Abstract
Oral sodium phenyl butyrate (SPB) is currently under investigation as a histone deacetylation (HDAC) inhibitor in Huntington disease (HD). Ongoing studies indicate that symptoms related to HD genetic abnormalities decrease with SPB therapy. In a recently reported safety and tolerability study of SPB in HD, we analyzed overall chromatographic patterns from a method that employs gradient Liquid Chromatography with series Electrochemical array, UV and Fluorescence (LCECA/UV/F) for measuring SPB and its metabolite phenylacetate (PA). We found that plasma and urine from SPB-treated patients yielded individual-specific patterns of ca. 20 metabolites which may provide a means for the selection of subjects for extended trials of SPB. The structural identification of these metabolites is of critical importance, since their characterization will facilitate understanding the mechanisms of drug action and possible side effects. We have now developed an iterative process with LCECA, parallel LCECA/LCMS, and high performance tandem MS, for metabolite characterization. We report here the details of this method and its use for identification of 10 plasma and urinary metabolites in treated subjects, including indole species in urine that are not themselves metabolites of SPB. This approach thus contributes to understanding metabolic pathways that differ among HD individuals being treated with SPB.
Keywords: Sodium phenyl butyrate, Huntington disease, LCECA, metabolites, parallel LC-EC-array/LC-MS, tandem mass spectrometry
INTRODUCTION
Huntington disease (HD) is an inherited neurodegenerative disorder characterized by motor and psychiatric dysfunction such as choreic movements and dementia, with symptomatic onset occurring typically between 30 and 50 years of age. HD is caused by the expansion of an unstable CAG trinucleotide repeat located in the Huntington gene of affected individuals. This repeat transcribes a polyglutamine chain near the N-terminus of the huntingtin protein, and puts HD in the broader category of polyglutamine diseases. Polyglutamine chains longer than 36 glutamines result in a toxic, mutant form of the huntingtin protein and cause those individuals to invariably develop HD. Mutant huntingtin has been shown to disrupt activator-dependent transcription in the early stages of HD pathogenesis [1]. Additionally, transcriptional deregulation and functional loss of transcriptional co-activator proteins have been implicated in pathogenesis of HD [2; 3] in that they lead to neuronal loss and gliosis, particularly in the cortex and basal ganglia regions of the HD patient brain [4; 5].
Although there is no cure for HD, progress has been made in slowing the rate of neurodegeneration, as well as reducing or alleviating disease symptoms in genetic animal models. Studies carried out in cell culture, yeast, and Drosophila models of polyglutamine diseases have indicated that histone deacetylase (HDAC) inhibitors might provide a useful class of therapeutic agents for HD due to the association of histone acetylation with gene transcription [6; 7; 8; 9; 10]. Steffan and colleagues have shown that fragments of the mutant huntingtin protein interact with CREB-binding protein, decreasing the acetylation of histone 4 [7]. When a transgenic Drosophilia model that exhibited this protein-protein interaction was treated with an HDAC inhibitor, degeneration and early adult death were decreased [7]. These results suggest that reduced acetyl transferase activity may be an important component to polyglutamine disease pathogenesis. Thus, HDAC inhibitors may possibly be used to lessen the transcriptional changes in HD [11].
Sodium phenyl butyrate (SPB) has been explored as an HDAC inhibitor in clinical trials for cytostatic antineoplastic agents and has shown the ability to potentiate the effects of cytotoxic agents on tumors [12; 13]. SPB treatment has seemed promising due to its limited side effects in both phase I and phase II clinical trials, as well as its reported success in treating patients for urea cycle disorders, sickle cell anemia, thalassemia minor, and cystic fibrosis [14; 15; 16]. This evidence, in addition to recent results suggesting that the global reduction of HDAC activity slows the rate of neurodegeneration in in vivo models of HD [7; 9; 10; 17], suggested SPB as a therapeutic agent for HD.
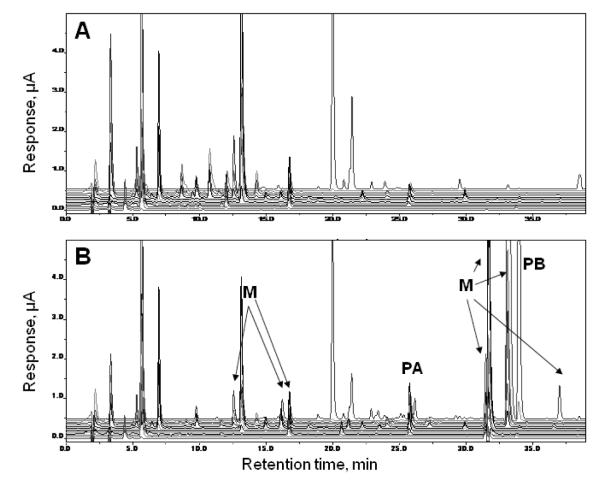
To understand the mechanisms of SPB drug action and possible individual-specific side effects in HD, it is essential to identify the structures of metabolites whose levels in urine and plasma samples differ between HD patients and/or controls. Results reported in published studies have identified metabolites of SPB in human plasma and urine [18; 19], and these studies have explored biochemical pathways SPB that could take in the body [18]. Literature data suggests that the patterns of SPB metabolites may vary for patients with different disorders. In recent studies of SPB therapy in Amyotrophic Lateral Sclerosis (ALS) and HD we developed an LCECA/UV/F (Liquid chromatography/Electrochemical array/Ultraviolet/Fluorescence) method targeted at SPB and its primary metabolite phenyl acetate (PA). We noted in the same subjects multiple quantifiable but qualitatively uncharacterized responses between subjects at baseline and on therapy. An example is shown in Figure 1. The evaluation of these qualitatively unknown compounds indicated significant differences between ALS and HD [20] subjects at similar dose levels. We undertook structural determinations of these compounds for two reasons. First, in order to devise possible biological mechanisms relating unknown peaks that correlate with drug effects, it is essential to know structures. Second, the LCECA/UV/F method is robust and relatively inexpensive and using it to capture additional information, beyond the simple pharmacokinetics of SPB and PA, would increase its utility in phenylbutyrate trials.
Figure 1.
LC-EC-array/UV/F method showing full 14-channel LC-EC-array/UV/Fluorescence-detected chromatograms of plasma from a patient, taken (A) before treatment and (B) on visit 6 after SPB-treatment. Metabolites of interest (M), phenylacetate (PA) and phenylbutyrate (PB) are labeled in B. This figure was generated directly from the CoulArray software.
Previous studies employed nuclear magnetic resonance (NMR) spectrometry to identify SPB metabolites in urine and included gas chromatography/mass spectrometry (GC/MS) analyses to confirm the assignments of several suspected metabolites [18]. Although these techniques have proven useful for identification of some novel metabolites, they have disadvantages in that they require large amounts of analyte (μg to mg of material, in the case of NMR), are limited to volatile analytes (GC/MS) or necessitate derivatization prior to GC/MS.
We recently demonstrated the use of parallel liquid chromatography electrochemical array with nanospray mass spectrometry (LC-EC-array-MS) to identify several SPB metabolites in the plasma of SPB-treated HD subjects [21]. HPLC coupled with electrochemical array detection (EC-Array) is a very sensitive technique used for profiling [22], analyzing and quantifying redox active species, even those that are present at low levels, down to picomolar concentrations [23; 24]. In addition to its sensitivity, this technique has the ability to quantify over 1000 components in biological samples in one HPLC analysis [25; 26], while also being able to differentiate among co-eluting species based on their oxidation potentials; this feature adds specificity to the analysis. Series coupling of ECA with UV and florescence provides a further mode of detection.The major limitation of the EC-Array is its inability to provide structural information on the analytes. In our recently reported study, this limitation was overcome by parallel coupling of HPLC with EC-array and MS [21], specifically nano-electrospray (nanoESI) on a quadrupole ion trap mass spectrometer. Nanospray introduces the sample in very fine droplets and is less subject to competition among components for favorable localization at the surface, with the result that the potential for ion suppression effects is minimized, and thus the coverage of even low abundance components is better than has been observed for high-flow analysis of complex biological mixtures [27]. Although the initial coupling required consideration of a number of parameters including the redox activity of a compound, solvent properties such as pH of the supporting electrolyte, and the LC flow rate, our results showed that it is possible to create a method that effectively satisfies the requirements of both the EC array and the mass spectrometer.
We present here an approach utilizing on-line and off-line LC-EC-array analysis to identify potential SPB metabolites and other unknown components detected in plasma and/or urine, that are unique (or significantly changed in concentration) when compared to controls. Structural elucidation of these compounds is achieved through use of a parallel coupling of LC-EC-array and ESI-MS, followed by off-line nanospray MS and tandem MS on a high resolution, high mass accuracy instrument. Plasma and urine samples were obtained from patients undergoing SPB treatment during a 4-month, double-blind, placebo-controlled, multicenter phase II study of SPB in individuals with early symptomatic HD (PHEND-HD). Among the components whose abundances were significantly higher in treated subjects, three novel and seven previously reported metabolites, not all originating directly from SPB, were isolated and their structures were determined. This allows their quantification on the basis of their responses in the LCECA/UV/F method used for routine monitoring of subjects and assignment of the observed components to their metabolic pathways. It also provides a more comprehensive approach towards achieving a full understanding of the biochemical effects of SPB as a therapeutic agent for HD and to classifying individual responses to the drug.
MATERIALS AND METHODS
Plasma and urine samples
Two hundred sixty-eight serial plasma and urine samples were obtained from 60 HD subjects in the PHEND-HD phase II study [28]. Baseline samples were obtained before treatment and additional samples were collected at five subsequent visits. The majority of subjects received the dose of 15 g SPB per day, however, some subjects received decreased dosage as a result of lower tolerance. Half the subjects were maintained on placebo through visit 2 and afterwards all subjects were on active medication until washout.
Initial database creation and analysis
For pharmacokinetic and compliance studies, all samples were analyzed using gradient LC with the series ECA/UV/F configuration optimized for ECA to resolve SPB and PA from ca. 600 plasma or urine metabolites at ng/mL levels. Gradient LC-EC analyses were performed using ESA model 582 Pumps (ESA Biosciences Inc., Chelmsford, MA) and a 14-channel ESA model 5600 CoulArray detector. Channels 1-12 used series Coulometric electrodes set in equal increments of 70 mV from 0-840 mV. Channel 13 measured UV absorbance at λ 210 nm. Channel 14 recorded florescence with excitation λ 220 nm/ emission λ 320 nm (ESA Biosciences Inc., Chelmsford, MA). A 4.6 mm × 250 mm Shiseido C18 5-μm column (ESA Biosciences Inc, Chelmsford, MA) was used. The gradient employed was linear from 0% Phase A (0.1 M LiPO4 pH 3) to 100% Phase B (0.1 M LiPO4 pH 3, 55% acetonitrile) over 45 min at the flow rate of 1 mL/min.
Plasma samples were prepared by a standard method, as follows. Plasma (125 μL) was precipitated with 500 μL of acetonitrile/0.4% acetic acid, vortexed for 30 sec, and centrifuged at 21,000 x g for 25 min at 4 °C. The supernatant (500 μL) was centrifugally evaporated and reconstituted to 100 μL in mobile phase A; a 50-μL aliquot was injected onto the system. Urine samples were diluted 1:5 with mobile phase A and 50 μL was directly injected onto the system. Figure 1 shows a typical example of the EC analysis of plasma from a subject before treatment and at visit 6, with areas of metabolites identified. All urine assays in the original data set were normalized to creatinine. Correlation of creatinine versus individual compounds was performed to verify that it was an effective normalizer. Plasma and urine samples were obtained at Massachusetts General Hospital (MGH), under IRB approvals. The primary report was restricted to SPB and PA, however, the characteristics of all resolved peaks were organized into a database for future analysis.
Procedure for selecting peaks for identification
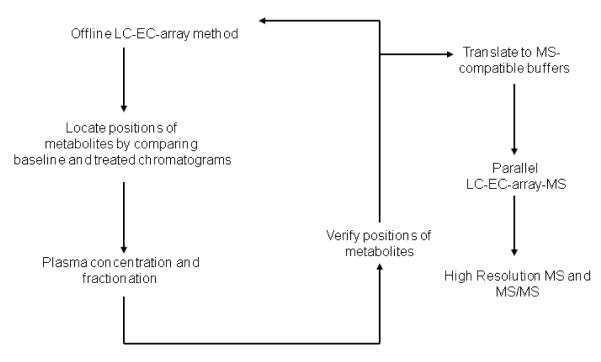
From the created database, components related to SPB and PA were selected for identification as follows: Patterns of all analytes were exported as peak tables and as digital maps, following protocols developed in a study of Parkinson Disease [22]. Peak tables contain entries for known compounds as concentrations (mol/L). Qualitatively unknown compounds are entered as relative values versus a pool of all samples in a study and identified by the channel on which they show maximum response and by time (e.g., channel 7 – 21.98 min). Digital maps are created by exporting data as digital values every 1.5 seconds and organizing these values in a column by ascending channel. These capture the entirety of the response of the platform [22] and allow definition of differences among classes (treated versus un-treated) in areas of the platform response where peak detection algorithms are not effective. Maps and peak tables were compared for baseline vs. dosed subjects to determine levels of direct (metabolite of the drug) or induced (metabolite significantly changed by the drug) metabolites. The criteria for selection of candidates for structural identification were that their chromatographic peaks (A) had levels 50 times greater in dosed subjects vs. subjects at baseline before administration of the drug, (B) showed correlation with either SPB or PA (Pearson coefficient of a compound greater than 0.65 versus SPB or PA), and (C) demonstrated consistency within an individual subject. Twenty peaks met these criteria. A scheme depicting the process used for concentration, isolation and identification of the peaks is presented in Figure 2.
Figure 2.
Flow chart showing the course of samples from initial offline LC-EC-array screening through sample fractionation, parallel LC-EC-array-MS analysis and high resolution and MS/MS characterization.
Preparation of plasma fractions for offline LC-EC-array metabolite identification
Pools of plasma from baseline and SPB-treated subjects were prepared from 200-μL sub-aliquots. Six mL of plasma pools from both baseline and treated HD subjects were precipitated with 24 mL of acetonitrile (ACN) containing 0.4% glacial acetic acid at 20 °C. Samples were vortexed for 30 sec and centrifuged for 30 min at 8000 x g at 4 °C. The supernatant was centrifugally vacuum evaporated and concentrated to 300 μL.
Fractionation of plasma samples
Concentrated plasma was fractionated using solid phase extraction (500 mg Diazem C-18 SPE, Diazem Corp. Midland, MI). Columns were equilibrated with 2 mL deionized water, 2 mL acetonitrile and 2 mL 2% acetic acid. Concentrated supernatant from the plasma preparation (300 μL) was loaded onto the SPE column. A single 300-μL collection was made to recover the void fraction and then 2 mL of each of the following eluants was collected: 0%, 10%, 20%, 30%, 40%, and 100% ACN. Each of the fractions was centrifugally evaporated and reconstituted in 200 μL of 0.02 M ammonium acetate, pH 4.2.
Fractionation of urine samples
Pools of urines (7.5 mL) created from the 15 subjects who showed the highest levels of SPB at visit 5 were compared to urine pools that had been obtained from the same subjects at baseline (before treatment). Since several peaks of interest were detected with electrochemical sensors at levels ca. 1-5 ng/mL, samples with highest levels of SPB were selected in order to obtain sufficient material for MS identification. These were concentrated by centrifugal evaporation to reduce the volume by 75%. As above, aliquots (300 μL) were separated on the same type of SPE columns using the same sequence of acetonitrile concentrations and volumes and the fractions were centrifugally evaporated and reconstituted.
Offline LC-EC-array Run of Fractions in MS Buffer
A 10-μL volume from each fraction was diluted to 100 μL. First, 20 μL were analyzed using the primary analytical method developed for initial database creation to confirm the position and signal intensity of the peaks selected for structural identification from the initial data analysis. Next, the solvent gradient developed for the LCECA/UV/F platform was modified to utilize MS-compatible buffer systems (1) Phase A: 0.02 M ammonium formate/formic acid, pH 3.1; Phase B: 0.02 M ammonium formate/formic acid, pH 3, 55% ACN; gradient 0% A-100% B over 45 min and (2) Phase A: 0.02 M ammonium acetate/acetic acid, pH 4.2; Phase B: 0.1 M ammonium acetate/acetic acid, pH 4.2, 55% ACN; gradient 0% A-100% B over 45 min). The flow rate was 1.0 mL/min. The selected peak locations and intensities were confirmed in this system, using 20- μL injections.
Due to the presence of numerous compounds of interest in urine fractions 0%, 10% and 30% ACN, these were re-extracted on Diazem C-18 SPE columns with smaller increments over the same range of ACN levels (0%-20% in 2% increments). These fractions were once again analyzed on the offline LCECA/UV/F to verify the presence of the metabolites of interest from their time and channel responses.
Parallel LC-EC-array-MS instrumentation
LC-MS analyses were performed using ESA model 582 Pumps (ESA Biosciences Inc., Chelmsford, MA) and an ESA model 5600 CoulArray detector; channels 1-12, 0-840 mV in 70- mV increments (ESA Biosciences Inc., Chelmsford, MA) coupled on-line to a QStar quadrupole orthogonal time-of-flight (Q-o-TOF) mass spectrometer (Sciex/Applied Biosystems, Foster City, CA) equipped with an ESI ion source. We sequentially used positive- and negative-ion scan modes (m/z 100-2000, ionspray voltage 4.5-5.5 kV). Metabolite mixtures were separated on a 4.6 mm x 250 mm (5-μm Shiseido C18) column. The flow rate was varied between 0.5-1.0 mL/min to optimize peak width and minimize noise levels for MS detection. The HPLC eluent was then split at a ratio of 9:1, with 90% being directed to the EC-array and 10% being delivered to the MS.
LC-EC-array-MS method
The 20% and 30% plasma fractions were diluted between 1:10 and 1:100 in 60:40 mobile phase A:B. Dilutions were based on the relative amounts of the compounds of interest and the requirement to (1) perform multiple different runs with varying LC and MS parameters and (2) to preserve material for subsequent MS/MS studies. Samples were assayed using a 20-min isocratic method with a total flow rate of 0.5 mL/min. Mobile phase A (0.1 M ammonium acetate) was delivered at 0.3 mL/min and mobile phase B (60% ACN/0.1 M ammonium acetate) was delivered at 0.2 mL/min. An Information Dependent Acquisition (IDA) MS method was designed to monitor the two most intense ion signals in the range m/z 100-500 and to fragment each of these components with the collision energy set to 20 eV and the quadrupole set to low resolution. With this method, it was possible to monitor the retention times of the metabolites of interest as they passed through the mass spectrometer, to compare the times to those of the peaks detected by the simultaneous EC-array analysis, to obtain initial values for parent masses of the compounds and to obtain the relevant MS/MS fragmentation. The above method was also used for analysis of the 0 and 10% urine fractions, and for the 20% ACN re-extracted portion from the 0% urine fraction and the 15% ACN re-extracted portion from the 10% urine fraction.
Additionally, the plasma fraction from the 40% ACN fractionation was diluted as above and assayed using a 60-min isocratic method. A total flow rate of 1.0 mL/min delivered at 0.9 mL/min A (0.1 M ammonium acetate) and 0.1 mL/min B (75% ACN/0.1 M ammonium acetate). The specific mobile phase compositions were chosen to optimize the stability of the electrospray in the Q-o-TOF MS. The IDA MS method was similar to that used above, but the collision energy was set at 50 eV. The higher collision energy was necessary to fragment the compound of interest in this fraction.
Assignment of fragmentation by MS and MS/MS
The parallel LCEC-LCMS method performed on the Q-o-TOF mass spectrometer allowed us to determine the mass of the metabolites of interest qualitatively located in the offline LCECA/UV/F method. This preliminary MS data allowed us to focus on these particular masses while obtaining exact masses using a high resolution, high mass accuracy instrument.
Plasma samples
The assignments for the compounds found in plasma samples were made on the basis of high resolution MS measurements of the molecular ions with the Orbitrap and MS/MS fragmentation in the quadrupole ion trap of the LTQ Orbitrap “Discovery” instrument (Thermo-Fisher, Inc., San Jose, CA) equipped with a with NanoMate TriVersa robot (Advion Corp., Ithaca, NY). The MS and MS/MS data sets were obtained for samples from the 20%, 30%, and 40% ACN fractions that were diluted 1:10 in 50/50 methanol/water and analyzed using nanoelectrospray in the negative-ion mode. Detection of intact molecular ions in the Orbitrap was obtained with mass resolution 1:30,000. The mass measurement accuracy was better than 5 ppm. Fragmentation was performed in the linear ion trap with the normalized collision energy for collision-induced decomposition (CID) set at 50.
Urine samples
As indicated in Figure 1, the results from after the initial LC-EC-array screening of the sample fractions were used to select the fractions containing peaks not present in baseline samples for analysis via parallel LC-EC-array-MS. Preliminary MS/MS spectra for the components in the urine fractions were acquired during the earlier study that used an LCQ Classic quadrupole ion trap MS (Thermo-Fisher, Inc., San Jose, CA) [21]. In the present study, samples re-extracted from the 0% and 10% urine fractions were diluted 1:10 in 70/30 methanol/water and run by nanoelectrospray in the negative-ion mode. High resolution MS/MS spectra for these compounds were measured using the QStar and LTQ Orbitrap mass spectrometers.
RESULTS
The overall analytical method developed here provides a comprehensive evaluation of plasma and urine samples obtained from SPB-treated HD patients, by using on-line EC-array and MS detection both separately and in parallel, and off-line high performance mass spectrometry. A parallel LC-EC-array-MS detection system was used to analyze samples obtained from SPB-treated HD patients taking part in the PHEND-HD tolerability trial study. The system consists of a binary HPLC pump connected to a normal bore C-18 column followed by a 9:1 passive flow splitter used to divide the eluant between the EC-array and MS detectors. The MS flow rate was maintained <100 μL/min in order to minimize possible ion suppression effects from both the biological samples and the high salt containing EC-array buffers and facilitate efficient ion transfer. Additionally, the flow split was important for preserving agreement of the retention times between the EC-array and the MS chromatograms in order to allow confident comparison between the two instruments and identification of SPB metabolites [21]. As in prior work, the transfer lines were set up such that arrival times at the EC detector and the MS detector would be simultaneous (within 1-2 sec). The components determined to be of interest were further characterized through the combination of high resolution MS measurements and CID tandem mass spectrometry (MS/MS).
The entire metabolite characterization schema, from sample processing and initial offline LC-EC-array profiling to parallel LC-EC-array-MS analyses and final high resolution MS and MS/MS characterization, is illustrated in Figure 2. As shown in this figure, the first step after preparing the samples with a simple protein precipitation and concentration of the supernatant is efficient separation of all electrochemically active compounds and distinction between the possible SPB metabolites and other components whose concentrations are enhanced in response to SPB treatment, by comparing baseline, or pre-treatment patient samples with treated patient samples. In addition to profiling the EC-active metabolites, the system used for this offline LC-EC-array analysis also monitored the samples with UV and fluorescence (F), out of consideration for SPB’s lack of electrochemical activity, but strong UV and F signals.
Figure 1 shows a typical comparison of two offline LC-EC-array chromatograms from the set used to detect SPB metabolites from HD patients. The upper panel of the figure, indicated as baseline, shows the LC-EC-array chromatogram of pooled plasma from HD patients prior to SPB administration. The bottom trace, marked as Visit 6, shows the LC-EC array chromatogram of pooled plasma obtained at the patients’ sixth visit, approximately four months into the study of the results from SPB administration. There are distinct changes in the chromatograms; peaks marked with the letter “M” in Figure 1 represent new metabolites formed as a result of the SPB drug treatment. In order to facilitate structural characterization of these metabolites, it was necessary to reduce the number of compounds being studied at one time. Thus, in the next stage of our methodology (Figure 2), the concentrated and pooled HD patient samples were fractionated using SPE. The 10%, 20%, 30%, 40% and 100% ACN fractions were collected from a Diazam SPE C-18 column and were subsequently analyzed with both the parallel LC-EC-array-MS and offline LC-EC-array systems. The fractions were re-run on the offline LC-EC-array system in order verify the positions of the metabolite peaks in comparison to the un-fractionated sample chromatogram.
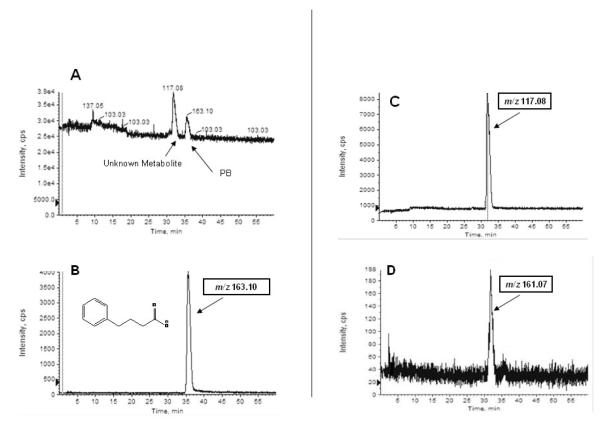
After the initial LC-EC-array screening of the plasma fractions, a potential SPB metabolite previously observed in the un-fractionated sample was found to be isolated in the 40% ACN sample. This compound eluted at 32 min in the chromatogram shown in Figure 1B. To identify this component, a parallel LC-EC-array-MS analysis was then performed on the 40% fraction. Figure 3 shows the LC-EC-array chromatogram recorded for the 40% ACN fraction obtained from the parallel LC-EC-array-MS system. The LC-EC-array retention time of the 40% peak was again 32 min, and this corresponded exactly to the retention time of a peak observed by MS. (Panel A of Figure 4). The mass spectrum obtained at 32 min in the IDA mode (Figure 4) showed peaks at m/z 117.08 and at m/z 161.07. Together with the extracted ion chromatograms (XIC) of these m/z values (Panels C and D Figure 4), this mass spectrum indicated that the [M-H]− of the compound was m/z 161.07 and that the peak observed at m/z 117.08 was a product obtained by fragmentation of this parent ion. Although, as indicated previously, SPB itself is not electrochemically active and thus no SPB peak was observed the LC-EC-array chromatogram, the elution of this component was detected as a maximum in the MS chromatogram of its [M-H]−m/z 163.08, at 35.4 min (Panel B Figure 4) and a signal at m/z 163 was detected in mass spectra obtained on either side of the maximum. The combined capabilities for monitoring both the EC-active and inactive compounds via parallel EC-array and MS detection makes the metabolite detection methodology developed here a powerful tool for unknown metabolite identification.
Figure 3.
LC-EC-array chromatograms of the 40% ACN fraction collected from the Diazam C-18 column. Elution of a metabolite of interest is indicated at the retention time of 32 minutes. PB is not EC active.
Figure 4.
(A) QStar Q-o-TOF MS total ion chromatogram of the 40% ACN plasma fraction containing phenylbutyrate (PB) and an unknown SPB metabolite. Other panels show single ion chromatograms of (B) m/z 163, corresponding to the [M-H]− of PB, and (C) m/z 117 and (D) m/z 161) which both displayed maxima at the retention time (32 min) of the unknown metabolite.
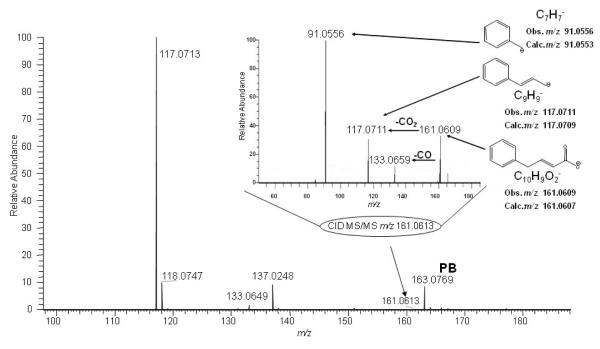
To obtain high resolution MS data to enable determination of the structure of the metabolite that has apparent [M-H]−m/z 161.07, on the basis of its accurate mass and MS/MS fragmentation pattern, the 40% fraction was analyzed via infusion using nanoelectrospray on an LTQ-Orbitrap MS. Figure 5 shows the full mass spectrum of the 40% fraction in the range m/z 100-200. The peak at m/z 163.0769 corresponds to the [M-H]− of the parent drug PB ([C10H11O2]−, calc. m/z 163.0764), which had its elution maximum in the following fraction but was present at as a low level contaminant in this fraction. For the spectrum shown in Figure 5, the ions observed at m/z 117.0713 and 161.0613 were initially attributed to unknown metabolite(s). The inset of Figure 5, shows the MS/MS spectrum obtained upon selection and fragmentation of the ion observed at m/z 161.0613. Accurate mass measurement of the abundant fragment at m/z 117.0711 showed that it corresponds to loss of CO2 from the [M-H]− and reinforced the proposition that the peak observed at m/z 161.0 in the parallel LC-EC-array-MS experiment corresponded to a metabolite of SPB. Additional fragment ions were observed; taken together, these allowed elucidation of the detailed structure of the metabolite. At the right side of the inset are shown the structures proposed for these abundant fragments, including the fragment observed at m/z 133.0659 that results from loss of a CO group, and the ion at m/z 91.0556, which can be assigned as the result from cleavage adjacent to the carbon located in the α-position with respect to the benzene ring. The elemental compositions determined here for the precursor and product ions of metabolite b led to its assignment as another known SPB metabolite, 4-phenyl-trans-crotonate, whose structure had been determined in SPB studies that investigated the mechanism of 3-hydroxy-4- phenylbutyrate formation in SPB-perfused rat livers [18]. However, this metabolite has not previously been reported in any human studies; its detection here provides new insight into the metabolism of SPB by these HD patients.
Figure 5.
Orbitrap nanospray MS spectrum of 40% ACN fraction collected from Diazam C-18 column at 32 min. Ions assigned to the unknown SPB metabolite (m/z 161.0613, 117.0713) are circled. The ([M-H]−m/z 163.0769) peak indicates presence of a the leading edge of the peak corresponding to the parent drug PB which reached its maximum in the following LC fraction. Inset shows Orbitrap MS/MS spectrum of the unknown metabolite, [M-H]−m/z 161.0609. Proposed structures are indicated for the metabolite selected as precursor and its abundant fragment ions.
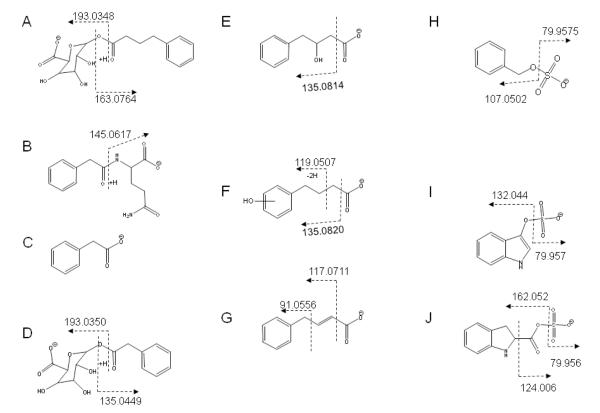
The same combined approaches were then applied to the remaining fractions of interest, originating from both plasma and urine. As described above for the 40% ACN plasma fraction, these samples were injected on the parallel LC-EC-array-MS system, with the MS being operated in the negative-ion mode. Since these fractions were more polar than both the identified 4-phenyl-trans-crotonate metabolite and the parent SPB drug, they eluted earlier than these compounds and it was only necessary to record data for 20 min. Each subsequent fraction was analyzed in the same manner and the results were examined with regard to the presence of components showing identical EC-array and MS retention times. The Orbitrap mass spectra recorded at the corresponding retention times provided high accuracy m/z values for the [M-H]− signals. All the unknown components of interest were subjected to high resolution MS analysis and/or CID MS/MS fragmentation. Tables 1 and 2 show the structures and other data pertinent to each of the unique metabolites found in the selected ACN fractions recovered by SPE from the total collected HD patient samples, plasma and urine, respectively. Using the approaches described in this study, we assigned these peaks as seven metabolites that have been previously reported in the literature [18; 21] and three previously undescribed metabolites (H, I, J in Figure 6). The approach used for identification of these metabolites and determinations of their molecular weights allowed correlation in time and EC response with the screening method. In the absence of authentic standard reference materials, the estimates of their concentrations were be made from the Coulometric response in the LCECA [17].
Table 1.
Metabolites found in SPB-treated HD Patient Plasma
| Plasma Metabolite, Instrument used for analysis |
[Ml-H]− | Plasma Fraction | Major Fragments observed m/z, El. Comp., error (ppm) |
|---|---|---|---|
|
(A) Phenylbutyryl-β-glucuronate [18] LTQ Orbitrap (MS and MS/MS) |
339.1083 (Obs) 339.1086 (Calc) −0.9 ppm |
20% ACN | 193.0348 [C6H9O7]− −2.6 175.0243 [C6H7O6]− −2.3 163.0764 [C10H11O2]− 0.0 113.0243 [C5H5O3]− −0.9 |
|
(B) Phenylacetylglutamine [18] LTQ Orbitrap (MS and MS/MS) |
263.1031 (Obs) 263.1037 (Calc) −2.3 ppm |
20% ACN | 245.0928 [C13H13N2O] −0.8 145.0617 [C5H9N2O3]− +0.8 |
|
(C) Phenylacetate [18] LTQ Orbitrap (MS) |
135.0455 (Obs) 135.0452 (Calc) + 2.2 ppm |
20% ACN | (solvent interference precluded MS/MS) |
|
(D) Phenylacetyl-p-glucuronate [18] LTQ Orbitrap (MS and MS/MS) |
311.0771 (Obs) 311.0773 (Calc) −0.6 ppm |
20% ACN | 193.0350 [C6H9O7]− −1.6 175.0245 [C6H7O6]− −1.1 135.0449 [C8H7O2]− −1.5 113.0244 [C5H5O3]− 0.0 |
|
(E) 3-hydroxy-4-phenylbutyrate [18] LTQ Orbitrap (MS and MS/MS) |
179.0717 (Obs) 179.0713 (Calc)+2.2 ppm |
20% ACN, 30% ACN | 161.0605 [C10H9O2]− −1.2 135.0814 [C9H11O]− 0.7 133.0658 [C9H9O]− 0.0 119.0502 [C8H7O]− 0.0 117.0717 [C9H9]− +6.8 |
|
(F) Hydroxyphenylbutyric acid(s) 3 isomers [21] LTQ Orbitrap (MS and MS/MS) |
179.0719 (Obs) 179.0713 (Calc) + 3.4 ppm |
30% ACN | 135.0820 [C9H11O]− +9.6 119.0507 [C8H7O]− +4.2 106.0430 [C6H5O]− +5.7 59.0143 [C2H3O2]− +8.5 |
|
(G) 4-phenyl-frans-crotonate [18] LTQ Orbitrap (MS and MS/MS) |
161.0609 (Obs) 161.0607 (Calc) + 1.5 ppm |
40% ACN | 133.0659 [C9H9O] −0.7 117.0711 [C9H9]− +1.7 91.0556 [C7H7]− +3.3 |
|
(H) Benzyloxyl sulfate LTQ Orbitrap (MS and MS/MS) |
187.0071 (Obs) 187.0070 (Calc) + 0.5 ppm |
20% ACN | 107.0502 [C7H7O]− 0.0 79.9575 [SO3]− +2.5 |
Table 2.
Metabolites found in SPB-treated HD Patient Plasma
| Urine Metabolite Instrument used for analysis |
[M-H]− | Urine Fraction | Major Fragments Obs. m/z, El.Comp., error (ppm) |
|---|---|---|---|
|
(B) Phenylacetylglutamine [18] LTQ Orbitrap (MS), QStar (MS/MS) |
263.1036 (Obs) 263.1037 (Calc) −0.4 ppm |
0% ACN (20% re-extracted) |
m/z 145.0619 [C5H9N2O3]− +0.7 m/z 127.0545 [C5H7N2O2]− +26.0 |
|
(E) 3-hydroxy-4-phenylbutyrate [18] LTQ Orbitrap (MS and MS/MS) |
179.0715 (Obs) 179.0714 (Calc) +0.6 ppm |
10% ACN |
m/z 161.0605 [C10H9O2]− −1.2 m/z 135.0814 [C9H11O]− −0.7 m/z 133.0658 [C9H9O]− 0.0 m/z 119.0502 [C8H7O]− 0.0 m/z 117.0717 [C9H9]− +6.8 |
|
(H) Benzyloxyl sulfate LTQ Orbitrap (MS and MS/MS) |
187.0071 (Obs) 187.0070 (Calc) + 0.5 ppm |
10% ACN (15% re-extracted) |
m/z 107.0502 [C7H7O]− 0.0 m/z 79.9575 [SO3]− +2.5 |
|
(I) 3-lndoxyl sulfate LTQ Orbitrap (MS), QStar (MS/MS) |
212.0023 (Obs) 212.0022 (Calc) + 0.5 ppm |
10% ACN (15% re-extracted) |
m/z 132.044 [C8H6NO]−−7.6 m/z 79.957 [SO3]− 0.0 |
|
(J) 3-carboxy sulfate indoline LTQ Orbitrap (MS), QStar (MS/MS) |
242.0125 (Obs) 242.0128 (Calc) −1.2 ppm |
10% ACN (15% re-extracted) |
m/z 162.052 [C9H8NO2]− −24.7 m/z 79.956 [SO3]−−12.5 |
Figure 6.
Structures and fragments of metabolites found in SPB-treated HD patient plasma and urine. Structures and fragments correspond to those listed in Tables 1 and 2.
DISCUSSION
Disease- and individual-specific metabolism of SPB
The process of utilizing parallel LC-EC-array/LC-MS to qualitatively identify both directly derived metabolites of SPB and metabolites that are significantly modified by SPB provided a the further qualification of the standard LCECA/UV/F method used in the ALS and HD trials. Evidence suggests that different individuals and subjects with different disorders tolerate and respond differently to the drug. The ability to monitor these differences biochemically expands the utility of the method in trials of SPB.
The metabolites presented in Tables 1 and 2 reflect various types of SPB metabolism. These include the pathways for enzymatic conversion to PA, enzymatic and non-enzymatic pathways related to oxidative stress, kidney and liver function that result in sulfonation and glucuronidation and suggest a possible effect on indole metabolism. Each of these processes can be postulated to be different among disorders and within individuals over time. Compounds E and G are side products of CoA-mediated metabolism to PA. The hydroxyphenyl butyrates (F in Figure 6) can be enzymatically derived or can result from the direct hydroxylation that occurs as a result of the high level of free radical production and oxidative stress in HD [29]. The glucuronide levels (A and D in Figure 6) and sulfonated compounds (H, I, J in Figure 6) reflect modified patterns of kidney and liver function.
Documentation of either positive or adverse patient reactions to SPB treatment and correlation of these reactions to each patient’s individual metabolite patterns can lead to definition of biomarkers for drug response. Once defined, these biomarkers may assist in predicting how individual patients will react to SPB treatment based on the presence and concentration of metabolites shown to uniquely correlate with a positive or negative response to the drug. These biomarkers can be identified as either metabolites of SPB itself, such as glucuronide conjugates, or molecules from other metabolic pathways that are being affected by SPB treatment.
The indole sulfonates (I and J in Figure 6), found in the urine of only the HD subjects treated with SPB, point to a possible role of SPB in modulating indole pathways. The formation of oxidized indoles has been shown to lead to toxicity in neurodegenerative diseases [30; 31; 32]; for example, aberrations in the tryptophan pathway are well known to occur in HD [33; 34]. Ultimately, the neurotoxicity is caused by aggregates formed from proteins crosslinked by oxidized indoles [35; 36; 37; 38; 39]. In particular, the intermediate free radical indole species formed by hydroxyl radical attack have been implicated in protein aggregation [32; 40], and may be involved in crosslinking mechanisms similar to those of the polyglutamines which are elevated in HD [41]. While mechanistic studies were not the primary focus of this work, the excretion of elevated levels of indoles from SPB-treated HD patients may indicate a secondary mechanism of SPB which modulates the potentially neurotoxic effect of indolic species.
CONCLUSIONS
We have demonstrated a systematic process for unknown metabolite identification using EC-array and MS detection, both separately and in parallel. The process was applied to the study of SPB metabolism in HD patients. By applying this process, we have expanded the capability of a method, originally developed for simply evaluating SPB and primary PA metabolite levels, to now include compounds that reflect multiple modes of SPB metabolism and effects on metabolic pathways which may result from both disease and individual-specific processes. Most importantly, application of the process to this sample set yielded the unexpected outcome of finding increased excretion of indole species as a consequence of therapy. The previously unreported elevation of these metabolites as a result of SPB therapy may reflect both the disease processes in HD and secondary effects of the therapeutic intervention in combination with HDAC processes.
ACKNOWLEDGEMENTS
This research was supported by NIH grants P41 RR010888 (CEC), S10 RR020946 (J. Zaia), R33 DK070326 (WRM), P01 NS045242 (SH), and the HIQ Foundation (SH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296:2238–43. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- [2].Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–33. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- [3].Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–8. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- [4].Imarisio S, Carmichael J, Korolchuk V, Chen CW, Saiki S, Rose C, Krishna G, Davies JE, Ttofi E, Underwood BR, Rubinsztein DC. Huntington’s disease: from pathology and genetics to potential therapies. Biochem J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- [5].Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–84. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- [6].McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci U S A. 2001;98:15179–84. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–43. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- [8].Hughes RE, Lo RS, Davis C, Strand AD, Neal CL, Olson JM, Fields S. Altered transcription in yeast expressing expanded polyglutamine. Proc Natl Acad Sci U S A. 2001;98:13201–6. doi: 10.1073/pnas.191498198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, Marsh JL, Thompson LM, Lewis CM, Marks PA, Bates GP. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100:2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, Smith K, Kowall NW, Ratan RR, Luthi-Carter R, Hersch SM. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;23:9418–27. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, Gao F, Fitzgerald KM, Borok JF, Herman D, Geschwind DH, Gottesfeld JM. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105:15564–9. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Samid D, Hudgins WR, Shack S, Liu L, Prasanna P, Myers CE. Phenylacetate and phenylbutyrate as novel, nontoxic differentiation inducers. Adv Exp Med Biol. 1997;400A:501–5. doi: 10.1007/978-1-4615-5325-0_67. [DOI] [PubMed] [Google Scholar]
- [13].Gilbert J, Baker SD, Bowling MK, Grochow L, Figg WD, Zabelina Y, Donehower RC, Carducci MA. A phase I dose escalation and bioavailability study of oral sodium phenylbutyrate in patients with refractory solid tumor malignancies. Clin Cancer Res. 2001;7:2292–300. [PubMed] [Google Scholar]
- [14].Shin HJ, Baek KH, Jeon AH, Kim SJ, Jang KL, Sung YC, Kim CM, Lee CW. Inhibition of histone deacetylase activity increases chromosomal instability by the aberrant regulation of mitotic checkpoint activation. Oncogene. 2003;22:3853–8. doi: 10.1038/sj.onc.1206502. [DOI] [PubMed] [Google Scholar]
- [15].Dover GJ, Brusilow S, Charache S. Induction of fetal hemoglobin production in subjects with sickle cell anemia by oral sodium phenylbutyrate. Blood. 1994;84:339–43. [PubMed] [Google Scholar]
- [16].Rubenstein RC, Zeitlin PL. A pilot clinical trial of oral sodium 4-phenylbutyrate (Buphenyl) in deltaF508-homozygous cystic fibrosis patients: partial restoration of nasal epithelial CFTR function. Am J Respir Crit Care Med. 1998;157:484–90. doi: 10.1164/ajrccm.157.2.9706088. [DOI] [PubMed] [Google Scholar]
- [17].Sadri-Vakili G, Cha JH. Histone deacetylase inhibitors: a novel therapeutic approach to Huntington’s disease (complex mechanism of neuronal death) Curr Alzheimer Res. 2006;3:403–8. doi: 10.2174/156720506778249407. [DOI] [PubMed] [Google Scholar]
- [18].Kasumov T, Brunengraber LL, Comte B, Puchowicz MA, Jobbins K, Thomas K, David F, Kinman R, Wehrli S, Dahms W, Kerr D, Nissim I, Brunengraber H. New secondary metabolites of phenylbutyrate in humans and rats. Drug Metab Dispos. 2004;32:10–9. doi: 10.1124/dmd.32.1.10. [DOI] [PubMed] [Google Scholar]
- [19].Comte B, Kasumov T, Pierce BA, Puchowicz MA, Scott ME, Dahms W, Kerr D, Nissim I, Brunengraber H. Identification of phenylbutyrylglutamine, a new metabolite of phenylbutyrate metabolism in humans. J Mass Spectrom. 2002;37:581–90. doi: 10.1002/jms.316. [DOI] [PubMed] [Google Scholar]
- [20].Cudkowicz ME, Andres PL, Macdonald SA, Bedlack RS, Choudry R, Brown RH, Jr., Zhang H, Schoenfeld DA, Shefner J, Matson S, Matson WR, Ferrante RJ. Phase 2 study of sodium phenylbutyrate in ALS. Amyotroph Lateral Scler. 2008:1–8. doi: 10.1080/17482960802320487. [DOI] [PubMed] [Google Scholar]
- [21].Schiavo S, Ebbel E, Sharma S, Matson W, Kristal BS, Hersch S, Vouros P. Metabolite identification using a nanoelectrospray LC-EC-array-MS integrated system. Anal Chem. 2008;80:5912–23. doi: 10.1021/ac800507y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bogdanov M, Matson Wayne R, Wang L, Matson T, Saunders-Pullman R, Bressman Susan S, Flint Beal M. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain FIELD Full Journal Title:Brain : a journal of neurology. 2008;131:389–96. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- [23].Gamache PH, McCabe DR, Parvez H, Parvez S, Acworth IN. The measurement of markers of oxidative damage, antioxidants and related compounds using HPLC and coulometric array analysis. Progress in HPLC-HPCE. 1997;6:99–126. [Google Scholar]
- [24].Milbury PE. CEAS generation of large multiparameter metabolic databases for determining categorical process involvement of biological molecules. Progress in HPLC-HPCE. 1997;6:127–144. [Google Scholar]
- [25].Kristal BS, Vigneau-Callahan K, Matson WR. Simultaneous analysis of multiple redox-active metabolites from biological matrices. Methods Mol Biol. 2002;186:185–94. doi: 10.1385/1-59259-173-6:185. [DOI] [PubMed] [Google Scholar]
- [26].Kristal BS, Vigneau-Callahan KE, Matson WR. Simultaneous analysis of the majority of low-molecular-weight, redox-active compounds from mitochondria. Anal Biochem. 1998;263:18–25. doi: 10.1006/abio.1998.2831. [DOI] [PubMed] [Google Scholar]
- [27].Gangl ET, Annan M, Spooner N, Vouros P. Reduction of signal suppression effects in ESI-MS using a nanosplitting device. Anal Chem. 2001;73:5635–5644. doi: 10.1021/ac010501i. [DOI] [PubMed] [Google Scholar]
- [28]. http://www.clinicaltrials.gov/ct2/show/NCT00212316?term=00212316&rank=1.
- [29].Hersch SM, Gevorkian S, Marder K, Moskowitz C, Feigin A, Cox M, Como P, Zimmerman C, Lin M, Zhang L, Ulug AM, Beal MF, Matson W, Bogdanov M, Ebbel E, Zaleta A, Kaneko Y, Jenkins B, Hevelone N, Zhang H, Yu H, Schoenfeld D, Ferrante R, Rosas HD. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2′dG. Neurology. 2006;66:250–2. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- [30].Volicer L, Chen JC, Crino PB, Vogt BA, Fishman J, Rubins J, Schenepper PW, Wolfe N. Neurotoxic properties of a serotonin oxidation product: possible role in Alzheimer’s disease. Prog Clin Biol Res. 1989;317:453–65. [PubMed] [Google Scholar]
- [31].Crino PB, Vogt BA, Chen JC, Volicer L. Neurotoxic effects of partially oxidized serotonin: tryptamine-4,5-dione. Brain Res. 1989;504:247–57. doi: 10.1016/0006-8993(89)91364-4. [DOI] [PubMed] [Google Scholar]
- [32].Volicer L, Langlais PJ, Matson WR, Mark KA, Gamache PH. Serotoninergic system in dementia of the Alzheimer type. Abnormal forms of 5-hydroxytryptophan and serotonin in cerebrospinal fluid. Arch Neurol. 1985;42:1158–61. doi: 10.1001/archneur.1985.04060110040013. [DOI] [PubMed] [Google Scholar]
- [33].Beal MF, Matson WR, Swartz KJ, Gamache PH, Bird ED. Kynurenine pathway measurements in Huntington’s disease striatum: evidence for reduced formation of kynurenic acid. J Neurochem. 1990;55:1327–39. doi: 10.1111/j.1471-4159.1990.tb03143.x. [DOI] [PubMed] [Google Scholar]
- [34].Beal MF, Matson WR, Storey E, Milbury P, Ryan EA, Ogawa T, Bird ED. Kynurenic acid concentrations are reduced in Huntington’s disease cerebral cortex. J Neurol Sci. 1992;108:80–7. doi: 10.1016/0022-510x(92)90191-m. [DOI] [PubMed] [Google Scholar]
- [35].Tamir H, Liu KP. On the nature of the interaction between serotonin and serotonin binding protein: effect of nucleotides, ions, and sulfhydryl reagents. J Neurochem. 1982;38:135–41. doi: 10.1111/j.1471-4159.1982.tb10864.x. [DOI] [PubMed] [Google Scholar]
- [36].Korlimbinis A, Hains PG, Truscott RJ, Aquilina JA. 3-Hydroxykynurenine oxidizes alpha-crystallin: potential role in cataractogenesis. Biochemistry. 2006;45:1852–60. doi: 10.1021/bi051737+. [DOI] [PubMed] [Google Scholar]
- [37].Korlimbinis A, Truscott RJ. Identification of 3-hydroxykynurenine bound to proteins in the human lens. A possible role in age-related nuclear cataract. Biochemistry. 2006;45:1950–60. doi: 10.1021/bi051744y. [DOI] [PubMed] [Google Scholar]
- [38].Parker NR, Jamie JF, Davies MJ, Truscott RJ. Protein-bound kynurenine is a photosensitizer of oxidative damage. Free Radic Biol Med. 2004;37:1479–89. doi: 10.1016/j.freeradbiomed.2004.07.015. [DOI] [PubMed] [Google Scholar]
- [39].Fishman JB, Rubins JB, Chen JC, Dickey BF, Volicer L. Modification of brain guanine nucleotide-binding regulatory proteins by tryptamine-4,5-dione, a neurotoxic derivative of serotonin. J Neurochem. 1991;56:1851–4. doi: 10.1111/j.1471-4159.1991.tb03440.x. [DOI] [PubMed] [Google Scholar]
- [40].P.A.R. Broderick DN, Kolodny EH, editors. Bioimaging in Neurodegeneration. Humana Press; 2005. [Google Scholar]
- [41].Jeitner TM, Matson WR, Folk JE, Blass JP, Cooper AJ. Increased levels of gamma-glutamylamines in Huntington disease CSF. J Neurochem. 2008;106:37–44. doi: 10.1111/j.1471-4159.2008.05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]