Abstract
Metallic nanopore arrays have emerged as optofluidic platforms with multifarious sensing and analytical capabilities such as label-free surface plasmon resonance (SPR) sensing of molecular binding interactions and surface-enhanced Raman spectroscopy (SERS). However, directed delivery of analytes through open nanopores using traditional methods such as external electric fields or pressure gradients still remains difficult. We demonstrate that nanopore arrays have an intrinsic ability to promote flow through them via capillary flow and evaporation. This passive “nano-drain” mechanism is utilized to concentrate biomolecules on the surface of nanopores for improved detection sensitivity or create ordered nanoscale arrays of beads and liposomes. Without using any external pump or fluidic interconnects, we can concentrate and detect the presence of less than a femtomole of streptavidin in 10 µL of sample using fluorescence imaging. Liposome nanoarrays are also prepared in less than 5 minutes and used to detect lipid-protein interactions. We also demonstrate label-free SPR detection of analytes using metallic nanopore arrays. This method provides a fast, simple, transportable and small-volume platform for labeled as well as label-free plasmonic analysis while improving the detection time and sensitivity.
Keywords: Nanohole array, nanopore, microfluidics, nanofluidics, surface plasmon resonance, flow-through sensing, diffusion limits, optofluidics, plasmonics, liposome, fluidic self-assembly
The emerging field of optofluidics has facilitated on-chip integration of micro- and nanofluidics with optical sensing and manipulation techniques for a wide variety of applications.1–3 The analytical tools mainly utilized by these optofluidic platforms have been fluorescence detection, surface plasmon resonance (SPR), and surface-enhanced Raman spectroscopy (SERS), which are often combined with trapping and manipulation of biomolecules and small particles.4–8 The potential advantages offered by optofluidic platforms include reduced sample consumption as well as improved sensitivity and detection time. However, demonstrating such benefits using highly miniaturized sensors has been challenging. In micro- and nanoscale sensors, diffusion-limited binding of low-concentration analytes to the sensing surface results in reduced sensitivity, prolonged detection time, artifacts in measured binding kinetics and excessive analyte consumption.9, 10 Thus new techniques to enhance targeted delivery of analytes beyond the conventional diffusion limit are needed. Likewise, rapid transport and site-specific trapping of particles such as beads, liposomes, or cells is also highly desirable, as it facilitates interrogation of complex biological molecules and structures over long experimental time scales, improving the depth and breadth of information obtainable.11 Cells or liposomes trapped on a pre-defined sensing surface can also be utilized for label-free analysis of ligands interacting with membrane receptors, which are important targets for drug discovery.12
Among various optofluidic systems, metallic nanohole arrays have been widely investigated for optical biosensing and spectroscopy13–26 because extraordinary optical transmission (EOT) through nanohole arrays27 can be exploited to develop SPR biosensors, which can detect changes in the interfacial refractive index imparted by analyte binding in a real-time label-free manner. While dead-ended nanohole arrays were initially used, subsequent development of open-ended nanohole arrays in a free-standing, gold-coated silicon nitride (Si3N4) membrane enabled “flow-through” plasmonic sensing to address mass transport limitations.21, 23, 24 Solid-state nanopores have also been useful in DNA sequencing28 and single-molecule spectroscopy.29, 30 In such open-ended nanopore systems, samples are typically injected through the nanopores using pressure-driven flow,21, 23 electrokinetic flow31 or electric field gradient focusing.32 These methods require external sources for generating pressure gradients, which can often damage the fragile membrane, or a bulky external power supply for creating an electric field.
In this work, we show that such external driving mechanisms are not required at all for small-volume concentration of molecules or particle trapping, since each nanopore in the metallic membrane can act as a nano-capillary and has an intrinsic ability to induce trans-nanopore solution flow. Techniques based on capillary flow have shown potential in generating passive flow and improving the sensitivity of assays.6, 33–36 Here we harness such effects in engineered metallic nanopores to demonstrate a small-volume concentration and integrated optical detection, which is enabled by metallic nanopore arrays simply acting as passive “nano-drains” without external pumps. Moreover, because there is no need for microfluidic interconnects, the system operates with zero dead volume, which drastically reduces sample consumption. This simple technique also helps prevent damage to the thin suspended nanopore membrane. Working on the principles of capillary flow and evaporation, our approach leads to rapid accumulation and local concentration, which enables arraying and analysis of proteins, polymer and silica beads, as well as phospholipid vesicles.
EXPERIMENTAL SECTION
Nanopore Array Fabrication
Low-pressure chemical vapor deposition (LPCVD) was used to coat low-stress Si3N4 films (100 nm) on both sides of silicon wafers. Photolithography and anisotropic Si etching with potassium hydroxide (KOH) created freely suspended Si3N4 membranes. Electron-beam evaporation was used to deposit a 200 nm-thick gold layer on the nitride surface, along with a 5 nm-thick Cr adhesion layer. Holes were milled through the suspended gold/nitride stack using a focused ion beam (FIB). For binding experiments, atomic layer deposition (ALD) was used to conformally deposit a 10 nm-thick silica layer on the chips. Additional fabrication details were described by Im et al.22
Sample Injection
During the experiments, the nanopore sensor chip was supported by glass slides, which acted as spacers as shown in Figure 1(e–f). The chip was placed with the flat surface (trans side) facing down. A 1–10 µL drop of solution containing particles or streptavidin R-phycoerythrin conjugate (SAPE) was placed on the cis side of the chip directly over a single reservoir. The sample was left undisturbed for 5 to 10 minutes and then covered with a cover slip for imaging.
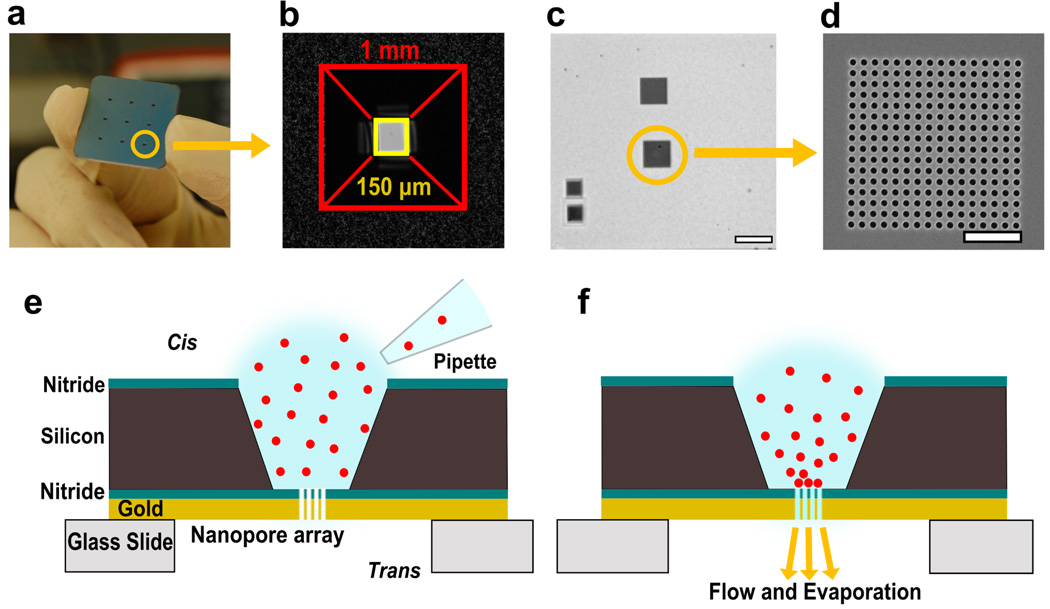
Figure 1.
Nanopore array chips and schematic of assembly process. (a) A 1 inch × 1 inch chip with nine etched reservoirs leading to suspended nitride sensing regions. A single reservoir has been circled. (b) Bright-field image showing a single truncated pyramidal reservoir leading to suspended membrane. The opening on the cis side has sides ~1 mm leading to a Si3N4 membrane with sides of ~150 µm. (c) Brightfield image of the suspended membrane after milling 4 nanopore arrays. Scale bar is 20 µm. (d) Scanning electron micrograph of a nanopore array (pore diameter: 600 nm, periodicity: 1 µm) milled using focused ion beam. Scale bar is 5 µm. (e, f) Schematics of a single reservoir and suspended Si3N4 membrane region showing spontaneous accumulation of particles onto the nanopore array.
Phospholipid Vesicle Preparation
Vesicles were formed by rehydration of dried lipids. Solutions of lipids in chloroform were obtained from Avanti polar lipids. These solutions were dried in a desiccator for at least 3 hours. Dried lipids were then rehydrated with 0.1 M NaCl overnight at a final concentration of 0.1 mg/ml. The lipid mixture contained 99% (w/w) Egg-PC (Phosphatidylcholine) and 1% (w/w) 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl, ammonium salt (Rho-PE) as a fluorescent component. After rehydration, the vesicles were vortex mixed for 30 seconds and sonicated for 10 minutes in a room temperature water bath. The vesicles were then extruded through a polycarbonate filter (100 nm) to obtain approximately 100 nm-diameter vesicles. For recording the time-lapse movie (Movie file S1), 10 µL of vesicle solution was added to the chip and a coverslip was placed on top. Images were recorded at an interval of 5 seconds with an acquisition time of 1 second.
Fluidic Self-Assembly of Beads on Nanopore Arrays
Silica and polystyrene beads were used to observe and confirm the generic and size-independent nature of the nanopore-induced fluidic self-assembly process. Silica beads (mean diameter: 700 nm) and fluorescent polystyrene beads – poly (Styrene/2%DiVinylBenzene/Vinyl-COOH), mean diameter 2.19 µm, were obtained from Bangs Labs and used at a final concentration of 107 beads/mL in deionized (DI) water. For the experiment with silica beads, Figure 2(f–h), 5 µL of solution was placed on back of a single reservoir for 5 minutes and the remaining solution was rinsed off. Four arrays (16 µm × 16 µm) with a 600 nm pore diameter and 1 µm periodicity were used. The sample was then dried to record images with a scanning electron microscope. For polystyrene beads, (Figure S1), 10 µL of solution was placed on back of a reservoir for 10 minutes and imaged in solution. Two arrays were milled on the suspended membrane. The larger array (32 µm × 32 µm) had a 2 µm periodicity and 1.2 µm pore diameter, whereas the smaller one (16 µm × 16 µm) had a 1 µm periodicity and 600 nm pore diameter.
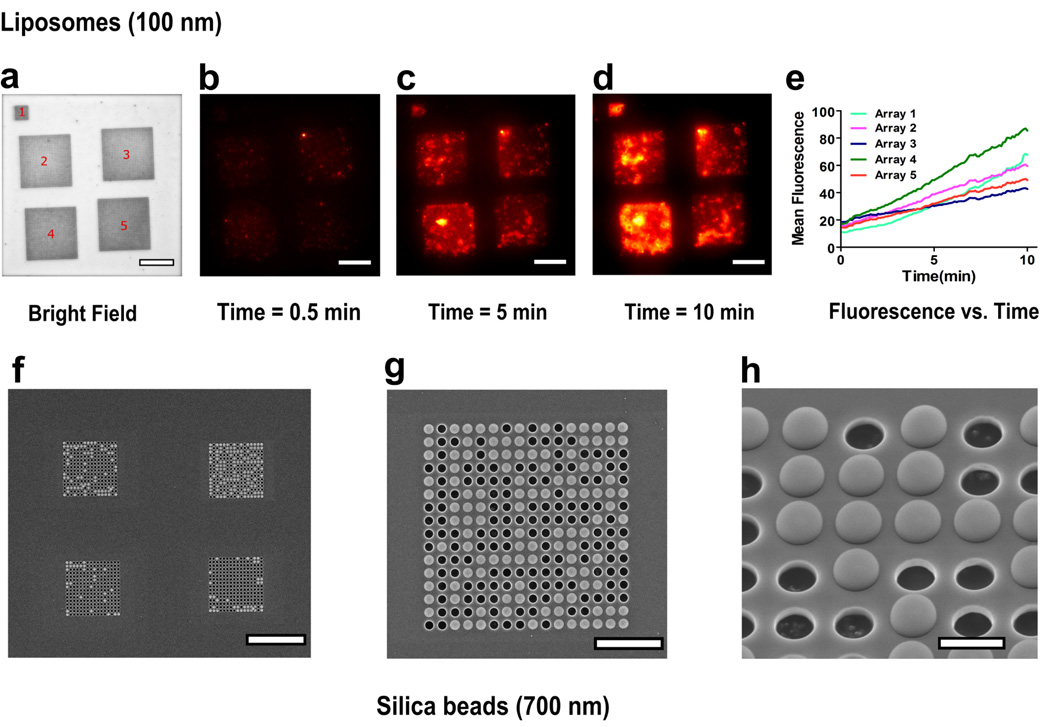
Figure 2.
Rapid, highly specific, and materials-general particle assembly over nanopore arrays, demonstrated using lipid vesicles and silica beads. (a–d) Bright field and time-lapse images of a suspended nitride area showing assembly of fluorescent liposomes over the array region. The larger 4 arrays (16 µm × 16 µm) have a 400 nm pore diameter and 1 µm periodicity, whereas the smaller array (8 µm × 8 µm) has a 200 nm pore diameter and 500 nm periodicity. (e) Graph showing the rate of increase of mean fluorescence of each array with time. (f–h) Scanning electron micrographs showing assembly of 700 nm silica beads over nanopore arrays. The arrays (16 µm × 16 µm) have a 600 nm pore diameter and 1 µm periodicity. (f) Suspended nitride membrane with silica beads assembled over four nanopore arrays with no beads visible elsewhere on the membrane. (g) SEM image demonstrating alignment of particles on an array having 600 nm pores with 1 µm periodicity. (h) A tilted and zoomedin image of the array. Scale bar is 20 µm for figures (a–f), 5 µm for figure (g) and 1 µm for figure (h).
SAPE and Silane-PEG-biotin Binding Assay
Streptavidin R-phycoerythrin conjugate (SAPE, mol. wt. 292.8 kD) was obtained from Invitrogen and was diluted in PBS to the desired concentration. Silane-Polyethylene glycol (PEG)-biotin was obtained from NanoCS Inc. Silane-PEG-biotin was diluted in distilled water to make a 1 mM solution. The nanopore array chips with silica layer were left submersed in the solution overnight to form a biotinylated self-assembled monolayer on the surface. The chips were then gently rinsed to remove excess biotin from the sample surface by submersing in distilled water for 5 minutes. This step was repeated thrice with fresh DI water.
For SAPE binding, 10 µL of sample solution was added to a reservoir and left for 10 minutes. A cover slip was placed on the backside of the chip and it was placed on a glass slide to be quickly imaged. The chip was then gently rinsed by submersing in fresh DI water for 5 minutes. This step was repeated thrice with fresh DI water to remove unbound SAPE before fluorescence imaging of the sensor chip.
Label-free SPR Detection
For SPR sensing, nanopore chips with a gold layer deposited on the cis side were used. The suspended nitride area had 4 nanopore arrays, two (16 µm × 16 µm) and two (8 µm × 8 µm), each with nanopore diameter 200 nm and period 500 nm. A monolayer of silane-PEG-biotin was formed on the chips after silica coating, as mentioned above. Optical transmission spectra through the nanopores were measured in phosphate buffered saline (PBS) before addition of SAPE. 10 µL SAPE solution was added to the chip and left undisturbed for 10 minutes. This solution was then replaced with fresh PBS thrice to remove unbound SAPE molecules. Spectra through the nanopore arrays were measured again and used to calculate the shift caused by binding of SAPE.
Choleratoxin Binding Assay
For preparation of phospholipid vesicles containing ganglioside functional group, vesicles were prepared as stated but with 1% (w/w) GM1 and extruded to 200 nm. For formation of a vesicle array, 5 µL of 0.1 mg/mL vesicle solution was placed over a single reservoir for 5 minutes. The cis side of the sample was rinsed with PBS to wash off excess solution and then blocked against nonspecific binding by adding bovine serum albumin (BSA; 2 mg/mL). The setup was left undisturbed for 30 minutes. The chip was rinsed with PBS to wash off excess BSA and 50 nM choleratoxin (CTX) was added to the cis side of the chip. Again the chip was left undisturbed for 30 minutes to allow the CTX to bind with GM1. The chip was washed with PBS again to remove unbound CTX. An upright microscope was used to capture fluorescence images through the trans side of the chip.
Microscope and Software
The images and optical transmission spectra were recorded using a Nikon Eclipse LV 100 upright microscope and a 50× objective. In the SPR experiments, a MATLAB™ script was used which fits a second-order polynomial function around the measured peaks and calculates their centroid. This script is used to track the shift of the desired peak. Photometrics CoolSNAP HQ2 CCD was used in combination with Voodoo software to record the images. ImageJ software was used for analysis and coloring of the images. Data plots were prepared using GraphPad Prism version 5.04. (GraphPad Software, Inc.)
RESULTS AND DISCUSSION
The chip design and schematic of action are shown in Figure 1. A 1 inch × 1 inch chip containing 9 sensing areas was fabricated in a silicon wafer by photolithography and anisotropic etching with KOH (Figure 1a). The visible openings form a truncated pyramid reservoir with sides of ~1 mm which converge to a suspended Si3N4 area with sides of approximately 150 µm (Figure 1b). For clarity, we refer to the surface of the chip with visible reservoir openings as the cis side and the opposite surface as the trans side. A single chip contains 16 sensing areas, each of which can be used independently. A 200 nm gold layer was deposited on the Si3N4 film to make the platform amenable to analytical techniques such as SPR,22 SERS,37 and electrochemistry.38 Figure 1c shows the suspended nitride area with 4 arrays milled using FIB. Scanning electron micrograph (SEM) of an array is shown in Figure 1d. The schematic (Figure 1e) shows a single drop of solution (1–10 µL in volume) placed on a single etched reservoir leading to nitride and gold layers with milled nanopore arrays. Figure 1f demonstrates the proposed passive convection mechanism for local concentration and assembly of particles at the nanopore array site. Evaporation of solution through the open end of the nanopores from the trans side of the chip can act as a passive pump to drive flow of solution towards the nanopores. The spontaneous flow of liquid towards the nanopore arrays directs particle migration and accumulation at the array site. Any loss of solution volume in the chamber due to evaporation and fluid flow also concentrates the particles and brings them closer to the nanopore arrays. Once the particles are close to the nanopore arrays, convection directs them to the array region where they are trapped (Figure 1f).
Particle Array Assembly
We investigated the time course of particle concentration by taking timelapse images. A drop of a solution of 0.1 M NaCl containing ~100 nm-diameter phospholipid vesicles was added on a single reservoir and images were taken every 5 seconds. A movie created using the time-lapse images can be viewed in the supplementary section (Movie S1). Figure 2a shows a bright-field image of the suspended nitride area with five nanopore arrays. The larger 4 arrays (16 µm × 16 µm) have a 400 nm pore diameter and 1 µm periodicity (pore center-to-center distance), whereas the smaller array (8 µm × 8 µm) has a 200 nm pore diameter and 500 nm periodicity. Figures 2b–d were taken from the time-lapse sequence and demonstrate the fluorescence increase over the nanopore arrays as more vesicles aggregate on them. The mean fluorescence intensity for each array area was measured and plotted as a function of time (Figure 2e). There was no significant change in fluorescence from the area in between the arrays. The plot also indicates that the rate of increase of mean fluorescence was approximately linear over the course of 10 minutes. The process is independent of the physical and chemical nature of the particle, as similar results were obtained for lipid vesicles extruded to 100 nm, silica beads (mean diameter: 700 nm) (Figure 2f–h) and two samples of fluorescently tagged polystyrene beads (mean diameters: 2.19 µm and 1.8 µm) (Figure S1 and S3). These results were obtained consistently with our experiments. Furthermore, during the observed interval, there was a steady flow of particles towards the arrays, which indicates that while the nanopores may be obscured by previously arriving particles, convective particle flux is not drastically reduced.
Effective particle aggregation was observed for pore diameters ranging from 200 to 1200 nm. 8 µm × 8 µm arrays with a 200 nm pore diameter and 500 nm periodicity showed favorable results. Metallic nanopore arrays with these dimensions were previously used for SPR sensing.22
Dependence of the passive assembly process on evaporation was supported by a test where particle accumulation over the nanopore arrays was significantly reduced upon placing the chip in a humidity chamber. (Figure S3) The effect of cis/trans addition as well as surface modification by deposition of silica on particle accumulation was studied. The samples were incubated with 10 µL solution of fluorescent polystyrene beads for 10 minutes and then imaged. (Figure S4) Although changing between cis and trans addition of solutions resulted in relatively small variations in particle accumulation, changing the surface characteristics had a much more prominent influence. The nanopore chips with conformal ALD silica coating demonstrated increased particle concentration as compared to unmodified Au/Si3N4 chips. This is likely due to improved wetting of the nanopores and increased surface area of the sample droplet on the more hydrophilic silica surface, which promotes faster evaporation.
Nanopore-directed convection can also be used for passive, spontaneous self-assembly of particles using the right combination of array parameters, concentration of particles used and time allowed for aggregation. (Figure 2f–h) For optimal packing of the particles into arrays, the pore diameter was tuned to be slightly smaller than the particle diameter and the edge-to-edge distance between pores was less than the pore diameter. The silica beads had a mean diameter of 700 nm whereas the nanopores had a diameter of 600 nm and periodicity of 1 µm. The resultant bead arrays were very stable and they remained immobilized after overnight water submersion. Furthermore, the areas outside of the nanopore arrays were completely devoid of adsorbed beads.
Time-lapse sequences, which recorded accumulation of 1.8 µm polystyrene beads over nanopore arrays, were used to generate an estimate of solution flow rate through the arrays. Trans addition of solution was used for two kinds of chips, type A with nitride surface exposed on the trans side and type B with 10 nm-thick silica deposited all over the chip. For an array (size 32 µm × 32 µm) with a 1.2 µm pore diameter and 2 µm periodicity, we obtained an estimated flow rate of approximately 6 nL/min for type A and 14 nL/min for type B. Individual arrays (size 16 µm × 16 µm) with a 600 nm pore size and 1 µm periodicity were estimated to have a flow rate of ~3 nL/min for samples type A and ~5 nL/min for type B. Similarly, arrays (8 µm × 8 µm) with a 200 nm pore size and 500 nm periodicity had an approximate flow rate of ~1 nL/min for sample type A and ~2 nL/min for type B. Using this mechanism and by increasing the number of arrays, it should be straightforward to achieve passive flow through speeds in the order of 0.1 µL/min through suspended membrane nanopore arrays. Patterning large-area nanopore arrays can further increase the flow rates.
Protein Preconcentration for Fluorescence and SPR Analysis
Protein preconcentration for biochemical assays is highly desired for early detection of low-concentration disease biomarkers. Micro- and nanofluidic channels in combination with applied electric field have been previously utilized for preconcentration and increased sensitivity for immunoassays.39, 40 We utilized our setup to demonstrate passive and rapid detection of low concentration analytes in small-volume droplets. The experiment was performed on a nanopore array chip with ALD silica coating. The chips were placed in a 1 mM silane-PEG-biotin solution overnight to form a monolayer of exposed biotin over the entire surface. The silane moiety covalently links the PEG-biotin to the silica surface and leaves the biotin free to interact with fluorescent SAPE (Figure 3a). A homogeneous monolayer of PEG-biotin ensures that SAPE can bind anywhere on the chip, not only on the nanopore array.
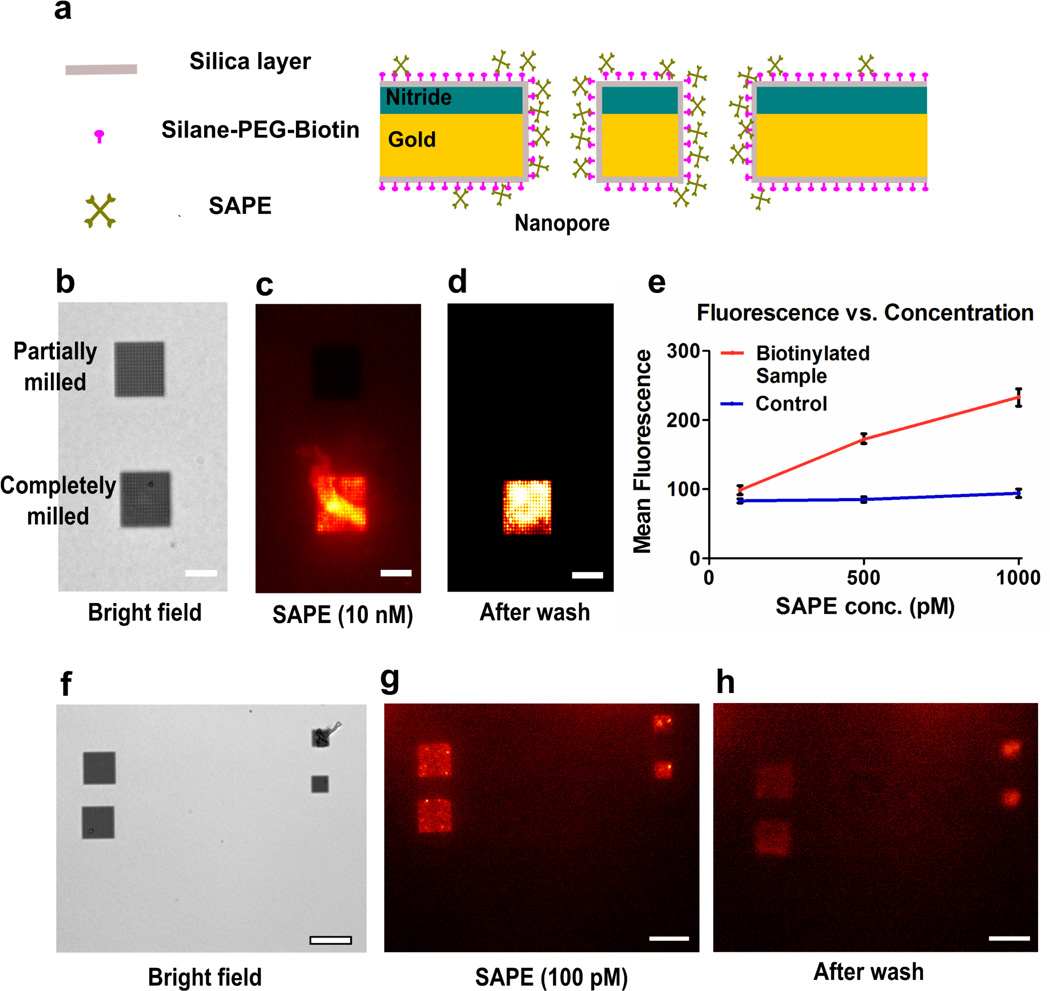
Figure 3.
Schematic, flow dependence and low concentration analysis of SAPE-biotin binding. (a) Schematic showing all surfaces of the chip covered with silica and a monolayer of silane-PEG-biotin. SAPE is directed towards the nanopore arrays where it binds with the biotin. (b) Bright-field image of a suspended nitride region with one partially milled nanopore array and an array with completely milled pores. The arrays (16 µm × 16 µm) have a 600 nm pore diameter and 1 µm periodicity. Scale bar is 10 µm for (b–d). (c) Fluorescence image of the sample after addition of 10 nM SAPE solution demonstrating nanopore-induced pre-concentration. The partially milled pores are not open on the trans side of the chip and hence cannot act as capillaries. (d) Fluorescence image of the sample after rinsing the chip with DI water. (e) A graph showing linear relationship between the mean fluorescence over the nanopore arrays as a function of the concentration of SAPE. (f) Bright-field image of a suspended nitride region with 4 nanopore arrays. Larger arrays (16 µm × 16 µm) have a 600 nm pore diameter and 1 µm periodicity. Smaller arrays (8 µm × 8 µm) have a 200 nm pore diameter and 500 nm periodicity. (g) Fluorescence image of the sample after addition of 100 pM SAPE solution. (h) Fluorescence image of the area after rinsing the chip with DI water. Scale bar is 20 µm for (f–h).
To confirm that the preconcentration process originates from trans-nanopore solution flow, we used an array of partially milled, dead-ended pores as control. (Figure 3b–d) The dead-ended pores were exposed to the focused ion beam for around two-thirds of the time compared to the completely milled arrays. Upon adding a 10 nM SAPE solution, intense fluorescence was exclusive to the completely milled arrays. As the dead-ended pores could not display capillary action, SAPE binding in them was dependent on diffusion of molecules. Comparison of the fluorescence intensities in the completely milled and partially milled pores demonstrates that the directed convection in the open nanopores significantly increased the analyte concentration and binding (Figure 3b–d).
To quantify the concentration response of the nanopore arrays, we used SAPE concentrations of 1 nM, 500 pM and 100 pM and measured the fluorescence of the nanopore array region after an allowed binding time of 10 minutes and rinsing with DI water. We were able to detect fluorescence from nanopore arrays for all the samples. (Figure 3e–h) For 100 pM, the small concentration combined with the small volume (10 µL) means that 1 femtomole of SAPE molecules were added to the cis side of the chip. (Figure 3f–h) A fraction of the solution was transported through the nanopores and detected within 10 minutes of concentration. For all the samples used, the background fluorescence from the region surrounding the nanopore arrays was not significantly larger than that observed before addition of SAPE. Also, a control sample, which had not been treated with silane-PEG-biotin solution, did not exhibit fluorescence from any region of the suspended membrane for all concentration values, indicating that SAPE does not strongly adsorb to silica. The mean fluorescence value per nanopore array was plotted against concentration of SAPE for the samples (Figure 3e). The biotinylated sample shows a linear response in terms of measured fluorescence to the concentration of the SAPE used. The observed fluorescence intensity was not dependent on the pore diameter and periodicity, so we can negate the possibility of the fluorescence enhancement being due to plasmonic effects.41
Analysis of SAPE binding to the nanopore arrays as compared to diffusion-limited dead-ended nanohole arrays has been further demonstrated in Figure S2. Directed convection of analyte solution to the nanopore arrays leads to accumulation of molecules and enhancement of fluorescence at the array site with time (Figure S2a). In comparison, there is no significant change in fluorescence measurements from the dead-ended nanohole arrays. For concentration-dependent binding analysis, SAPE concentrations of 1, 10, and 25 nM were added to the nanopore samples whereas the dead-ended nanoholes were used with SAPE concentrations of 25, 50, and 100 nM. The mean fluorescence values per nanohole array were obtained after a 10-minute binding interval and subsequent washing with DI water. Analysis of fluorescence measurements clearly demonstrates the enhanced binding response obtained from the nanopore array samples (Figure S2b).
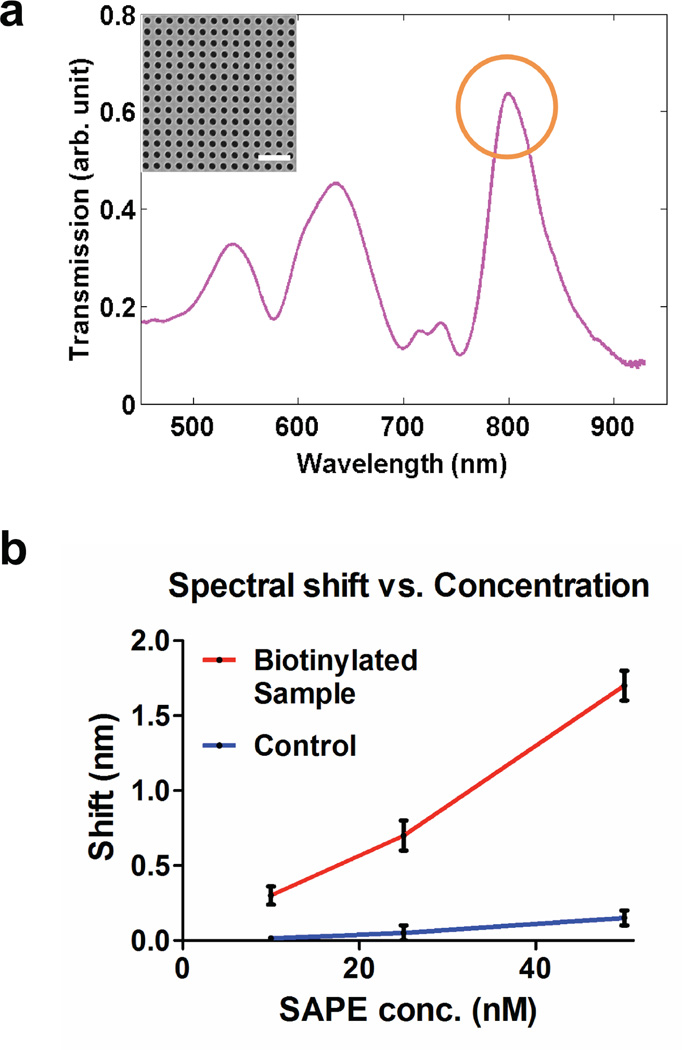
Nanopore arrays with gold deposited on the cis side were used to measure SAPE binding by SPR. The diameter of the nanopores and periodicity of the array are 200 and 500 nm, respectively (Figure 4a inset). These dimensions were chosen to position optical transmission peaks through nanopore arrays in the visible regime for spectral measurements. As previously shown, the nanopore array made in metallic films can couple the incident light to surface plasmons, giving optical transmission peaks at specific wavelengths determined by the geometry of the array as well as the interfacial refractive index. The peak transmission wavelength is sensitive to the interfacial refractive index changes caused by molecular binding, and thus enables label-free SPR biosensing.22 Normalized transmission spectra obtained through the nanopore array is shown in Figure 4a. A transmission peak at approximately 800 nm, which is associated with surface plasmons at the gold-water interface, was used for sensing. The plot (Figure 4b) shows an increase in spectral shift with SAPE concentration for the biotinylated sample, whereas the control sample with no silane-PEG-biotin shows very little shift because of negligible nonspecific binding of SAPE to the surface, which agrees with the fluorescence measurements.
Figure 4.
Surface plasmon resonance detection of SAPE-biotin binding. (a) Normalized transmission spectra as recorded from an array of 200 nm nanopores. The transmission peak at approximately 800 nm corresponds to the gold-water interface and was used to measure the shift in transmission. (Inset) SEM image of nanopores (diameter: 200 nm, periodicity: 500 nm) used for spectral measurements. Scale bar is 1.5 µm. Four arrays were milled on the suspended nitride area for measuring the transmission spectra, two large (16 µm × 16 µm) and two smaller (8 µm × 8 µm). (b) A graph showing shift of transmission spectra through the nanopore arrays upon SAPE binding as a function of SAPE concentration.
Liposome Nanoarray
The results obtained with particles demonstrate that this method can be used to create particle nano- or microarrays without the need for chemical modification of the surface or particle. The successful application of microarray technology in high throughput analysis of biological particles has now led to tremendous interest in development of nanoarrays, which focus on immobilization of biomolecules in a small volume with very high spatial density.42–45 In the method presented here, once the particles are in position and adhered to the walls of the nanopores, they could be utilized to perform statistical studies or create random particle arrays for multiplex biosensing experiments.46, 47 Accumulation of a monolayer of particles over 300 nm-deep nanopores enables study of the arranged particles from both the cis and trans sides of the chip. (Figure 5a) As seen in the schematic viewed from the trans side, each pore acts as a window with the ability to analyze the particle immobilized inside a nanopore. Thus we end up with an orderly assembly of particles in a nano- or microarray format without the requirement of any external chemical modification, which may alter the properties and behavior of the objects being tested. This method of analysis, where a thin membrane is used as a shadow mask to create an impression of arrays, can be utilized for single particle studies.
Figure 5.
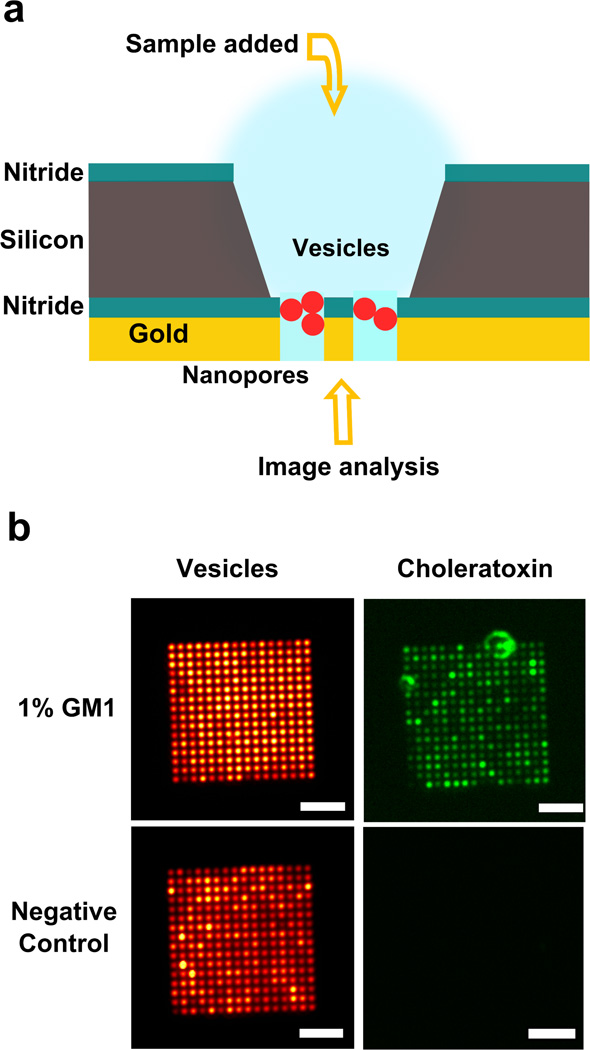
Suspended nanopore arrays for biological assays. (a) Schematic showing that lipid vesicles immobilized in nanopores are accessible from both sides for analysis. (b) Two suspended nanopore arrays on a chip where vesicles were added from the cis reservoir side and images were collected from the trans side. The vesicles contained ganglioside GM1, which is bound by fluorescently tagged cholera toxin. The arrays (16 µm × 16 µm) have a 600 nm pore diameter and 1 µm periodicity. Scale bar: 5 µm.
For analysis of an array of single particles from both sides, the simplest case would be as visible in Figure 2f–h where the particle size is homogeneous, larger than the pore diameter and results in a monolayer thus enabling single particle analysis. The process is also efficient for physiologically relevant cases where the particle immobilization might be less uniform owing to size variability. We demonstrate this method using lipid vesicles and test them for a membrane-bound receptor. Lipid bilayer vesicles act as models for cell membranes and this method could be utilized for study of membrane components including membrane-bound proteins, which are targets for the action of many drugs.12, 48 Vesicles with ganglioside GM1 (monosialotetrahexosylganglioside) were extruded to 200 nm and used for this assay. The B-subunit of cholera toxin (CTX) binds GM1 with high affinity. Nanopore arrays with a 600 nm pore diameter and 1 µm periodicity were used. The vesicle solution and subsequently Alexa 488-conjugated CTX were both added from the cis side. The images were obtained from the trans side utilizing each nanopore as a viewing window. Figure 5b shows that we can utilize the advantages of two open surfaces with this design, with the vesicles and their reaction with CTX being recorded in a visible array format.
CONCLUSION
We have demonstrated a fast, passive, and small-volume method of directed immobilization of particles in orderly arrays using a suspended Au/Si3N4 membrane perforated with nanopores. Our experiments demonstrate that nanopore arrays can act as nano-drains, spontaneously draining solutions from the reservoir via capillary flow and evaporation at the open end of the nanopores. This process, in turn, promotes molecular concentration and particle accumulation around the nanopores. This simple setup improves analyte transport by directing convection to the sensor area, obviating the need for other complicated techniques or external power sources. The ease of experimentation, analysis and minimal sample consumption (less than 10 µL) make these nanopore arrays a powerful optofluidic platform for point-of-care sensing of biomarkers, which occur in low concentration. The simplicity of the passive sample injection and concentration makes the platform potentially useful for diagnostic applications in the developing world, as particle array formation or molecular concentration and subsequent fluorescence readout and analysis requires minimal setup and expertise. In addition, we have shown that the nanopore-directed binding of molecules at the array site can be detected via SPR. The setup can also be used to obtain a periodic arrangement of monolayer of particles, aimed to gather statistical information from small-volume assays. These results make a strong case for nanopore arrays as integrated optofluidic platforms, with their application demonstrated in label-free sensing, fluorescence assays, concentration and particle trapping. Efficient and large-scale fabrication of the nanopore platform is possible based on recent advances in high-throughput fabrication of metallic nanoholes.17, 25, 49 Our platform can readily be combined with nanohole-based SPR,13, 15, 19, 50 SERS,14, 37 and electrochemical sensors38 for improved chemical selectivity and lower detection limits for molecules of biological and physiological relevance.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by grants from the National Institutes of Health (R01 GM092993), the National Science Foundation (NSF CAREER Award and DBI 0964216), the Office of Naval Research (ONR) Young Investigator Program (N00014-11-1-0645), and the Minnesota Partnership Award for Biotechnology and Medical Genomics.
REFERENCES
- 1.Psaltis D, Quake SR, Yang C. Nature. 2006;442:381–386. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 2.Monat C, Domachuk P, Eggleton BJ. Nat. Photonics. 2007;1:106–114. [Google Scholar]
- 3.Erickson D, Sinton D, Psaltis D. Nat. Photonics. 2011;5:583–590. [Google Scholar]
- 4.Myers FB, Lee LP. Lab Chip. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 5.Fan X, White IM. Nat. Photonics. 2011;5:591–597. doi: 10.1038/nphoton.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Li H, Reddy K, Shelar HS, Nittoor VR, Fan X. Appl. Phys. Lett. 2011;98:041104. [Google Scholar]
- 7.Dimov IK, Kijanka G, Park Y, Ducrée J, Kang T, Lee LP. Lab Chip. 2011;11:2701. doi: 10.1039/c1lc20105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo P, Hall EW, Schirhagl R, Mukaibo H, Martin CR, Zare RN. Lab Chip. 2012;12:558–561. doi: 10.1039/c2lc21092d. [DOI] [PubMed] [Google Scholar]
- 9.Sheehan PE, Whitman LJ. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 10.Squires TM, Messinger RJ, Manalis SR. Nat. Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 11.Cherney DP, Harris JM. Annu. Rev. Anal. Chem. 2010;3:277–297. doi: 10.1146/annurev-anchem-070109-103404. [DOI] [PubMed] [Google Scholar]
- 12.Cooper MA. J. Mol. Recognit. 2004;17:286–315. doi: 10.1002/jmr.675. [DOI] [PubMed] [Google Scholar]
- 13.Brolo AG, Gordon R, Leathem B, Kavanagh KL. Langmuir. 2004;20:4813–4815. doi: 10.1021/la0493621. [DOI] [PubMed] [Google Scholar]
- 14.Brolo AG, Arctander E, Gordon R, Leathem B, Kavanagh KL. Nano Lett. 2004;4:2015–2018. [Google Scholar]
- 15.Dahlin AB, Zäch M, Rindzevicius T, Käll M, Sutherland DS, Höök F. J. Am. Chem. Soc. 2005;127:5043–5048. doi: 10.1021/ja043672o. [DOI] [PubMed] [Google Scholar]
- 16.Tetz K, Pang L, Fainman Y. Opt. Lett. 2006;31:1528–1530. doi: 10.1364/ol.31.001528. [DOI] [PubMed] [Google Scholar]
- 17.Henzie J, Lee MH, Odom TW. Nat. Nanotechnol. 2007;2:549–554. doi: 10.1038/nnano.2007.252. [DOI] [PubMed] [Google Scholar]
- 18.Lesuffleur A, Im H, Lindquist NC, Oh SH. Appl. Phys. Lett. 2007;90:243110. [Google Scholar]
- 19.Yang JC, Ji J, Hogle JM, Larson DN. Nano Lett. 2008;8:2718–2724. doi: 10.1021/nl801043t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindquist NC, Lesuffleur A, Im H, Oh SH. Lab Chip. 2009;9:382–387. doi: 10.1039/b816735d. [DOI] [PubMed] [Google Scholar]
- 21.Eftekhari F, Escobedo C, Ferreira J, Duan X, Girotto EM, Brolo AG, Gordon R, Sinton D. Anal. Chem. 2009;81:4308–4311. doi: 10.1021/ac900221y. [DOI] [PubMed] [Google Scholar]
- 22.Im H, Wittenberg NJ, Lesuffleur A, Lindquist NC, Oh SH. Chem. Sci. 2010;1:688–696. doi: 10.1039/C0SC00365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonsson MP, Dahlin AB, Feuz L, Petronis S, Höök F. Anal. Chem. 2010;82:2087–2094. doi: 10.1021/ac902925e. [DOI] [PubMed] [Google Scholar]
- 24.Yanik AA, Huang M, Kamohara O, Artar A, Geisbert TW, Connor JH, Altug H. Nano Lett. 2010;10:4962–4969. doi: 10.1021/nl103025u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brolo AG. Nat. Photonics. 2012;6:709–713. [Google Scholar]
- 26.Wittenberg NJ, Im H, Xu X, Wootla B, Watzlawik J, Warrington AE, Rodriguez M, Oh SH. Anal. Chem. 2012;84:6031–6039. doi: 10.1021/ac300819a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebbesen TW, Lezec HJ, Ghaemi HF, Thio T, Wolff P. Nature. 1998;391:667–669. [Google Scholar]
- 28.McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A. Nano Lett. 2010;10:2237. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chansin GAT, Mulero R, Hong J, Kim MJ, deMello AJ, Edel JB. Nano Lett. 2007;7:2901–2906. doi: 10.1021/nl071855d. [DOI] [PubMed] [Google Scholar]
- 30.Miles BN, Ivanov AP, Wilson KA, Doğan F, Japrung D, Edel JB. Chem. Soc. Rev. 2013;42:15–28. doi: 10.1039/c2cs35286a. [DOI] [PubMed] [Google Scholar]
- 31.Chen HM, Pang L, Gordon MS, Fainman Y. Small. 2011;7:2750–2757. doi: 10.1002/smll.201100264. [DOI] [PubMed] [Google Scholar]
- 32.Escobedo C, Brolo AG, Gordon R, Sinton D. Nano Lett. 2012;12:1592–1596. doi: 10.1021/nl204504s. [DOI] [PubMed] [Google Scholar]
- 33.Cesaro-Tadic S, Dernick G, Juncker D, Buurman G, Kropshofer H, Michel B, Fattinger C, Delamarche E. Lab Chip. 2004;4:563–569. doi: 10.1039/b408964b. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Jing N, Chou IH, Cote GL, Kameoka J. Lab Chip. 2007;7:630–632. doi: 10.1039/b618105h. [DOI] [PubMed] [Google Scholar]
- 35.Lynn NS, Dandy DS. Lab Chip. 2009;9:3422. doi: 10.1039/b912213c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamblin MN, Xuan J, Maynes D, Tolley HD, Belnap DM, Woolley AT, Lee ML, Hawkins AR. Lab Chip. 2010;10:173–178. doi: 10.1039/b916746c. [DOI] [PubMed] [Google Scholar]
- 37.Yu Q, Braswell S, Christin B, Xu J, Wallace PM, Gong H, Kaminsky D. Nanotechnology. 2010;21:355301. doi: 10.1088/0957-4484/21/35/355301. [DOI] [PubMed] [Google Scholar]
- 38.Dahlin AB, Sannomiya T, Zahn R, Sotiriou GA, Vörös J. Nano Lett. 2011;11:1337. doi: 10.1021/nl104424q. [DOI] [PubMed] [Google Scholar]
- 39.Wang YC, Stevens AL, Han J. Anal. Chem. 2005;77:4293–4299. doi: 10.1021/ac050321z. [DOI] [PubMed] [Google Scholar]
- 40.Wang YC, Han J. Lab Chip. 2008;8:392–394. doi: 10.1039/b717220f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brolo AG, Kwok SC, Moffitt MG, Gordon R, Riordon J, Kavanagh KL. J. Am. Chem. Soc. 2005;127:14936–14941. doi: 10.1021/ja0548687. [DOI] [PubMed] [Google Scholar]
- 42.Bruckbauer A, Zhou D, Kang DJ, Korchev YE, Abell C, Klenerman D. J. Am. Chem. Soc. 2004;126:6508–6509. doi: 10.1021/ja0317426. [DOI] [PubMed] [Google Scholar]
- 43.Sinensky AK, Belcher AM. Nat. Nanotechnol. 2007;2:653–659. doi: 10.1038/nnano.2007.293. [DOI] [PubMed] [Google Scholar]
- 44.Lee BK, Lee HY, Kim P, Suh KY, Kawai T. Lab Chip. 2009;9:132–139. doi: 10.1039/b809732a. [DOI] [PubMed] [Google Scholar]
- 45.Wittenberg NJ, Im H, Johnson TW, Xu X, Warrington AE, Rodriguez M, Oh SH. ACS Nano. 2011;5:7555–7564. doi: 10.1021/nn202554t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walt DR. Chem. Soc. Rev. 2010;39:38–50. doi: 10.1039/b809339n. [DOI] [PubMed] [Google Scholar]
- 47.Goodey A, Lavigne JJ, Savoy SM, Rodriguez MD, Curey T, Tsao A, Simmons G, Wright J, Yoo SJ, Sohn Y, Anslyn EV, Shear JB, Neikirk DP, McDevitt JT. J. Am. Chem. Soc. 2001;123:2559–2570. doi: 10.1021/ja003341l. [DOI] [PubMed] [Google Scholar]
- 48.Drews J. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 49.Im H, Lee SH, Wittenberg NJ, Johnson TW, Lindquist NC, Nagpal P, Norris DJ, Oh SH. ACS Nano. 2011;5:6244–6253. doi: 10.1021/nn202013v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Im H, Sutherland JN, Maynard JA, Oh SH. Anal. Chem. 2012;84:1941–1947. doi: 10.1021/ac300070t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.