofloxacin; MOX, moxifloxacin; NAL, nalidixic acid.
Additional amino acid changes were detected in the QRDR of GyrB in two strains for which the nalidixic acid MIC was 32 μg/ml: Asp489 to Asn and Thr472 to Ile.
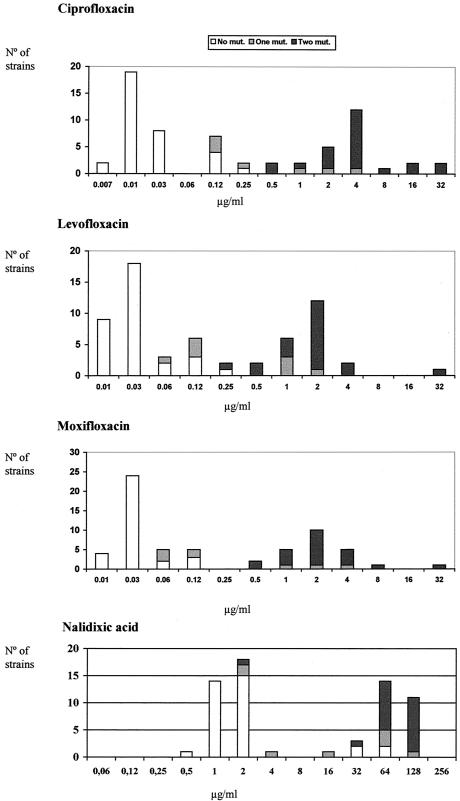
The distribution of the microbial population according to the susceptibilities of the isolates to ciprofloxacin, levofloxacin, moxifloxacin, and nalidixic acid, combined with data on the amino acid changes in the QRDRs of GyrA and ParC, are depicted in Fig. 1. All strains for which ciprofloxacin, levofloxacin, or moxifloxacin MICs were ≥0.5 μg/ml had amino acid modifications, while the opposite (no amino acid substitutions) was true for strains for which ciprofloxacin MICs were ≤0.06 μg/ml and levofloxacin and moxifloxacin MICs were ≤0.03 μg/ml. All but four strains for which nalidixic acid MICs were ≥4.0 μg/ml had amino acid modifications (see below).
FIG. 1.
Distribution of quinolone MICs for the H. influenzae strain collection according to the presence or absence of amino acid modifications in the QRDRs of GyrA and ParC. One mutation was always detected in GyrA, and two mutations were detected in GyrA or both GyrA and ParC.
For all 21 strains with two amino acid changes in the QRDRs of GyrA and ParC, ciprofloxacin and moxifloxacin MICs were ≥0.5 μg/ml, levofloxacin MICs were ≥0.25 μg/ml, and nalidixic acid MICs were ≥32 μg/ml (Fig. 1).
Laboratory screening of strains with amino acid changes in the QRDRs of GyrA and ParC.
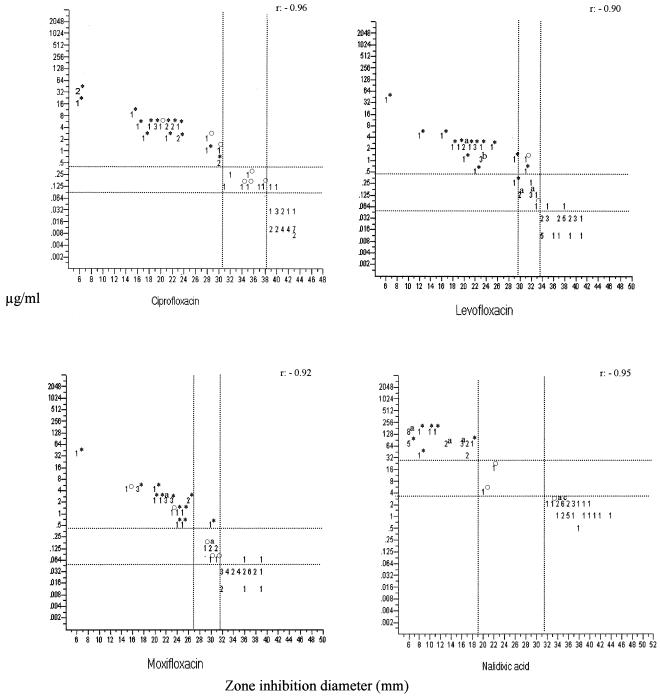
Figure 2 shows the scattergrams obtained by comparing the inhibition zone diameters with the disks of each quinolone and the MICs obtained by the standard microdilution method. The correlation coefficients of the data obtained by microdilution and disk diffusion were −0.96 for ciprofloxacin, −0.90 for levofloxacin, −0.92 for moxifloxacin, and −0.95 for nalidixic acid. The tentative MICs and disk diffusion zone diameters for ciprofloxacin, moxifloxacin, levofloxacin, and nalidixic acid obtained by using the susceptibility data obtained by microdilution and disk diffusion, which allowed us to determine the presence or absence of amino acid changes in GyrA and ParC as a mechanism of resistance to quinolones, are provided in Table 2 and Fig. 2. There was an interval that included strains either with one amino acid change or without any amino acid changes in the QRDR of GyrA. For this interval the percentage of strains with one amino acid change in the QRDR of GyrA was different for each quinolone tested, as follows: for ciprofloxacin, microdilution method, 44.5%; disk diffusion method, 57.1%; for levofloxacin, microdilution method, 27.2%; disk diffusion method, 40%; for moxifloxacin, microdilution method, 44.4%; disk diffusion method, 50%; and for nalidixic acid, microdilution and disk diffusion methods, 100% (two strains). The percentages of very major, major, and minor errors derived from the proposed screening values are indicated in Table 2.
FIG. 2.
Tentatively proposed microbiological criteria for ciprofloxacin, moxifloxacin, levofloxacin, and nalidixic acid for the microdilution and disk diffusion tests for H. influenzae strains with and without amino acid modifications in the QRDR of GyrA. Symbols: *, strains with two amino acid modifications in the QRDR of GyrA and ParC; ○, one amino acid change in the QRDR of GyrA. Letters: a, of these strain groups, only one had one amino acid modification in the QRDR of GyrA; b, of this strain group, two had one amino acid modification in the QRDR or GyrA; c, of this strain group, one had two amino acid modifications in the QRDRs of GyrA and ParC.
TABLE 2.
Tentative microbiological criteria for ciprofloxacin, moxifloxacin, levofloxacin, and nalidixic acid for microdilution and disk diffusion tests for H. influenzae with and without amino acid changes in QRDRs of GyrA and ParC
| Quinolone | MIC (μg/ml) criteria (correlated zone diameter [mm])
|
% Test error
|
||||
|---|---|---|---|---|---|---|
| No amino acid changes | None or one amino acid change in QRDR of GyrA | Two amino acid changes in QRDRs of GyrA and ParC | Very major | Major | Minor | |
| Ciprofloxacin | ≤0.06 (≥39) | 0.12-0.25 (31-38) | ≥0.5 (≤30) | 0.0 | 0.0 | 3.2 |
| Levofloxacin | ≤0.03 (≥34) | 0.06-0.25 (30-33) | ≥0.5 (≤29) | 0.0 | 0.0 | 8.5 |
| Moxifloxacin | ≤0.03 (≥31) | 0.06-0.25 (27-31) | ≥0.5 (≤26) | 0.0 | 0.0 | 5.0 |
| Nalidixic acid | ≤2.0 (≥32) | 4.0-16 (19-31) | ≥32 (≤18) | 0.0 | 0.0 | 0.0 |
We made an attempt to correlate current NCCLS criteria for fluoroquinolone-susceptible H. influenzae (23) with the inhibition zone diameters and amino acid changes obtained in this study (Table 3).
TABLE 3.
Distribution of the strain collection according to current NCCLS criteria for fluoroquinolone-susceptible H. influenzae and comparison with data on amino acid modifications in the QRDRs of GyrA and ParC
| Quinolone | NCCLS criteria (μg/ml)a | Proposed zone diameter (mm)b | No. (%) of strains with:
|
||
|---|---|---|---|---|---|
| No mutation | One mutation in GyrA QRDR | Two mutations in either GyrA or ParC QRDR | |||
| Ciprofloxacin | ≤1 | ≥28 | 34 (81) | 5 (12) | 3 (7) |
| Levofloxacin | ≤2 | ≥18 | 33 (60) | 7 (12.7) | 15 (27.3) |
| Moxifloxacin | ≤1 | ≥23 | 33 (76.7) | 5 (11.6) | 5 (11.6) |
Use of nalidixic acid and other fluoroquinolones for screening.
For four strains in which no amino acid modifications in the QRDR of GyrA were detected, the nalidixic acid MICs were between 32 and 64 μg/ml (Fig. 1). The corresponding ciprofloxacin MICs ranged from 0.06 to 0.12 μg/ml. These results were confirmed in at least three separate experiments. We considered the possibility of the existence of another resistance mechanism in these strains, and for this reason we sequenced a DNA fragment that included the QRDRs of GyrB and ParE. We found one amino acid change in ParC outside the QRDR (Ser133 to Ala) in one of the strains, and we detected two amino acid changes in the QRDR of GyrB (Asp489 to Asn and Thr472 to Ile) in two strains; we noticed no additional amino acid changes in the remaining strain. There were no differences in the ciprofloxacin and norfloxacin MICs for these four strains, as determined in the presence and absence of CCCP or reserpine.
Three additional strains presented amino acid changes in the QRDR of GyrA, but the nalidixic acid MIC was 2 μg/ml for all of them. The strains were therefore grouped with the nalidixic acid-susceptible strains (Fig. 2). These three strains were from the United States (1) and were sent to our reference laboratory; the ciprofloxacin MICs for the three strains were 0.12, 4, and 32 μg/ml, respectively; the levofloxacin MICs were 0.12, 2, and 32 μg/ml, respectively; and the moxifloxacin MICs were 0.06, 4, and 32 μg/ml, respectively.
According to the proposed screening criteria (Table 2), we estimated that the sensitivity and specificity of susceptibility tests with nalidixic acid for the discrimination of strains of H. influenzae with amino acid changes in the QRDRs of GyrA and ParC were 95.2 and 80%, respectively, which represents a sensitivity lower than that reported for Salmonella but a similar specificity (18). For the other fluoroquinolones tested, the sensitivities and specificities for the discrimination of strains with a resistance mechanism due to at least two amino acid changes in the QRDRs of GyrA and ParC were 95 to 100% and 90.4 to 93.3%, respectively.
DISCUSSION
Fluoroquinolone resistance and therapeutic failures have been described in S. pneumoniae (10, 12, 19) and other pathogens, including H. influenzae (3). The development of resistance and therapeutic failure during quinolone treatment might be due to the presence of infecting strains that have already undergone the initial step in the process of acquiring a mechanism of resistance, generally, the acquisition of GyrA mutations in gram-negative bacteria (11, 14, 26). We have recently reported on the first case of levofloxacin treatment failure in a patient with community-acquired pneumonia due to levofloxacin resistance in an H. influenzae strain isolated from the patient's blood (3). The patient had previously been treated with moxifloxacin for 5 days. Most general surveillance studies have shown that the rate of resistance to fluoroquinolones in H. influenzae is low (15). However, this resistance rate could be higher among isolates recovered from patients with chronic respiratory infections, as the clinical management of these patients often includes repetitive treatments with oral quinolones (7, 17) and quinolone resistance could be induced as a consequence of this strong antibiotic pressure.
An increase in the rate of fluoroquinolone use, particularly for the treatment of respiratory tract infections, has been recorded in recent years. In Spain, the numbers of prescriptions of quinolones for adult outpatients increased by 266.6% between 1985 and 2000 (5, 20), probably as a consequence of the introduction of fluoroquinolones with enhanced activities against S. pneumoniae. This increment may speed the future emergence of fluoroquinolone resistance in respiratory pathogens, including H. influenzae.
There have been several attempts to propose resistance breakpoints for quinolones in H. influenzae (2, 4, 8, 29), but the major weakness of those studies was that most of them lacked representative nonsusceptible populations from clinical infections. Our screening proposal criteria are intended to identify microorganisms belonging to subpopulations with a resistance mechanism (mutations in the QRDR of GyrA) that may be responsible for the spread of quinolone resistance (22). In this study, mutations in the QRDR of GyrA were consistently found in strains for which ciprofloxacin, levofloxacin, and moxifloxacin MICs were ≥0.5 μg/ml and in about 40% of strains for which ciprofloxacin MICs were between 0.12 and 0.25 μg/ml and moxifloxacin MICs were between 0.06 and 0.12 μg/ml and in 27% of strains for which levofloxacin MICs were between 0.06 and 0.25 μg/ml. In the case of nalidixic acid, the majority of strains for which MICs were ≥4.0 μg/ml had mutations in GyrA. When we include the data for the second target (ParC), our criteria can distinguish between strains with one amino acid change in GyrA from another group with two amino acid changes (either one change each in GyrA and ParC or two changes in GyrA). The fluoroquinolone MICs were the highest for the latter group of strains: ≥0.5 μg/ml for ciprofloxacin and moxifloxacin, ≥0.25 μg/ml for levofloxacin, and ≥32 μg/ml for nalidixic acid. In our study, the group of strains with two amino acid changes in the QRDR of GyrA also had an additional mutation in the QRDR of ParC and corresponded to the strains for which the MICs of all quinolones tested were the highest (Table 1).
Levofloxacin and moxifloxacin are recommended for the treatment of community-acquired pneumonia. The criteria suggested in this study should facilitate the laboratory detection of strains with decreased susceptibilities to quinolones. We have previously suggested that cross-resistance to a large group of quinolones is usually found in H. influenzae (25); for that reason, the use of the proposed screening for decreased susceptibility to one of the quinolones may be sufficient to detect decreased susceptibilities to all of them in the majority of cases.
Classically, nalidixic acid has been proposed for use in tests for screening for susceptibility in members of the family Enterobacteriaceae, especially Salmonella (9, 18). Brenwald et al. (6) described two strains of H. influenzae with decreased susceptibilities to ciprofloxacin (MICs, 0.06 and 2.0 μg/ml, respectively); neither strain presented an inhibition zone in tests with a 30-μg nalidixic acid disk, and both had mutations in GyrA (Ser64 to Phe) similar to those described in this study. According to our data, nalidixic acid would also be appropriate for use in tests of decreased susceptibility in H. influenzae, although there are several exceptions. In our series, three U.S. isolates would have erroneously been considered to be free of mutations in the QRDR of GyrA. This resistance phenotype was not detected in Spanish isolates, and isolates with this phenotype may represent a different quinolone-resistant subpopulation. Strains with similar resistance phenotypes have been reported among other bacteria, such as Staphylococcus aureus (26) and Escherichia coli (21); but this is a new phenomenon in H. influenzae. Novel mechanisms of resistance have been proposed for isolates of this phenotype, such as multidrug efflux (26) and decreased outer membrane permeability (21).
Nalidixic acid MICs were between 32 and 64 μg/ml for four Spanish isolates lacking modifications in GyrA; however, one strain had an one amino acid change outside the QRDR of ParC, and two strains each had one amino acid change in the QRDR of GyrB. One strain had no mutations in any of the DNA gyrases or topoisomerases assayed. In addition, the initial phenotypic characterization of an eventual efflux mechanism was negative for these four strains. Other alternative resistance mechanisms, such as an alteration in outer membrane permeability, may operate in these strains (21). The nalidixic acid-resistant (MICs, >32 μg/ml), ciprofloxacin-susceptible phenotype has been described to be strongly prevalent among clinical E. coli isolates in Spain (27). This phenotype is linked to a mutation in the Ser83 amino acid codon of gyrA. We have also found several H. influenzae strains with a mutation in the QRDR of GyrA (Asp88 to Asn) for which nalidixic acid MICs were ≥32 μg/ml and ciprofloxacin MICs were 0.12 to 0.25 μg/ml.
In summary, in this study we have shown that decreased susceptibility to quinolones in H. influenzae is usually associated with amino acid changes in the QRDRs of GyrA and ParC. We have suggested MIC and inhibition zone criteria that may be useful for the separation of clinical isolates of H. influenzae that are fully susceptible to ciprofloxacin, levofloxacin, moxifloxacin, and nalidixic acid from others with various degrees of decreased susceptibilities to these quinolones as a result of target modifications in GyrA and ParC. Nalidixic acid and other oral fluoroquinolones could be used in tests to screen for the detection of decreased susceptibility to quinolones in H. influenzae with a sensitivity of >95% and a specificity of >80%.
Acknowledgments
We thank Maite Camacho and Enrique Moguel for collaboration in this study, as well as the Biopolymers Unit of ISCIII. We thank Janet Hinder for providing us with two quinolone-resistant H. influenzae isolates.
This work was supported by a grant from the Fondo de Investigacion Sanitaria (grant 99/0304), Ministerio de Sanidad of Spain. M. Pérez-Vazquez is a recipient of a grant from the Instituto de Salud Carlos III (grant 02/0056). We also thank the Fundacion Ciencias Microbianas (Hospital Ramon y Cajal, Madrid, Spain) for its contribution.
REFERENCES
- 1.Barriere, S. L., and J. Hindler. 1993. Ciprofloxacin resistant Haemophilus influenzae in a patient with chronic lung disease. Ann. Pharmacother. 27:309-310. [DOI] [PubMed] [Google Scholar]
- 2.Barry, A. L., J. H. Jorgensen, D. J. Hardy, S. D. Allen, C. N. Baker, P. C. Fuchs, and J. C. McLaughlin. 1992. Interpretive criteria and quality control parameters for testing susceptibility of Haemophilus influenzae to enoxacin, ofloxacin, and temafloxacin. J. Clin. Microbiol. 30:3013-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastida, T., M. Pérez-Vázquez, J. Campos, M. C. Cortés-Lletget, F. Román, F. Tubau, A. G. de la Campa, and C. Alonso-Tarrés. 2003. Therapeutic failure in a case of pneumonia due to levofloxacin-resistant Haemophilus influenzae. Emerg. Infect. Dis.91:1476-1478. [DOI] [PubMed]
- 4.Biedenbach, D. J., and R. N. Jones. 2002. Evaluation of in vitro susceptibility testing criteria for gemifloxacin when tested against Haemophilus influenzae strains with reduced susceptibility to ciprofloxacin and ofloxacin. Diagn. Microbiol. Infect. Dis. 43:323-326. [DOI] [PubMed] [Google Scholar]
- 5.Bremón, A. R., M. Ruiz-Tovar, B. P. Gorricho, P. D. de Torres, and R. L. Rodríguez. 2000. Non-hospital consumption of antibiotics in Spain: 1987-1997. J. Antimicrob. Chemother. 45:395-400. [DOI] [PubMed] [Google Scholar]
- 6.Brenwald, N., J. M. Andrews, G. Jevons, and R. Wise. 2003. Detection of ciprofloxacin resistance in Haemophilus influenzae using nalidixic acid and BSAC methodology. J. Antimicrob. Chemother. 51:1311-1312. [DOI] [PubMed] [Google Scholar]
- 7.Campos, J., F. Román, M. Georgiou, C. García, R. Gómez-Lus, R. Cantón, H. Escobar, and F. Baquero. 1996. Long-term persistence of ciprofloxacin-resistant Haemophilus influenzae in patients with cystic fibrosis. J. Infect. Dis. 174:1345-1347. [DOI] [PubMed] [Google Scholar]
- 8.Corkill, J. E., A. Percival, P. McDonald, and A. I. Bamber. 1994. Detection of quinolone resistance in Haemophilus ssp. J. Antimicrob. Chemother. 34:841-844. [DOI] [PubMed] [Google Scholar]
- 9.Crump, J. A.,, T. J. Barret, J. T. Nelson, and F. J. Angulo. 2003. Reevaluating fluoroquinolone breakpoints for Salmonella enterica serotype Typhi and for non-Typhi salmonellae. Clin. Infect. Dis. 37:75-81. [DOI] [PubMed] [Google Scholar]
- 10.Davidson, R., R. Calvancanti, J. Brunton, D. J. Bast, J. C. de Acevedo, P. Kibsey, C. Fleming, and D. E. Low. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:747-750. [DOI] [PubMed] [Google Scholar]
- 11.Deguchi, T., T. Kawamura, M. Yasuda, M. Nakano, H. Fukuda, H. Kato, N. Kato, Y. Okano, and Y. Kawada. 1997. In vivo selection of Klebsiella pneumoniae strains with enhanced quinolone resistance during fluoroquinolone treatment of urinary tract infections. Antimicrob. Agents Chemother. 41:1609-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of H. influenzae Rd. Science 269:449-604. [DOI] [PubMed] [Google Scholar]
- 14.Galanakis, N., H. Giamarellou, T. Moussas, and E. Dounis. 1997. Chronic osteomyelitis caused by multi-resistant gram-negative bacteria: evolution of treatment with newer quinolones after prolonged follow-up. J. Antimicrob. Chemother. 39:241-246. [DOI] [PubMed] [Google Scholar]
- 15.García-Rodriguez, J. A., F. Baquero, J. Garcia de Lomas, and L. Aguilar. 1999. Antimicrobial susceptibility of 1422 H. influenzae isolates from respiratory tract infections in Spain results of 1-year multicenter surveillance study. Infection 27:265-267. [DOI] [PubMed] [Google Scholar]
- 16.Georgiou, M., R. Muñoz, F. Román, R. Cantón, R. Gómez-Lus, J. Campos, and A. De la Campa. 1996. Ciprofloxacin-resistant Haemophilus influenzae possesses mutations in analogous positions of GyrA and ParC. Antimicrob. Agents Chemother. 40:1741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groeneveld, K., L. van Alphen, P. P. Eijk, G. Vischers, H. M. Jansen, and H. C. Zanen. 1990. Endogenous and exogenous reinfections by Haemophilus influenzae in patients with chronic obstructive pulmonary disease: the effect of antibiotic treatment of persistence. J. Infect. Dis. 161:512-517. [DOI] [PubMed] [Google Scholar]
- 18.Hakanen, A., P. Kotilainen, J. Jalava, A. Siitonen, and P. Huovinen. 1999. Detection of decreased fluoroquinolone susceptibility in salmonellas and validation of nalidixic acid screening test. J. Clin. Microbiol. 37:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, M. E., A. M. Staples, I. Critchley, C. Thornsberry, P. Heize, H. D. Engler, and D. F. Sahm. 2000. Benchmarking the in vitro activities of moxifloxacin and comparator agents against recent respiratory isolates from 377 medical centers throughout the United States. Antimicrob. Agents Chemother. 44:2645-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lázaro, E., M. Madurga and F. J. de Abajo. 2002. Evolución del consumo de antibióticos en España, 1985-2000. Med. Clin. (Barcelona) 118:561-568. [DOI] [PubMed] [Google Scholar]
- 21.Moniot-Ville, N., J. Guigert, N. Moreau, F. Acar, E. Collatz, and L. Gutman. 1991. Mechanisms of quinolone resistance in a clinical isolate of Escherichia coli highly resistant to fluoroquinolones but susceptible to nalidixic acid. Antimicrob. Agents Chemother. 35:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouton, J. W. 2002. Breakpoints: current practice and future perspectives. Int. J. Antimicrob. Agents 19:323-331. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing. Eleventh informational supplement M100-S12. NCCLS, Wayne, Pa.
- 24.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A5. NCCLS, Wayne, Pa.
- 25.Pérez-Vázquez, M., F. Román, M. C. Varela, R. Cantón, and J. Campos. 2003. Activities of thirteen quinolones by three susceptibility testing methods against a collection of Haemophilus influenzae isolates with different levels of susceptibility to ciprofloxacin: evidence for cross-resistance. J. Antimicrob. Chemother. 51:147-151. [DOI] [PubMed] [Google Scholar]
- 26.Piddock, L. J., Y. Fang Jin, M. A. Webber, and M. J. Everett. 2002. Novel ciprofloxacin-resistant, nalidixic acid-susceptible mutant of Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2276-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz, J., J. Gómez, M. N. Navia, A. Ribera, J. M. Sierra, F. Marco, J. Mensa, and J. Vila. 2001. High prevalence of nalidixic resistant, ciprofloxacin susceptible phenotype among clinical isolates of Escherichia coli and other Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 42:257-261. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, L., W. Pan, M. Viñas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 179:6855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sewell, D. L., A. L. Barry, S. D. Allen, P. C. Fuchs, J. H. Jorgensen, and F. Tenover. 1995. Tentative criteria for determining the in vitro susceptibilities of Haemophilus influenzae, including quality control parameters, to two fluoroquinolones (grepafloxacin and PD 131628). Diagn. Microbiol. Infect. Dis. 21:175-179. [DOI] [PubMed] [Google Scholar]
- 30.Vila, J., J. Ruiz, F. Sánchez, F. Navarro, B. Mirelis, M. T. Jiménez de Anta, and G. Prats. 1999. Increase in quinolone resistance in Haemophilus influenzae strain isolated from a patient with recurrent respiratory infections treated with ofloxacin. Antimicrob. Agents Chemother. 43:161-162. [DOI] [PMC free article] [PubMed] [Google Scholar]