Abstract
The copper(I)-catalyzed azide-alkyne cycloaddition, the most widely recognized reaction of click chemistry, is accelerated by tris(triazolylmethyl)amine-based ligands. Here, we compared two new ligands in this family, BTTP and the corresponding sulfated ligand BTTPS, for three bioconjugation applications: (1) labeling of alkyne-tagged glycoproteins in crude cell lysates, (2) labeling of alkyne/azide-tagged glycoproteins on the surface of live mammalian cells, and (3) labeling of azides in surface proteins of live Escherichia coli. Though BTTPS exhibits faster kinetics than BTTP in accelerating the CuAAC in in vitro kinetic measurements, its labeling efficiency is slightly lower than BTTP in conjugating biomolecules bearing a significant amount of negative charges due to electrostatic repulsion. Nevertheless, the negative charge conferred by the sulfate at physiological conditions significantly reduced the cellular internalization of the coordinated Cu(I), thus making BTTPS-Cu(I) a better choice for live cell labeling.
Keywords: click chemistry, copper, bioconjugation, glycoconjugates
Introduction
The past 30 years have witnessed a growing synergy between advanced organic chemistry and biomedical research.[1] Featuring exquisite selectivity and bioorthogonality, the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC),[2-4] the most widely recognized reaction of click chemistry,[5] is one of the prominent chemical transformations that have brought new technique advances in numerous biological fields.[6, 7] For example, the CuAAC has offered simpler and highly specific methods for bioorthogonal conjugation in the covalent labeling of biomolecules.[8-11] It also allows straightforward derivatization of natural products to generate new activities.[12, 13] However, the current Cu(I) catalyst formulation has two major problems: toxicity, hindering its use in living systems,[14, 15] and slow kinetics,[16] hampering its use in quantitative functionalization of biomolecules of limited quantities.[17]
Our initial screening of a small library consisting of 14 water-soluble tris(triazolylmethyl)amine-based ligands has led to the discovery of a potent ligand, 2-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl) ethyl hydrogen sulfate (BTTES), that dramatically accelerated the rate of the azide-alkyne cycloaddition by coordinating with the in situ generated Cu(I), and also rendered the CuAAC nontoxic.[18] The new catalyst formulation is: BTTES-CuSO4 (for live cell labeling, [ligand]:[CuSO4] = 6:1, [CuSO4] = 50–75 μM and 2.5 mM sodium ascorbate to reduce CuII to CuI in situ). Although this catalytic system holds great promise for biocompatible applications, further improvement of its activity and lowering of the Cu loading are desirable for broader biomedical applications. Building upon this discovery, we performed a structure-activity relationship study, in which we identified a new ligand, 3-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)propyl hydrogen sulfate (BTTPS), that shows better kinetics in accelerating the CuAAC.
Herein, we compare the activity of BTTPS and the corresponding unsulfated ligand—3-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)propanol, BTTP—in a set of bioconjugation studies: (1) labeling of alkyne-tagged glycoproteins in crude cell lysates, (2) labeling of azide- and alkyne-tagged glycoproteins on the surface of live mammalian cells, and (3) labeling of azide-bearing proteins on the surface of live E. coli. We discover that both ligands are highly efficient for all these bioconjugation applications with biocompatibility of the catalyst further improved by the ligand sulfation.
Kinetic Evaluation of the Ligand-Accelerated CuAAC
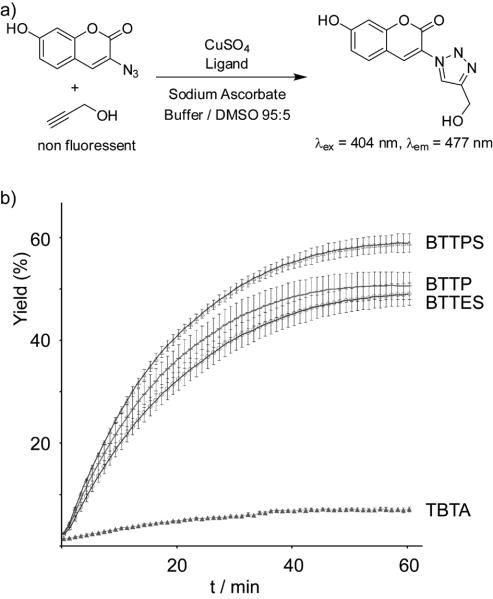
The relative reactivity of the Cu(I) catalysts in the form of TBTA-Cu(I), BTTES-Cu(I), BTTP-Cu(I), and BTTPS-Cu(I) complexes was determined in a fluorogenic assay by reacting propargyl alcohol with 3-azido-7-hydroxycoumarin (Scheme 2a).[19] In all studies, CuSO4·5H2O was used as the copper source to form complexes with the ligands. The active catalysts were generated in situ using 2.5 mM of sodium ascorbate. BTTPS showed the highest activity in accelerating the CuAAC among the four tris(triazolylmethyl)amine-based ligands evaluated, in which BTTP and BTTES showed comparable rate-acceleration, with TBTA exhibiting the lowest activity. BTTPS-Cu(I) gave the largest slope of the reaction curve, yielding greater than 50% cycloaddition product within the first 30 minutes when 75 μM CuSO4 was used and the ligand-CuSO4 ratio was 6:1. By contrast, the TBTA-mediated reaction was significantly slower, yielding less than 20% cycloaddition product (Scheme 2b).
Scheme 2.
Comparison of CuAAC kinetics in the presence of various accelerating ligands. a) A fluorogenic assay for qualitative measurement of CuAAC kinetics. b) Conversion-time profiles of CuAAC in the presence of various ligands. Reaction conditions: propargyl alcohol (50 μM), 3-azido-7-hydroxy-coumarin (100 μM), CuSO4 (75 μM), ([Ligand]:[CuSO4] = 6:1), 0.1 M potassium phosphate buffer (pH 7.0)/DMSO 95:5, sodium ascorbate (2.5 mM), room temperature. Error bars represent the standard deviation of three replicate experiments.
Labeling of Glycoproteins in Crude Cell Lysates
To compare the efficacy of BTTPS- and BTTP-mediated CuAAC in labeling glycoproteins, we sought to investigate bioconjugation of affinity probes to alkyne-tagged sialylated glycoproteins in crude cell lysates, the first step in enriching these proteins for glycoproteomic analysis. We cultured Jurkat cells, a human T lymphocyte cell line, in medium supplemented with peracetylated N-(4-pentynoyl)mannosamine (Ac4ManNAl) to introduce terminal alkyne to the cell-surface sialylated glycoconjugates.[20] After 3 days, we lysed the cells and reacted the cell lysates with either biotin-azide or FLAG-azide via the CuAAC. In these labeling reactions, 100 μM of biotin-azide or FLAG-azide was used as the coupling partner, and the ratio of azide, ligand, CuSO4 and sodium ascorbate was held at 1:5:2.5:25, a labeling condition optimized in our lab. After a one-hour reaction, we probed the modified cell lysates with anti-biotin or anti-FLAG Western blot. Robust labeling was observed in both cases for lysates isolated from cells treated with Ac4ManNAl, whereas no signals were detectable for lysates obtained from cells cultured in the absence of the sugar (Figure 1). Consistent with the kinetic measurements, stronger signals were detected for BTTPS-Cu(I)-treated lysates than the BTTP-Cu(I)-treated counterparts when biotin was used as the probe (Figure 1a). Interestingly, when FLAG was used as the probe, the BTTP-Cu(I) catalyst afforded stronger signals (Figure 1b). At neutral pH, the FLAG peptide bears a high amount of negative charges, which may interfere with the approach of BTTPS that is functionalized with a negatively charged arm, thus lowering the labeling efficiency.
Figure 1.
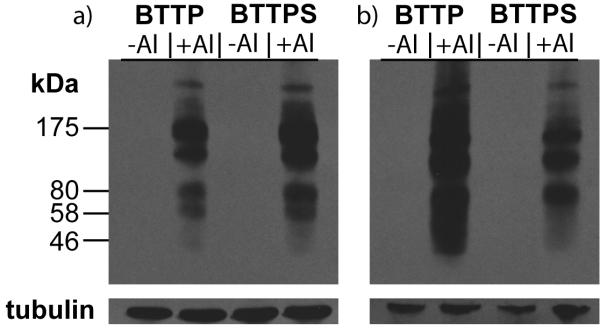
Comparison of the efficiency of the BTTPS-Cu(I) and BTTP-Cu(I)-mediated azide-alkyne cycloaddition in labeling glycoproteins in crude cell lysates. Cell lysates prepared from Ac4ManNAl-treated or untreated Jurkat cells were reacted with a) biotin azide (100 μM) or b) FLAG-azide (100 μM) in the presence of sodium ascorbate (2.5 mM), and CuSO4 (250 μM) premixed with ligands BTTPS or BTTP (500 μM). Reactions were allowed to proceed for 1 h at room temperature, and analyzed by Western blot using an HRP-conjugated anti-biotin antibody (a, top panel) or an HRP-conjugated anti-FLAG antibody (b, top panel). Anti-tubulin Western blots were perform to confirm equal protein loadings (bottom panel)
Labeling of Live Mammalian Cells
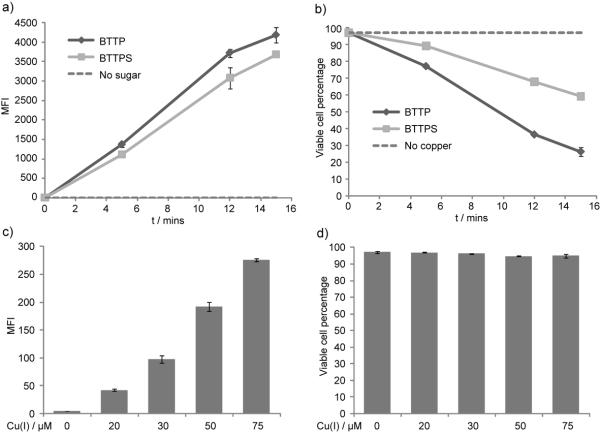
In our ligand design, sulfation of BTTP is used to generate a negatively charged ligand, BTTPS, to minimize the cellular internalization of the coordinated copper. To evaluate if sulfation confers the ligand the desired property, we compared BTTPS-Cu(I) and BTTP-Cu(I) in live cell labeling experiments. We metabolically labeled Jurkat cells with Ac4ManNAl and reacted the treated cells with biotin-azide (50 μM) in the presence of BTTPS-Cu(I) or BTTP-Cu(I) for 5–15 min at room temperature (catalyst formulation: [ligand]:[CuSO4] = 6:1, [CuSO4] = 75 μM). After the reaction was quenched with BCS, the biotinylated cells were incubated with Alexa Fluor 488-streptavidin. The cells were then stained with 7-aminoactinomycin D (7-AAD), a fluorescent molecule with strong affinity for double-stranded DNA, and analyzed by flow cytometry. 7-AAD does not pass through intact membrane, but it readily enters damaged cells with compromised membrane. Therefore, healthy and damaged cells can be easily distinguished. As shown in Figure 2a, cell-associated Alexa Fluor 488 fluorescence were detected for both BTTPS-Cu(I) or BTTP-Cu(I) treated cells, and the fluorescence increased along with the increase of the reaction time. Although both catalytic systems showed comparable labeling efficiency, BTTPS-Cu(I) was significantly better in protecting cells from the Cu(I)-associated toxicity, especially for labeling with extended reaction course (15 minutes). Greater than 60% cells were still undamaged after treatment with BTTPS-Cu(I) (7-AAD negative), whereas only 26% cells treated with BTTP-Cu(I) reminded normal (Figure 2b). Furthermore, we noticed that the labeling temperature also plays a significant role in modulating the Cu-associated toxicity. When both Jurkat cells and the labeling reagents were pre-cooled to 4 °C before triggering the CuAAC, greater than 85% cells remained viable after a 15 min BTTPS-Cu(I)-mediated reaction with high labeling efficiency achieved (mean fluorescence intensity 2200, for reactions performed at 4 °C–room temperature vs 3679, for reactions performed at room temperature).
Figure 2.
Comparison of the efficiency of the BTTPS-Cu(I) and BTTP-Cu(I)-mediated azide-alkyne cycloaddition in labeling sialylated glycoconjugates in live cells. Jurkat cells were cultured in the presence or absence of Ac4ManNAl for 3 days. Cells were then reacted with biotin azide (50 μM) in the presence of sodium ascorbate (2.5 mM), and CuSO4 premixed with ligands BTTPS or BTTP ([ligand]:[CuSO4] = 6:1). Reactions were quenched with BCS, stained with Alexa Fluor 488-streptavidin, 7-AAD, and analyzed by flow cytometry. (a) Mean fluorescence intensity (MFI) of cells treated with the BTTPS-Cu(I) or BTTP-Cu(I) catalyst ([CuSO4] = 75 μM) in the course of 5-15 min reactions. (b) Percentage of viable cells without cell-membrane damage post the click reactions with the BTTPS-Cu(I) or BTTP-Cu(I) catalyst ([CuSO4] = 75 μM) in the course of 5-15 min reactions. (c) MFI of cells treated with the BTTPS-Cu(I) catalyst ([CuSO4] = 20–75 μM) in a one-minute reaction. (d) Percentage of viable cells without cell-membrane damage post the click reactions with the BTTPS-Cu(I) catalyst ([Cu] = 20–75 μM) in a one-minute reaction.
Our previous studies showed that significant labeling of Ac4ManNAl-treated Jurkat cells was achieved with BTTES-Cu(I)-mediated click chemistry when 50–75 μM CuSO4 was used as the copper source. With the observation that BTTPS confers CuAAC faster kinetics than BTTES, we were eager to test if efficient cell labeling could be achieved with BTTPS using lower copper loading. Toward this end, we reacted alkyne-bearing Jurkat cells with biotin-azide (50 μM) in the presence of BTTPS-Cu(I) for 1 min at room temperature (catalyst formulation: [ligand]:[CuSO4] = 6:1, [CuSO4] = 20–75 μM). After the reaction was quenched with bathocuprioine disulfate (BCS), the biotinylated cells were incubated with Alexa Fluor 488-streptavidin, stained with 7-aminoactinomycin D (7-AAD), and analyzed by flow cytometry. As shown in Figure 2c, significant labeling was realized with as low as 30 μM copper loading.
The BTTPS-Cu(I) catalyst is equally active in detecting cell surface azides in Jurkat cells metabolically treated with peracetylated N-azidoacetylmannosamine (Ac4ManNAz).[21] To evaluate if the faster kinetic behavior of BTTPS-Cu(I) vs. BTTES-Cu(I) can be transferred in vivo, we compared these two catalysts directly in biotinylation of azido sialic acid (SiaNAz)-bearing Jurkat cells. Under exact same labeling conditions, ~15% higher labeling efficiency was achieve using the BTTPS-Cu(I) catalyst (Figure 3).
Figure 3.
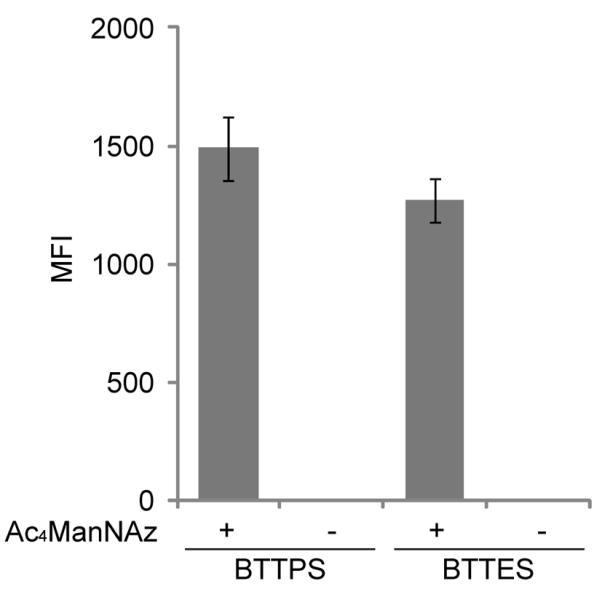
Comparison of the efficiency of the BTTPS-Cu(I) and BTTES-Cu(I) in labeling azido sialic acids in live cells. Jurkat cells were cultured in the presence or absence of Ac4ManNAz for 3 days. Cells were then reacted with biotin alkyne (50 μM) in the presence of sodium ascorbate (2.5 mM), and CuSO4·5H2O premixed with ligands BTTPS or BTTES ([CuSO4] = 75 μM, [ligand]:[CuSO4] = 6:1). After 3 min, reactions were quenched with BCS, stained with Alexa Fluor 488-streptavidin, 7-AAD, and analyzed by flow cytometry.
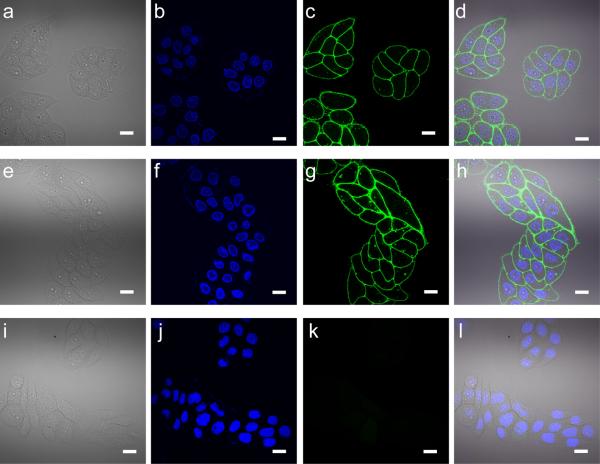
We next evaluated the use of BTTP- and BTTPS-mediated CuAAC for direct fluorescence imaging of glycans on live cell surface. HeLa cells, a human epithelial carcinoma cell line, were incubated with 50 μM Ac4ManNAz to metabolically incorporate the corresponding SiaNAz into their cell surface glycoconjugates. The resulting HeLa cells bearing azides on their cell surface were reacted with Alexa Fluor 488-alkyne (50 μM) in the presence of BTTP-Cu(I) or BTTPS-Cu(I) ([ligand]:[ CuSO4] = 6:1, [CuSO4] = 50 μM). After 5 min, the reaction was quenched with BCS. As shown by confocal fluorescence microscopy, robust Alexa Fluor 488 fluorescence was detected in the cell membrane (Figure 4). Under exactly the same imaging condition, comparable labeling efficiency was achieved by using BTTPS-Cu(I) and BTTP-Cu(I). No obvious cytotoxicity was observed post the Cu(I)-treatment as confirmed by trypan blue stain (data not shown).
Figure 4.
Fluorescent imaging of SiaNAz-containing glycans on cell surface using biocompatible CuAAC. HeLa cells were incubated with (a-h) or without (i-l) 50 μM Ac4ManNAz for 3 days. The cells were subsequently labeled with Alexa Flour 488-alkyne using BTTP-Cu(I) and BTTPS-Cu(I)-catalyzed AAC for 5 min with 50 μM CuSO4, [ligand]:[CuSO4] = 1:6, sodium ascorbate 2.5 mM (a-d ligand: BTTP, e-l ligand: BTTPS). The cell nuclei were stained with Hoechst 33342 prior to microscopy analysis. The first column: bright field; the second column, Hoechst 33342 channel; the third column, Alexa Flour 488 channel; the fourth column, overlay. Scale bars: 20 μm.
Labeling of azide-bearing surface proteins in E. Coli
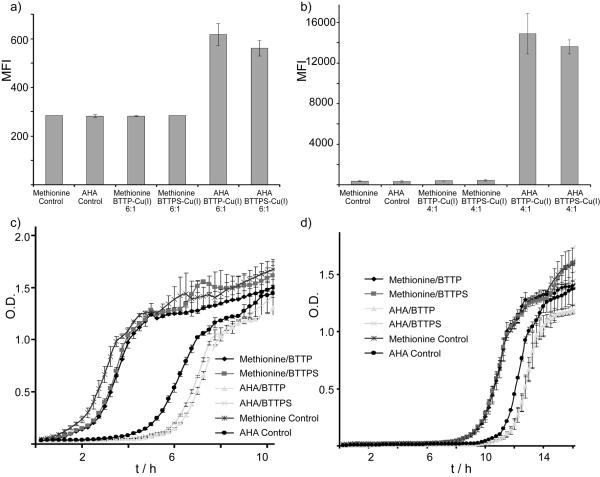
The development of the genetically-encoded unnatural amino acid technology based on the amber nonsense codons[22] and the complementary metabolic approach[23] allows the incorporation of functional groups beyond those found in the 20 canonical amino acids into proteins of live cells. As demonstrated elegantly by the Tirrell group, azide- or alkyne-bearing groups can be introduced to the surface of E. coli, and be further derivatized by the CuAAC.[24, 25] However, the bacteria subjected to the overexpression of outer membrane protein C (OmpC) containing azidohomoalanine (AHA) and then treated with the canonical CuAAC reagents (i.e. CuSO4, TBTA[26], tris(2-carboxyethyl)phosphine), were unable to divide after being transferred to rich medium. [24, 25] To evaluate if the BTTP-Cu(I) or BTTPS-Cu(I) catalyst could improve the viability of the treated bacteria, we reacted E. coli cells expressing OmpC with biotin-alkyne. Here, two [ligand]:[CuSO4·5H2O] ratios, 6:1 and 4:1, were used. After a 10-minute reaction, the bacteria were stained with Alexa Fluor 488-streptavidin and subjected to flow cytometry analysis. As expected, bacteria expressing OmpC and induced in the presence of 20 natural amino acids have the same mean fluorescence as the unlabeled cells. By contrast, bacteria induced in the presence of AHA showed 2.2-fold increase in the fluorescence of Alexa Fluor 488 over the background when the 6:1 ligand-Cu(I) complex was used (Figure 5a). When [ligand]:[Cu] ratio decreased to 4:1, greater than 30-fold fluorescence over the background was observed (Figure 5b). Notably, with the canonical CuAAC condition the comparable labeling level was only achieved after a 16-hour reaction.[25] In our E. Coli labeling experiments, BTTP-Cu(I) gave 10% stronger signal than BTTPS-Cu(I) (Figure 5b). In PBS buffer (pH = 7.4), the surface of E. coli is known to be negatively charged, which may have repulsive interactions with the negatively charged BTTPS, thus lowering the labeling efficiency of the corresponding BTTPS-Cu(I) complex.[27]
Figure 5.
Labeling E. coli using CuAAC. E. coli bearing mOmpC were cultured in the presence or absence of azidohomoalanine till O.D. reaching 1, and then reacted with biotin azide (50 μM) in the presence of sodium ascorbate (2.5 mM), and CuSO4 (75 μM) premixed with ligands BTTPS or BTTP at room temperature. Reactions were quenched by BCS after 10 minutes, and probed with Alexa Fluor 488-streptavidin and analyzed by flow cytometry. MFI of E. coli reacted with BTTPS-Cu(I) or BTTP-Cu(I) ([ligand]:[CuSO4] = 6:1 complex) (a), BTTPS-Cu(I) or BTTP-Cu(I) ([ligand]:[CuSO4] = 4:1 complex) (b). The corresponding E. coli growth curve post the click reactions (c and d).
To evaluate if the copper catalysts cause any long-term perturbations to the treated bacteria, we cultured the copper-treated and untreated E. coli. in nutrient-rich medium and followed their growth by O.D. reading. As shown in Figures 5c and 5d, the bacteria bearing AHA showed ~ 4-hour delay in their cell proliferation compared to the bacteria growing with all 20 natural amino acids through this series of experiments. Noteworthy, the Cu-treatment barely had any influence on the bacteria growing with 20 natural amino acids—the Cu(I)-treated and untreated bacteria showed similar proliferation trend. However, after the bacteria bearing AHA were subjected to the CuAAC—one-hour delay in their cell proliferation was observed. Nevertheless, both the Cu(I)-treated and untreated bacteria reached same cell density by 15 hours after being transferred to the rich medium, suggesting only minor toxicity was imparted to the labeled E. coli.
Conclusion
Our systematic investigation of BTTPS-Cu(I) and BTTP-Cu(I) in this series of labeling studies showed that both catalysts are highly efficient for bioconjugation. Consistent with the criteria of our ligand design, the negatively charged sulfate minimized the membrane permeability of the coordinated Cu(I) at physiological conditions, and significantly retained cell viability post the Cu(I)-treatment. Therefore, the BTTPS-Cu(I) catalyst is the clear choice for live cell surface labeling experiments. Importantly, ligand-CuSO4 ratio greater than one is critical to reduce the Cu(I)-associated toxicity, a phenomenon discovered by us and the Finn Lab previously.[18, 28] However, the negative charge conferred by the sulfate functionality may impose electrostatic repulsion with probes or biomolecules bearing significant negative charges, thus reducing the catalytic efficiency of the corresponding BTTPS-Cu(I) complex. For these reasons, BTTP-Cu(I) may be a better choice for in vitro labeling of negatively charged biomolecules.
Noteworthy, this study has identified highly reliable Cu(I)-catalyst formulations that can be easily scaled up and adopted to various bioconjugation applications. The new catalysts reported here also solved the longstanding problems of the canonical CuAAC, including toxicity and poor labeling efficiency at low substrate concentration, thus opening new possibilities for bioorthogonal click chemistry-based molecular imaging and proteomic analysis.
Experimental Section
Metabolic labeling of Jurkat cell and detection using CuAAC click chemistry and flow cytometry analysis
Jurkat cells were cultured for 3 days in untreated RPMI 1640 medium or medium containing 50 μM Ac4ManNAl (or Ac4ManNAz). The cells then were distributed into a 96-well round bottom tissue culture plate (0.4–0.5 million cells/well), pelleted (300 × g, 3 min), and washed 2 × with 200 μL of labeling buffer (PBS, pH 7.4, containing 1% FBS). Cells were then resuspended in 92 μL labeling buffer, followed by addition of 100 μM biotin-alkyne (or biotin-azide to react with SiaNAl), BTTP or BTTPS-CuSO4 complex ([ligand]:[CuSO4]=6:1) and 2.5 mM sodium ascorbate in labeling buffer at room temperature. The reactions were quenched by adding 2 μL of copper chelator BCS (50 mM). Then the cells were pelleted, washed 3 × with labeling buffer, and resuspended in the same buffer containing 1 μg/mL streptavidin-Alexa Fluor 488 and incubated in dark at 4 oC for 30 min. Following the incubation, cells were washed 3 × with labeling buffer and resuspended in 400 μL FACS buffer (Hank’s Balanced Salt Solution, pH = 7.4, 1% FBS, 2 μg/mL 7-AAD, 0.2% NaN3) for flow cytometry analysis. Flow cytometry experiments were performed on a Becton Dickinson FACScan flow cytometer using a 488 nm argon laser. At least 15,000 cells were recorded for each sample. Flow cytometry data were analyzed using FlowJo. Mean fluorescence intensity (MFI) was calculated for live cells.
Metabolic labeling of Jurkat cell glycoproteins and detection using CuAAC click chemistry and Western blot
Jurkat cells were incubated for 3 days in RPMI medium or RPMI medium containing 50 μM Ac4ManNAl. The cells were washed with PBS, harvested by centrifugation (300 × g, 3 min), and homogenized in lysis buffer (100 mM sodium phosphate, 150 mM NaCl, 1% NP-40, pH 7.4) containing protease inhibitors (Roche cOmplete tablets, EDTA-free) by 10 freeze-thaw cycles. Insoluble debris was removed by centrifugation (10,000 × g, 10 min) and the soluble protein concentration was determined using the DC protein assay kit. To label the sialylated glycoproteins, protein was resuspended at a concentration of 0.69 mg/mL and reacted with 100 μM FLAG-azide or 100 μM biotin-azide in a 100 μL reaction containing premixed ligand-CuSO4 complex ([CuSO4] = 250 μM, [ligand]:[CuSO4] = 2:1) and 2.5 mM freshly prepared sodium ascorbate. Ligands used included BTTP, and BTTPS. The samples were lightly vortexed and allowed to react for 1 hour (25 °C, 800 rpm in eppendorf Theromomixer R). Reacted samples (8.3 μg each) were heated in loading buffer at 95 °C for 5 min, and resolved on 4-20% Criterion™ XT Precast Gels. The samples were transferred to nitrocellulose, and incubated for 1 hour at room temperature in blocking buffer (5% non-fat milk in TBST (Tris buffered saline with 0.1% Tween-20, pH 7.5)). The blocked membrane was incubated for 1 hour at room temperature with an HRP-anti-FLAG antibody (1:3000 dilution) or an HRP-anti-biotin antibody (1:100,000 dilution) in blocking buffer, washed 3 × with TBST. The membranes were developed using SuperSignal® West Pico Chemiluminescent Substrate and imaged on film. To confirm equal protein loading, the primary antibody treated membranes were stripped (http://www.abcam.com/ps/pdf/protocols/Stripping%20for%20reprobing.pdf). The membranes were then incubated for 1 hour at room temperature in blocking buffer, and then incubated with monoclonal anti-tubulin clone AA13 (1:1000 dilution) for 1 hour at room temperature. After being washed 3 × with TBST, the membranes were incubated with an HRP-Goat-anti-mouse (1:60,000 dilution) for 1 hour at room temperature, then washed 3 × with TBST. The membranes were incubated with SuperSignal® West Pico Chemiluminescent Substrate and imaged on film.
HeLa cell labeling using CuAAC chemistry and Imaging with Confocal Microscopy
HeLa cells were cultured in DMEM medium supplemented with or without Ac4ManNAz (50 μM) on Lab-Tek™ Chambered Coverglass for 3 days. The cells were washed with 3 × PBS (100 μL), reacted with Alexa Flour 488-alkyne (50 μM) in a 100 μL reaction containing premixed ligand-CuSO4 complex ([CuSO4] = 5 0 μM, [ligand]:[CuSO4] = 6:1) and 2.5 mM freshly prepared sodium ascorbate for 5 min at room temperature. The reaction was quenched with 1 mM BCS. The cells were washed with 3 × PBS, stained with Hoechst 33342 at 4 °C for 15 min. Laser Scanning Confocal Microscope (Nikon, A1R-si) 60 × was used for imaging the Alexa Flour 488 on the Hela cell surface.
Metabolic labeling of E. Coli and detection with CuAAC chemistry and flow cytometry analysis
A single colony of M15MA[pQE-60/OmpC] was grown in M9 minimal medium supplemented with all twenty natural amino acids and with carbenicillin (100 mg/L) and kanamycin (35 mg/L) till O.D.600 = 1. The bacteria were pelleted (2500 × g, 10 min), resuspended in 50 mL M9 medium (supplemented with 19 amino acids), and agitated at 37 °C for 10 min. The cells were pelleted, resuspended in same volume M9 medium (supplemented with 19 amino acids), and divided into 2 equal portions (~25 mL each): one was added methionine (40 mg/L), and one added azidohomoanaline (AHA, 40 mg/L). To each culture was added IPTG (1 mM) and shaken at 37 °C for 3 h. The E. Coli culture (~25 mL each) were centrifuged at 2500 × g for 5 minutes, and washed once with 12.5 mL PBS. The cells were centrifuged again, and resuspended in 2.5 mL PBS. In a 96-well round bottom tissue culture plate, to each well containing an aliquot of 200 μL of these bacteria were added biotin-alkyne (100 μM), premixed ligand-CuSO4 complex ([CuSO4] = 75 μM, [ligand]:[CuSO4] = 4:1 or 6:1). After 10 min, BCS was added (1 mM) to the bacteria to quench the reaction. Bacteria were then washed 3 × with 200 μL PBS, and resuspended in 200 μL PBS, and devided into two portions. Portion one was diluted with PBS to 1 mL, from which 10 μL was taken and diluted with 190 μL M9 medium containing all 20 amino acids (40 mg/L each) as well as carbenicillin (100 mg/L) and kanamycin (35 mg/L), and shaken at 37 oC. O.D.600 was measured at every 15 min for 18 hour period time using Synergy Hybrid Plate Reader. Portion two was incubated with Alexa Fluor 488-streptavidin (final concentration 1 μg/mL) at 4 oC for 25 minutes. Bacteria were then washed 3 × with 200 μL PBS, resuspendeed in 200 μL PBS, and analyzed by flow cytometry. Flow cytometry experiments were performed on a Eclipese™ iCyt flow cytometer using a 488 nm argon laser. At least 20,000 cells were recorded for each sample. Flow cytometry data were analyzed using FlowJo.
Scheme 1.
CuACC is accelerated by the Cu(I)-stablizing ligands. TBTA = tris((1-benzyl-1H-1,2,3-triazol-4-yl)methyl)amine.
Acknowledgement
This work was partially supported by the National Institutes of Health grants GM080585, GM093282 (P.W.) and DuPont Young Professor Award (P.W.). Part of the ligand synthesis was performed as a User Project at the Molecular Foundry, Lawrence Berkeley National Laboratory, which was supported by the Office of Science, Office of Basic Energy Sciences, U.S. Department of Energy, under contract DE-AC02-05 CH11231. A. T. was supported by the Albert Einstein Summer Undergraduate Research Program. We thank Prof. Valery Fokin for discussions on the mechanism of CuAAC and Prof. A. James Link for providing M15MA[pQE-60/OmpC].
References
- [1].Corey EJ, Czako B, Kurti L. Molecules and Medicine. John Wiley & Sons; Hoboken: 2008. [Google Scholar]
- [2].Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- [3].Tornoe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- [4].Hein JE, Fokin VV. Chem. Soc. Rev. 2010;39:1302–1315. doi: 10.1039/b904091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kolb HC, Finn MG, Sharpless KB. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- [6].Kolb HC, Sharpless KB. Drug Discov. Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- [7].Wu P, Fokin VV. Aldrichimica Acta. 2007;40:7–17. [Google Scholar]
- [8].Speers AE, Cravatt BF. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- [9].Salic A, Mitchison TJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kaltgrad E, O’Reilly MK, Liao L, Han S, Paulson JC, Finn MG. J. Am. Chem. Soc. 2008;130:4578–4579. doi: 10.1021/ja077801n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hong V, Presolski SI, Ma C, Finn MG. Angew. Chem. Int. Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat. Biotechnol. 2003;21:1467–1469. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
- [13].Lin H, Walsh CT. J. Am. Chem. Soc. 2004;126:13998–14003. doi: 10.1021/ja045147v. [DOI] [PubMed] [Google Scholar]
- [14].Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- [15].Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Presolski SI, Hong V, Cho SH, Finn MG. J. Am. Chem. Soc. 2010;132:14570–14576. doi: 10.1021/ja105743g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kaltgrad E, Sen Gupta S, Punna S, Huang CY, Chang A, Wong CH, Finn MG, Blixt O. Chembiochem. 2007;8:1455–1462. doi: 10.1002/cbic.200700225. [DOI] [PubMed] [Google Scholar]
- [18].Soriano Del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J. Am. Chem. Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org. Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- [20].Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Angew. Chem. Int. Ed. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Laughlin ST, Agard NJ, Baskin JM, Carrico IS, Chang PV, Ganguli AS, Hangauer MJ, Lo A, Prescher JA, Bertozzi CR. Methods Enzymol. 2006;415:230–250. doi: 10.1016/S0076-6879(06)15015-6. [DOI] [PubMed] [Google Scholar]
- [22].Wang L, Schultz PG. Angew. Chem. Int. Ed. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- [23].Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Curr. Opin. Chem. Biol. 2010;14:774–780. doi: 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Link AJ, Tirrell DA. J. Am. Chem. Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- [25].Link AJ, Vink MK, Tirrell DA. J. Am. Chem. Soc. 2004;126:10598–10602. doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- [26].Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- [27].Li J, McLandsborough LA. Int. J. Food Microbiol. 1999;53:185–193. doi: 10.1016/s0168-1605(99)00159-2. [DOI] [PubMed] [Google Scholar]
- [28].Hong V, Steinmetz NF, Manchester M, Finn MG. Bioconjug. Chem. 2010;21:1912–1916. doi: 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]