Abstract
Human β-defensins 2 and 3 (HBD-2 and HBD-3) are inducible peptides present at sites of infection in the oral cavity. A few studies have reported broad-spectrum antimicrobial activity for both peptides. However, no comprehensive study has thoroughly investigated their potential against oral pathogens. The purpose of this study was to test the effectiveness of HBD-2 and HBD-3 against a collection of oral organisms (Actinobacillus actinomycetemcomitans, Fusobacterium nucleatum, Porphyromonas gingivalis, Peptostreptococcus micros, Actinomyces naeslundii, Actinomyces israelii, Streptococcus sanguis, Streptococcus mutans, Candida tropicalis, Candida parapsilosis, Candida krusei, Candida glabrata, and Candida albicans). Radial diffusion assays were used to test HBD-2 and HBD-3 activities against at least three strains of each species. There was significant variability in MICs, which was strain specific rather than species specific. MICs ranged from 3.9 to >250 μg/ml for HBD-2 and from 1.4 to >250 μg/ml for HBD-3. HBD-3 demonstrated greater antimicrobial activity and was effective against a broader array of organisms. Overall, aerobes were 100% susceptible to HBD-2 and HBD-3, whereas only 21.4 and 50% of the anaerobes were susceptible to HBD-2 and HBD-3, respectively. HBD-2 and HBD-3 also demonstrated strain-specific activity against the Candida species evaluated. Interestingly, an association between HBD-2 and HBD-3 activities was noted. This suggests that the two peptides may have similar mechanisms yet utilize distinct pathways. The lack of activity against specific anaerobic strains and Candida warrants further investigation of the potential resistance mechanisms of these organisms. Finally, the significant variability between strains underlies the importance of testing multiple strains when evaluating activities of antimicrobial peptides.
Innate immunity plays an important role in the battle against bacterial colonization as a part of the host protective defense against caries, periodontal diseases, and yeast infections in the oral cavity (43). To date, many factors are known to play a role in this response system, one of which is the β-defensin family.
Human β-defensins (HBDs) are small cationic peptides, produced by epithelial cells, that are believed to be part of both the innate and adaptive immune responses in that they demonstrate antimicrobial as well as chemotactic activities (21, 23, 39, 44). β-Defensin peptides are present in gingival epithelia (6, 7), saliva (32), and gingival crevicular fluid (9), placing them as a first line of defense in the oral cavity. HBD-1 is expressed constitutively by gingival epithelial cells (26, 32), whereas mRNA expression of HBD-2, -3, and -4 has been shown to be up-regulated by specific microorganisms (15, 16, 22, 23, 25, 27) and by proinflammatory cytokines such as interleukin-1β, tumor necrosis factor alpha, and gamma interferon in keratinocyte cell cultures (16, 23, 27, 28, 32). There is also evidence that their expression is increased during inflammation in vivo (23, 30, 32).
The β-defensin family has shown activities against gram-positive and gram-negative bacteria, fungi, and enveloped viruses in vitro (13, 39). The activities of HBD-1, -2, and -4 have been reported to be predominantly effective against gram-negative bacteria such as Escherichia coli and Pseudomonas aeruginosa (15, 39), with weak or no activity against gram-positive bacteria such as Staphylococcus aureus and Streptococcus pyogenes (15, 21, 36). HBD-2 was found to be 10-fold more potent than HBD-1 and exhibited activity against P. aeruginosa at physiological concentrations (100 ng/ml) (39). HBD-4 seemed to be less effective than HBD-2 against selected gram-positive and gram-negative bacteria and yeasts, with the exception of P. aeruginosa, for which HBD-4 displayed a greater antimicrobial activity than the other known defensin peptides (15). In contrast, HBD-3 has shown broad-spectrum activity against both gram-negative and gram-positive bacteria at concentrations much lower than those for other members of the β-defensin family (23). In addition, its activity appears to be less salt sensitive than those of HBD-1, -2, and -4 (1, 15, 17, 39). Therefore, HBD-3 is considered the most potent β-defensin peptide described thus far.
Although detailed analyses of the antimicrobial mechanism of action of the β-defensins are still incomplete, there is now circumstantial evidence that permeabilization of membranes is involved, either through the formation of multimeric pores as described for the α-defensins (42) or by an electrostatic charge-based mechanism as suggested by the structural and electrostatic properties of HBD-2 oligomer (24). The latter hypothesis seems to be in accordance with the morphological changes observed in S. aureus when it is treated with HBD-3 (23).
Several reports have evaluated the antimicrobial spectrum of the β-defensins; however, to date there is no comprehensive study which has thoroughly investigated the activity of defensins against oral microorganisms. Furthermore, few studies have assessed their antimicrobial properties against more than one, or at most two, strains of a given species. The purpose of this study, therefore, was to investigate the specific antimicrobial properties of the two inducible and reportedly most active peptides, HBD-2 and -3, against a panel of oral microorganisms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The activities of HBD-2, HBD-3, and SMAP29, a sheep cathelicidin (19), against aerobic and anaerobic, gram-positive and gram-negative oral bacteria and Candida species were determined (Table 1). For the purposes of this study, aerobic bacteria were defined as those grown with 5% CO2 and included Streptococcus sanguis, Streptococcus mutans, Actinomyces naeslundii, Actinomyces israelii, and E. coli. At least three strains of each bacterial and Candida species were tested with HBD-2, HBD-3, and SMAP29 (Table 1). Actinobacillus actinomycetemcomitans, E. coli, S. sanguis, and S. mutans were grown in tryptic soy broth (Difco Laboratories, Detroit, Mich.) with 0.6% yeast extract (Difco); Fusobacterium nucleatum was grown in Schaedler broth (Difco); Porphyromonas gingivalis was grown in tryptic soy broth (Difco) supplemented with 5 μg of hemin (Sigma, St. Louis, Mo.) per ml; Peptostreptococcus micros was grown in brain heart infusion (Difco) with 0.5% neopeptone and 5 μg of hemin (Sigma) per ml; and A. naeslundii and A. israelii were grown in brain heart infusion (Difco). Candida albicans, Candida tropicalis, Candida glabrata, Candida parapsilosis, and Candida krusei were grown in YPD (1% yeast, 2% peptone, 2% dextrose) broth (Difco). Cultures were grown overnight either aerobically in a 5% CO2 incubator or anaerobically in 85% N2-10% H2-5% CO2.
TABLE 1.
Susceptibilities of the bacteria and yeast to HBD-2, HBD-3, and SMAP29
| Species | Gram stain reaction | Strain | MIC (μg/ml) of:
|
||
|---|---|---|---|---|---|
| HBD-2 | HBD-3 | SMAP29 | |||
| A. actinomycetem- comitans | − | Y4 | >250.0 | 47.9 | 1.7 |
| 1200 | >250.0 | 9.6 | 1.3 | ||
| ATCC 29523 | >250.0 | >250.0 | 2.9 | ||
| F. nucleatum | − | 1594 | >250.0 | >250.0 | 1.3 |
| 1908 | >250.0 | 13.2 | 0.9 | ||
| 25598 | 6.5 | 32.7 | 1.0 | ||
| ATCC 49256 | 10.3 | 4.5 | 0.6 | ||
| P. gingivalis | − | W50 | >250.0 | >250.0 | 30.1 |
| ATCC 33277 | 34.6 | 5.7 | 25.0 | ||
| ATCC 49417 | >250.0 | >250.0 | 13.0 | ||
| P. micros | + | ATCC 33270 | >250.0 | >250.0 | 16.7 |
| 8050 | >250.0 | >250.0 | 5.7 | ||
| 2903-02 | >250.0 | 15.9 | 7.5 | ||
| 97-1502 | >250.0 | >250.0 | 11.8 | ||
| A. naeslundii | + | 14B01 | 14.0 | 7.0 | 1.4 |
| 11A01 | 8.2 | 7.2 | 1.3 | ||
| 14B4C | 14.0 | 4.1 | 1.2 | ||
| A. israelii | + | 9P04 | 10.0 | 10.8 | 2.0 |
| 1P04 | 10.0 | 9.0 | 1.9 | ||
| 5A40 | 9.1 | 9.0 | 1.7 | ||
| S. sanguis | + | AC59 | 25.0 | 21.3 | 3.0 |
| P695 | 10.0 | 7.9 | 2.9 | ||
| NP506 | 8.8 | 7.6 | 3.0 | ||
| S. mutans | + | ATCC 25175 | 4.1 | 5.0 | 1.9 |
| Ingbritt 162 | 4.4 | 2.6 | 1.7 | ||
| OMZ175 | 4.7 | 2.8 | 1.9 | ||
| 330-5 | 7.8 | 3.4 | 1.6 | ||
| C. tropicalis | II7 | 3.9 | 3.5 | 1.3 | |
| 5 | 13.1 | 3.3 | 2.5 | ||
| 362 | 11.9 | 14.4 | 6.4 | ||
| C. parapsilosis | Pf27 | 9.3 | 3.0 | 1.6 | |
| Pf5 | 14.6 | 1.4 | 1.4 | ||
| ATCC 22019 | 17.8 | 12.4 | 6.4 | ||
| C. krusei | ATCC 6258 | 12.2 | 2.0 | 1.6 | |
| P31 | 13.3 | 3.2 | 2.5 | ||
| 932638 | >250.0 | 13.7 | 13.6 | ||
| C. glabrata | 931010 | >250.0 | >250.0 | >250.0 | |
| 932474 | 22.7 | 33.8 | 12.0 | ||
| 1480.42 | >250.0 | >250.0 | >250.0 | ||
| C. albicans | FC16 | 9.4 | 3.2 | 1.6 | |
| FC5 | 4.6 | 2.8 | 1.9 | ||
| ATCC 820 | 59.2 | 7.1 | 3.7 | ||
| E. coli | − | ATCC 9637 | 3.7 | 5.1 | 2.8 |
Radial diffusion assay.
Radial diffusion assays (29) were performed to test the antimicrobial activities of HBD-2 and HBD-3. E. coli was used as a control organism, and SMAP29 was used as a control peptide. Briefly, cells were grown in their appropriate media overnight under the conditions described above to mid-log phase, centrifuged at 7,500 × g for 15 min, rinsed with fresh medium, and resuspended in 10 mM sodium phosphate, pH 7.4. An underlay gel was prepared, which consisted of a mixture of 1% agarose in 10 mM sodium phosphate (pH 7.4) containing 4 × 105 yeast cells or 4 × 106 bacteria. The mixture was immediately poured into a square petri dish and allowed to solidify. A series of 3-mm-diameter wells were punched in the agar, and 5 μl of recombinant HBD-2 or -3 (PeproTech, Rocky Hill, N.J.) or SMAP29 (Multiple Peptide Systems, San Diego, Calif.) diluted in 0.01% acetic acid-0.1% human serum albumin was added to each well at concentrations ranging from 250 to 0.25 μg/ml. A control well containing only 10 mM sodium phosphate buffer was also prepared on each plate. The plates were then incubated under the appropriate aerobic or anaerobic conditions at 37°C for 3 h to allow for peptide diffusion. Ten milliliters of a 1% agar overlay gel containing medium specific for the organism tested was then poured over the first agar layer to provide nutrients for the cells. The plates were incubated again for 12 to 18 h until zones of inhibition were visible. Antimicrobial activities were expressed as MICs, which were calculated as previously described (4, 40). Briefly, zones of inhibition were recorded with a Boley gauge as radial diffusion units (zone of inhibition − well diameter ×10). The x intercept obtained from the relationship between radial diffusion units versus log10 peptide concentration was determined after regression. Resistance was defined as an MIC of greater than 250 μg/ml. Each experiment was performed in triplicate from different inoculums, and the statistical significance of the assay was evaluated by using a Student two-tailed t test. Coefficients of correlations (Pearson coefficient) were calculated to test the association between peptides and were evaluated by a P value representing the chance that random sampling would result in a correlation coefficient as far from zero as (or farther than) the one observed in the experiment.
RESULTS
Activities of HBD-2 and -3 against oral bacteria.
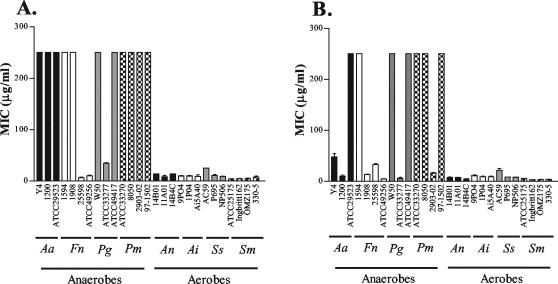
Overall, aerobes (S. sanguis, S. mutans, A. naeslundii, A. israelii, and E. coli) were more susceptible to HBD-2 and HBD-3 than anaerobes (A. actinomycetemcomitans, F. nucleatum, P. gingivalis, and P. micros) (Fig. 1 and Table 1). Of the two defensins tested, HBD-3 demonstrated greater activity against a broader array of organisms than HBD-2 (P < 0.05). Aerobic and anaerobic bacteria demonstrated strain rather than species specificity in their susceptibilities to HBD-2 and HBD-3. Both peptides were active against selected gram-positive and gram-negative bacteria (Fig. 1 and Table 1).
FIG. 1.
Susceptibility of aerobic and anaerobic oral bacteria to HBD-2 and -3. MICs, obtained from radial diffusion assays, of HBD-2 (A) and HBD-3 (B) against A. actinomycetemcomitans (Aa), F. nucleatum (Fn), P. gingivalis (Pg), P. micros (Pm), A. naeslundii (An), A. israelii (Ai), S. sanguis (Ss), and S. mutans (Sm) are shown. Values are means and standard deviations from triplicate assays.
For the anaerobes, differences in susceptibilities were seen for HBD-2 and HBD-3. Fifty percent of the isolates were susceptible to HBD-3, whereas only 21.4% of anaerobes were susceptible to HBD-2. HBD-3 was also more potent than HBD-2 (P < 0.005) against the four different anaerobic species tested. MICs ranged from 6.5 to >250 μg/ml for HBD-2 and from 4.5 to >250 μg/ml for HBD-3 (Table 1). Individual strains of each of the four species demonstrated resistance to both HBD-2 and HBD-3 (i.e., strains ATCC 29523, 1594, W50, and ATCC 33270 of A. actinomycetemcomitans, F. nucleatum, P. gingivalis, and P. micros, respectively). Overall, P. micros was the most resistant organism, with all strains resistant to HBD-2 and only one strain, 2903-02, susceptible to HBD-3. F. nucleatum was the most susceptible anaerobic organism, with two strains susceptible and one resistant to both HBD-2 and HBD-3; the fourth strain tested (strain 1908) was resistant to HBD-2 but susceptible to HBD-3 (Fig. 1 and Table 1).
Similar to the case for the anaerobes, HBD-3 demonstrated greater activity than HBD-2 against the aerobes (P < 0.05). However, the difference between the activities of HBD-2 and -3 was not as great as that observed with the anaerobes. All of the aerobes tested (S. sanguis, S. mutans, A. naeslundii, A. israelii, and E. coli) were susceptible to both HBD-2 and -3. MICs ranged from 4.1 to 25.0 μg/ml for HBD-2 and from 2.6 to 21.3 μg/ml for HBD-3 (Table 1).
SMAP29, the control peptide, displayed greater activity toward both the aerobes and anaerobes than either HBD-2 or HBD-3. The activities of SMAP29 paralleled those previously obtained in our laboratory for A. actinomycetemcomitans, F. nucleatum, P. gingivalis, and E. coli (19). Interestingly, those strains demonstrating resistance to the defensins were susceptible to SMAP29.
Activities of HBD-2 and -3 against Candida species.
The activities of HBD-2 and HBD-3 against Candida were similar to those observed for both the anaerobic and aerobic bacteria (Fig. 2). The antimicrobial variability observed was strain specific rather than species specific, and inhibition ranged from 3.9 to >250 μg/ml for HBD-2 and from 1.4 to >250 μg/ml for HBD-3. Like the anaerobes, individual Candida strains demonstrated resistance to both HBD-2 and -3. Two of the three C. glabrata isolates tested, 931010 and 1480.42, were resistant to HBD-2 and -3, and C. krusei isolate 932638 was resistant to HBD-2. Interestingly, the same two C. glabrata isolates were resistant to the control peptide, SMAP29 (Table 1). C. glabrata strain 932474, however, was susceptible to HBD-2, HBD-3, and SMAP29, with MICs of 22.7, 33.8, and 12.0 μg/ml, respectively (Table 1). Again, HBD-3 demonstrated greater antimicrobial activity at lower concentrations than did HBD-2 (P < 0.005). The MICs of HBD-2 and HBD-3 were similar for the Candida species and the aerobic bacteria (resistant strains C. glabrata 931010, C. glabrata 1480.42, and C. krusei 932638 were excluded from the analysis).
FIG. 2.
Susceptibility of Candida species to HBD-2 and -3. MICs, obtained from radial diffusion assays, of HBD-2 (A) and HBD-3 (B) against C. tropicalis (Ct), C. parapsilosis (Cp), C. krusei (Ck), C. glabrata (Cg), and C. albicans (Ca) are shown. Values are means and standard deviations from triplicate assays.
Association between antimicrobial activities of HBD-2, HBD-3, and SMAP29.
There was strong evidence of association between the MICs obtained with HBD-2 and HBD-3. For the bacterial population (including aerobes and anaerobes), the coefficient of correlation between HBD-2 and -3 was high (r = 0.7; P < 0.0001). SMAP29 antimicrobial activity was correlated to those of HBD-2 and -3 as well, with lesser strength (r = 0.37 [P = 0.03] and r = 0.50 [P = 0.046], respectively). When the bacterial population was split into the subgroups of aerobes and anaerobes, there was a better correlation between HBD-2 and -3 within the aerobic population (r = 0.83; P = 0.0002) than within the anaerobic population (r = 0.54; P = 0.024). Following this trend, SMAP29 antimicrobial activity correlated weakly with that of HBD-3 for the aerobic species (r = 0.56; P = 0.023) but not with that of HBD-2. No correlation between SMAP29 and HBD-2 or -3 was found within the anaerobic population. The weak correlation or lack of correlation observed for the three peptides with the anaerobes could be explained by the large number of isolates that were not susceptible to the peptides even at the highest concentrations.
Strong associations of antimicrobial activity were observed when the three peptides were tested against the Candida species. HBD-2 and HBD-3 activities were highly correlated (r = 0.79; P = 0.0002). SMAP29 showed a strong correlation as well with both HBD-2 and HBD-3 (r = 0.8 [P = 0.0002] and r = 0.99 [P < 0.0001], respectively).
DISCUSSION
In humans, β-defensins are found in oral tissues (6, 10, 26, 32), salivary glands (2, 21, 45), salivary secretions (32), and gingival crevicular fluid (9). In gingival tissue, HBD-1 and HBD-2 mRNAs are localized predominately in the suprabasal stratified epithelium and the peptides detected in upper epithelial layers, consistent with the formation of the stratified epithelial barrier (7), a region exposed to oral bacteria in plaque on the tooth surface (7). In this study, antimicrobial activities of HBD-2 and -3 were assessed for the first time against a panel of oral microorganisms, including several anaerobes. We showed that HBD-2 and -3 are antimicrobial against aerobic and anaerobic, gram-positive and gram-negative oral bacteria and Candida species and thus may likely play an important role in preventing the development of caries, periodontal infections, and candidiasis. Their importance as initial innate defenders coincides with other findings in our laboratory, where increased expression of HBD-3 was associated with health rather than periodontal disease (1a).
The antimicrobial activities of both peptides were strain specific rather than species specific, such that no particular delineation between species was observed regarding the efficacy of either peptide. The variability observed within a species in our study emphasizes the importance of evaluating several strains of a given species in order to better assess the susceptibility profile of a particular species.
Similar to that reported for nonoral microorganisms, HBD-3 demonstrated a broader spectrum of activity against the organisms tested and greater antimicrobial activity than HBD-2 (23). However, the activities of HBD-2 and HBD-3 still appeared to be associated. This suggests that while HBD-2 and -3 may share a similar target, they may also possess very specific mechanisms of action. This hypothesis is supported by the fact that the antimicrobial activity of HBD-3 is not affected by increased ionic strength (unlike that of HBD-2 [41]), suggesting that binding of HBD-3 to a negatively charged bacterial membrane may not be its only mechanism of action (23). Starner et al. demonstrated that unlike that of HBD-2, the activity of HBD-3 was not mediated by binding to the lipooligosaccharide of Haemophilus influenzae, suggesting that HBD-3 interacts with different binding sites or possesses a different mechanism of action than HBD-2 (40). It has been theorized that HBD-3 could be more active in part due to a higher net cationic charge than that observed for HBD-2 (23, 25, 40) or that it has the ability to form dimers (35). Interestingly, even though HBD-2 was less effective than HBD-3, it was still active against the gram-positive aerobes tested in the present study. These results are somewhat contradictory to the accepted notion that HBD-2 is not very effective against gram-positive bacteria (15, 39). This could be explained, however, by the limited number of organisms or strains tested in previous studies.
Our findings also suggest that there are anaerobic pathogens that are resistant to the β-defensins in the oral cavity. This is an unexpected finding, and the implications for the pathogenesis of periodontal disease are not yet known. One hypothesis to explain the resistance observed for the anaerobes is that the β-defensin targets may be absent or modified in anaerobes. As reported for other antimicrobial peptides (11, 33), the β-defensin activity could be linked to the respiratory mechanism. Interestingly, in preliminary experiments in our laboratory, growing selected isolates of E. coli, S. sanguis, and A. actinomycetemcomitans under both aerobic and anaerobic conditions did not affect HBD-2 or -3 antimicrobial activities (data not shown).
The resistance to antimicrobial β-defensins and other peptides may also be due to altered outer membrane proteins or altered lipopolysaccharide (LPS) structures (14, 31, 34). Recently, Brissette and Lukehart demonstrated that Treponema denticola, which lacks a traditional LPS (37), was naturally resistant to HBD-2 (3). The fact that LPS or lipooligosaccharide can be variable between strains of the same species (18) could partially explain the variable susceptibility pattern observed within a species, where, for example, F. nucleatum 49256 was very susceptible to the defensins tested compared to F. nucleatum 1594. Modification of such molecules by bacteria may be part of their strategy to evade the activities of antimicrobial peptides (18). For example, Starner et al. demonstrated that the susceptibility of H. influenzae to HBD-2 was influenced by lipooligosaccharide acylation of the membrane (40), which has been associated with P. aeruginosa resistance to cationic antimicrobial peptides (12).
Interestingly, P. gingivalis, one of the most resistant species in this study, is notorious for its production of a wide variety of proteolytic enzymes (5), which have been implicated in the inactivation of several known antimicrobial peptides (8).
The resistance demonstrated by certain anaerobic bacteria against the β-defensins could raise concern about the defensins' role as innate defenders in the development and progression of periodontitis. However, the strong antimicrobial activities displayed against the aerobic or early colonizers (i.e., S. sanguis, A. naeslundii, and A. israelii) could disrupt the environment and inhibit sequential colonization and development of the periodontal biofilm. In turn, this could then inhibit or prevent the colonization efforts of periodontal pathogens such as F. nucleatum, A. actinomycetemcomitans, P. gingivalis, and P. micros. In addition, the susceptibility of S. mutans and A. naeslundii to both defensins suggests that β-defensins may be important in preventing colonization of cariogenic organisms. Finally, the efficacy described for HBD-2 and -3 against multiple new medically important emerging species of Candida that are frequently isolated in immunosuppressed individuals shows promising application in the treatment of these opportunistic pathogens, which are becoming resistant to traditional antifungal therapy.
In summary, antimicrobial peptides are emerging as significant host defense and immune response molecules involved in mucosal defense. Their antimicrobial properties and the emergence of resistance to classical antibiotics have led investigators to study their potential as therapeutic agents. Most cationic peptides do not induce resistance in vitro and have the capacity to enhance the antimicrobial activity of classical antibiotics (20). Future studies evaluating individual strains among species with differing susceptibilities to the defensins could provide insight into the mechanisms of action for the specific defensins. This would be important, since their therapeutic applications will be based on their activities and limited by their resistance potential. It will also be important to evaluate the potential synergism of defensins and other innate molecules. Finally, evaluating individual functions of the β-defensins could assist in justifying the reported number of putative new defensins discovered in this family (38), since true redundancy of function is rare if not absent in nature.
Acknowledgments
This work was supported by Public Health Service grant 1RO1 DE13334.
We are grateful to David Beighton, Su Brailsford, and Robert Burne for providing some of the bacterial strains and to David Soll for providing some of the Candida strains. We are also grateful for scientific discussions with Kim Brogden and E. P. Greenberg about this project and for critical reading of the manuscript by Kim Brogden.
REFERENCES
- 1.Bals, R., X. Wang, Z. Wu, T. Freeman, V. Bafna, M. Zasloff, and J. M. Wilson. 1998. Human β-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Investig. 102:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Bissell, J., S. Joly, G. K. Johnson, C. C. Organ, D. Dawson, P. B. McCray, Jr., and J. M. Guthmiller. Expression of β-defensins in gingival health and in periodontal disease. J. Oral Pathol. Med., in press. [DOI] [PubMed]
- 2.Bonass, W. A., A. S. High, P. J. Owen, and D. A. Devine. 1999. Expression of beta-defensin genes by human salivary glands. Oral Microbiol. Immunol. 14:371-374. [DOI] [PubMed] [Google Scholar]
- 3.Brissette, C. A., and S. A. Lukehart. 2002. Treponema denticola is resistant to human β-defensins. Infect. Immun. 70:3982-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, A. M., T. Ganz, A. M. Liese, M. D. Burdick, L. Liu, and R. M. Strieter. 2001. IFN-Inducible ELR− CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623-627. [DOI] [PubMed] [Google Scholar]
- 5.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, and E. C. Reynolds. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 6.Dale, B. A., J. R. Kimball, S. Krisanaprakornkit, F. Roberts, M. Robinovitch, R. O'Neal, E. V. Valore, T. Ganz, G. M. Anderson, and A. Weinberg. 2001. Localized antimicrobial peptide expression in human gingiva. J. Periodontal Res. 36:285-294. [DOI] [PubMed] [Google Scholar]
- 7.Dale, B. A., and S. Krisanaprakornkit. 2001. Defensin antimicrobial peptides in the oral cavity. J. Oral Pathol. Med. 30:321-327. [DOI] [PubMed] [Google Scholar]
- 8.Devine, D. A., P. D. Marsh, R. S. Percival. M. Rangarajan, and M. A. Curtis. 1999. Modulation of antibacterial peptides activity by products of Porphyromonas gingivalis and Prevotella spp. Microbiology 145:965-971. [DOI] [PubMed] [Google Scholar]
- 9.Diamond, D. L., J. R. Kimball, S. Krisanaprakornkit, T. Ganz, and B. A. Dale. 2001. Detection of beta-defensins secreted by human oral epithelial cells. J. Immunol. Methods 256:65-76. [DOI] [PubMed] [Google Scholar]
- 10.Dunsche, A., Y. Acil, H. Dommisch, R. Siebert, J. M. Schroder, and S. Jepsen. 2002. The novel human beta-defensin-3 is widely expressed in oral tissues. Eur. J. Oral Sci. 110:121-124. [DOI] [PubMed] [Google Scholar]
- 11.Edgerton, M., S. E. Koshlukova, M. W. B. Araujo, R. C. Patel, J. Dong, and J. Bruenn. 2000. Salivary histatin 5 and human neutrophils defensins 1 kill Candida albicans via shared pathways. Antimicrob. Agents Chemother. 44:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 13.Ganz, T., and R. I. Lehrer. 1995. Defensins. Pharmacol. Ther. 66:191-205. [DOI] [PubMed] [Google Scholar]
- 14.Ganz, T. 2001. Fatal attraction evaded: how pathogenic bacteria resist cationic polypeptides. J. Exp. Med. 193:F31-F33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia, J. R., A. Krause, S. Schulz, F. J. Rodriguez-Jimenez, E. Kluver, K. Adermann, U. Forssmann, A. Frimpong-Boateng, R. Bals, and W. G. Forssmann. 2001. Human beta-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 15:819-1821. [PubMed] [Google Scholar]
- 16.Garcia, J. R., F. Jaumann, S. Schulz, A. Krause, J. Rodriguez-Jimenez, U. Forssmann, K. Adermann, E. Kluver, C. Vogelmeier, D. Becker, R. Hedrich, W. Forssmann, and R. Bals. 2001. Identification of a novel, multifunctional β-defensin (human β-defensin 3) with specific antimicrobial activity. Cell Tissue Res. 306:257-264. [DOI] [PubMed] [Google Scholar]
- 17.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human beta-defensin 1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 18.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides Cell. 95:189-196. [DOI] [PubMed] [Google Scholar]
- 19.Guthmiller, J. M., K. G. Vargas, R. Srikantha, L. L. Schomberg, P. L. Weistroffer, P. B. McCray, Jr., and B. F. Tack. 2001. Susceptibilities of oral bacteria and yeast to mammalian cathelicidins. Antimicrob. Agents Chemother. 45:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock, R. E. 1997. Peptide antibiotics. Lancet. 349:418-422. [DOI] [PubMed] [Google Scholar]
- 21.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 1997. A peptide antibiotic from human skin. Nature 387:861-862. [DOI] [PubMed] [Google Scholar]
- 22.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J. M. Schroder. 2000. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am. J. Respir. Cell. Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 23.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 24.Hoover, D. M., K. R. Rajashankar, R. Blumenthal, A. Puri, J. J. Oppenheim, O. Chertov, and J. Lubkowski. 2000. The structure of human β-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 42:32911-32918. [DOI] [PubMed] [Google Scholar]
- 25.Jia, H. P., B. C. Schutte, A. Schudy, R. Linzmeier, J. M. Guthmiller, G. K. Johnson, B. F. Tack, J. P. Mitros, A. Rosenthal, T. Ganz, and P. B. McCray Jr. 2001. Discovery of new human beta-defensins using a genomics-based approach. Gene 263:211-218. [DOI] [PubMed] [Google Scholar]
- 26.Krisanaprakornkit, S., A. Weinberg, C. N. Perez, and B. A. Dale. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect. Immun. 66:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kappaB transcription factor family. J. Immunol. 168:316-324. [DOI] [PubMed] [Google Scholar]
- 29.Lehrer, R. I., M. Rosenman, S. S. L. Harwig, R. Jackson, and P. Eisenhauer. 1991. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137:167-173. [DOI] [PubMed] [Google Scholar]
- 30.Liu, L., L. Wang, H. P. Jia, C. Zhao, H. H. Heng, B. C. Schutte, P. B. McCray, Jr., and T. Ganz. 1998. Structure and mapping of the human beta-defensin HBD-2 gene and its expression at sites of inflammation. Gene 222:237-244. [DOI] [PubMed] [Google Scholar]
- 31.Lysenko, E. S., J. Gould, R. Bals, J. M. Wilson, and J. N. Weiser. 2000. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expression in the upper respiratory tract. Infect. Immun. 68:1664-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray Jr. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyasaki, K. I., A. L. Bodeau, T. Ganz, M. E. Selsted, and R. I. Lehrer. 1990. In vitro sensitivity of oral, gram-negative, facultative bacteria to bactericidal activity of human neutrophils defensins. Infect. Immun. 58:3934-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rana, F. R., C. M. Sultany, and J. Blazyk. 1990. Interaction between Salmonella typhimurium lipopolysaccharide and the antimicrobial peptide, magainin 2 amide. FEBS Lett. 261:464-467. [DOI] [PubMed] [Google Scholar]
- 35.Schibli, D. J., H. N. Hunter, V. Aseyev, T. D. Starner, J. M. Wiencek, P. B. McCray, Jr., B. F. Tack, and H. J. Vogel. 2002. The solution structures of the human β-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 227:8279-8289. [DOI] [PubMed] [Google Scholar]
- 36.Schröder, J. M., and J. Harder. 1999. Hum. beta-defensin-2. Int. J. Biochem. Cell Biol. 31:645-651. [DOI] [PubMed] [Google Scholar]
- 37.Schultz, C. P., V. Wolf, R. Lange, E. Mertens, J. Wecke, D. Naumann, and U. Zahringer. 1998. Evidence for a new type of outer membrane lipid in oral spirochete Treponema denticola. Functioning permeation barrier without lipopolysaccharides. J. Biol. Chem. 273:15661-15666. [DOI] [PubMed] [Google Scholar]
- 38.Schutte, B. C., J. P. Mitros, J. A. Bartlett, J. D. Walters, H. P. Jia, M. J. Welsh, T. L. Casavant, and P. B. McCray, Jr. 2002. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh, P. K., H. P. Jia, K. Wiles, J. Hesselberth, L. Liu, B. D. Conway, E. P. Greenberg, E. V. Valore, M. J. Welsh, T. Ganz, B. F. Tack, and P. B. McCray. 1998. Production of beta defensins by human airway epithelia. Proc. Natl. Acad. Sci. USA 95:14961-14966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starner, T. D., W. E. Swords, M. A. Apicella, and P. B. McCray Jr. 2002. Susceptibility of nontypeable Haemophilus influenzae to human β-defensins is influenced by lipooligosaccharide acylation. Infect. Immun. 70:5287-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomita, T., S. Hitomi, T. Nagase, H. Matsui, T. Matsuse, S. Kimura, and Y., Ouchi. 2000. Effect of ions on antibacterial activity of human beta defensin 2. Microbiol. Immunol. 44:749-754. [DOI] [PubMed] [Google Scholar]
- 42.White, S. H., W. C. Wimley, and M. E. Selsted. 1995. Structure, function and membrane integration of defensins. Curr. Opin. Struct. Biol. 5:521-527. [DOI] [PubMed] [Google Scholar]
- 43.Whittaker, C. J., C. M. Klier, and P. E. Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]
- 44.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schröder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, C., I. Wang, and R. I. Lehrer. 1996. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 396:319-322. [DOI] [PubMed] [Google Scholar]