Abstract
As a reference laboratory, the Streptococcus Laboratory at the Centers for Disease Control and Prevention (CDC) is frequently asked to confirm the identity of unusual or difficult-to-identify catalase-negative, gram-positive cocci. In order to accomplish the precise identification of these microorganisms, we have systematically applied analysis of whole-cell protein profiles (WCPP) and DNA-DNA reassociation experiments, in conjunction with conventional physiological tests. Using this approach, we recently focused on the characterization of three strains resembling the physiological groups I (strain SS-1730), II (strain SS-1729), and IV (strain SS-1728) of enterococcal species. Two strains were isolated from human blood, and one was isolated from human brain tissue. The results of physiological testing were not consistent enough to allow confident inclusion of the strains in any of the known enterococcal species. Resistance to vancomycin was detected in one of the strains (SS-1729). Analysis of WCPP showed unique profiles for each strain, which were not similar to the profiles of any previously described Enterococcus species. 16S ribosomal DNA (rDNA) sequencing results revealed three new taxa within the genus Enterococcus. The results of DNA-DNA relatedness experiments were consistent with the results of WCPP analysis and 16S rDNA sequencing, since the percentages of homology with all 25 known species of Enterococcus were lower than 70%. Overall, the results indicate that these three strains constitute three new species of Enterococcus identified from human clinical sources, including one that harbors the vanA gene. The isolates were provisionally designated Enterococcus sp. nov. CDC Proposed New Species of Enterococcus 1 (CDC PNS-E1), type strain SS-1728T (= ATCC BAA-780T = CCUG 47860T); Enterococcus sp. nov. CDC PNS-E2, type strain SS-1729T (= ATCC BAA-781T = CCUG 47861T); and Enterococcus sp. nov. CDC PNS-E3, type strain SS-1730T (= ATCC BAA-782T = CCUG 47862T).
The enterococci have undergone considerable changes in taxonomy in the past few years. Since the recognition of Enterococcus as a separate genus (14), several new species have been described as a result of improvements in the methods for their identification combined with a growing interest in their role as opportunistic pathogens (4, 9, 16). However, the differentiation of some of the species belonging to the genus Enterococcus remains problematic because of the overlap of phenotypic characteristics (6, 17). Furthermore, some enterococcal bacteria do not possess all typical characteristics, and questions on their precise identification may arise (6, 8, 16). In an effort to confirm the identity of unusual or difficult-to-identify catalase-negative, gram-positive cocci, we have used analysis of whole-cell protein profiles, DNA-DNA reassociation experiments, and 16S ribosomal DNA (rDNA) sequencing, in conjunction with conventional physiological tests, to phylogenetically characterize three hitherto-unknown Enterococcus-like bacteria isolated from human clinical specimens and resembling, respectively, the physiological groups I, II, and IV of enterococcal species (16). On the basis of the results of a polyphasic taxonomic study, we considered that the three unknown cocci represent three new species within the genus Enterococcus. We propose to denominate them Enterococcus sp. nov. Centers for Disease Control and Prevention (CDC) Proposed New Species of Enterococcus 1 (CDC PNS-E1), Enterococcus sp. nov. CDC PNS-E2, and Enterococcus sp. nov. CDC PNS-E3, in light of the recommendation in minute 10 of the July 2002 meeting of the International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of staphylococci and streptococci (20) that refers to the description of new species based on a single isolate.
MATERIALS AND METHODS
Bacterial strains.
The three strains in this study were taken from the culture collection of the CDC Streptococcus Laboratory. Two strains were isolated from blood; one of these (SS-1728T) was isolated from a patient in Evanston, Ill., in 1991, and the other (SS-1729T) was isolated from a patient in Los Angeles, Calif., in 1997. The third strain (SS-1730T) was isolated from brain tissue obtained from an 11-month-old patient in Honolulu, Hawaii, in 2001. The type strains of all enterococcal species described to date were included for comparative purposes.
Characterization of strains.
The strains were tested for their phenotypic characteristics by conventional physiological tests and serological grouping as described previously (8, 16). Reactivity with the AccuProbe Enterococcus culture confirmation test (Gen-Probe, Inc., San Diego, Calif.) was assayed as directed by the manufacturer.
Susceptibility testing for vancomycin and detection of resistance gene.
Susceptibility testing for penicillin, ampicillin, vancomycin, chloramphenicol, doxycycline, linezolid, rifampin, gentamicin, and streptomycin was done with the Etest (AB Biodisk, Solna, Sweden) as described by the manufacturer and interpreted as recommended by the National Committee for Clinical Laboratory Standards (12). PCR for detection of the vancomycin resistance genes was done by the procedures suggested by Clark et al. (3).
Analysis of whole-cell protein profiles by SDS-PAGE.
Preparation of extracts and analysis of whole-cell protein profiles by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as described by Merquior et al. (11). Coefficients of similarity or Dice indices were determined for each isolate by using the Molecular Analyst Fingerprinting Plus software package, version 1.6 (Bio-Rad Laboratories, Hercules, Calif.), and a dendrogram was constructed by the unweighted pair-group method with arithmetic averages.
DNA reassociation studies.
Harvesting and lysis of the bacterial cells were performed by previously described methods (17). Extraction and purification of DNA and the determination of DNA relatedness by using the hydroxyapatite hybridization method were done as described by Brenner et al. (1). DNA-hybridization experiments were performed at 55°C for optimal DNA reassociation. The G+C content was determined by the optical melting temperature and equilibrium buoyant methods according to the procedures of Mandel et al. (10). All samples were run at least three times, using DNA from Escherichia coli K-12 as a control.
16S rRNA gene sequence and phylogenetic analysis.
DNA extraction and purification were done with a QIAamp DNA Mini Kit (Qiagen Inc., Valencia, Calif.) according to the manufacturer's instructions. Purified genomic DNA was amplified by using the Expand high-fidelity PCR system (Roche, Indianapolis, Ind.) with primers fL1 and rL1 (13). Amplification conditions were 94°C for 5 min; 35 cycles of 94°C for 15 s, 50°C for 15 s, and 72°C for 90 s; and finally a single extension of 72°C for 5 min followed by a 4°C hold. Products were confirmed by running 10-μl samples on a 1.0% (wt/vol) agarose gel for 30 min at 75 V. Excess deoxynucleoside triphosphates and primers were inactivated with the ExoSAP-IT method (U.S. Biochemical Corp., Cleveland, Ohio). Cycle sequencing was performed with Big Dye version 3.1 dye terminator chemistry (Applied Biosystems, Foster City, Calif.) by standard protocols with a 3.2 pM concentration of small subunit primers. Excess dyes were removed with magnetic carboxylate beads (Agencourt Bioscience, Beverly, Mass.), and reaction products were sequenced on an ABI 3100 sequencer (Applied Biosystems). Sequences were assembled with Seqmerge (Genetics Computer Group) and trimmed to at least two confirming reads. Novel sequences were gapped, and unique sequences were subjected to a Blast search against GenBank. Related entries were aligned with Pileup (5) and trimmed to consensus, and further analysis was performed with Bioedit and Treecon. In Bioedit, the sequences were realigned with Clustal W (18) with 1,000 bootstraps, and a distance matrix was created. In Treecon, distances of aligned sequences were estimated by the Jukes-Cantor method and bootstrapped 1,000 times, and tree topology was determined by the neighbor-joining method. The final phylogenetic tree was rooted with an outgroup, and bootstrap values of above 85% were displayed as percentages.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rDNA sequences of Enterococcus sp. nov. CDC PNS-E1, Enterococcus sp. nov. CDC PNS-E2, and Enterococcus sp. nov. CDC PNS-E3 are AY321375, AY321376, and AY321377, respectively.
RESULTS AND DISCUSSION
All three enterococcus-like isolates were nonmotile, catalase-negative, gram-positive cocci, occurring in short chains, in pairs, and singly. Colonies in blood agar were circular and grey, with alpha-hemolysis. They all were positive in the Enterococcus AccuProbe assay and leucine aminopeptidase activity test and grew in 6.5% NaCl and at 10°C. Strains SS-1728 and SS-1729 were also positive for hydrolysis of esculin in the presence of bile and for pyrrolidonyl arylamidase activity and also grew at 45°C. Strain SS-1730 did not grow at 45°C and was negative for hydrolysis of esculin in the presence of bile and for pyrrolidonyl-α-naphthylamide hydrolysis. The last result put that strain together with a group composed of four other enterococcal species, i.e., E. cecorum, E. columbae, E. pallens, and E. saccharolyticus, which are also negative for pyrrolidonyl-α-naphthylamide hydrolysis; these strains emphasize the need for caution when using such a test as rapid preliminary identification in clinical laboratories, especially considering the fact that most of these species have been reported to be isolated from human specimens (8, 16). Additional physiological characteristics of the three isolates are shown in Table 1. However, the results of physiological testing were not consistent enough to allow confident inclusion in any of the already-known enterococcal species.
TABLE 1.
Characteristics of the three new enterococcal speciesa
| Test or characteristics | Enterococcus sp. nov. CDC PNS-E1 (SS-1728T) | Enterococcus sp. nov. CDC PNS-E2 (SS-1729T) | Enterococcus sp. nov. CDC PNS-E3 (SS-1730T) |
|---|---|---|---|
| Hydrolysis | |||
| BE | + | + | − |
| LAP | + | + | + (w) |
| PYR | + | + | − |
| Arginine | − | + | − |
| Hippurate | − | − | − |
| Growth | |||
| At 10 °C | + | + | + |
| At 45 °C | + | + | − |
| In 6.5% NaCl | + | + | + |
| Gas production in Man- Rogosa-Sharpe broth | − | − | − |
| Susceptibility to vanco- mycin | S | R | S |
| Group D antigen | + | + | − |
| Motility | − | − | − |
| Pigment production | − | − | − |
| Tolerance to tellurite | − | − | − |
| Pyruvate utilization | + | − | + |
| Voges-Proskauer | + | + | − |
| AccuProbe | + | + | + |
| Acid production from: | |||
| Arabinose | − | − | + |
| Glycerol | − | + | + |
| Inulin | − | − | − |
| Lactose | + | + | − |
| Maltose | + | + | + |
| Mannitol | − | + | + |
| Melibiose | − | − | − |
| MGP | + | − | − |
| Raffinose | − | − | − |
| Ribose | − | + | + |
| Sorbitol | − | − | + |
| Sorbose | − | − | + |
| Sucrose | + | + | − |
| Trehalose | − | + | + |
BE, hydrolysis of esculin in presence of bile; LAP, leucine aminopeptidase activity; PYR, pyrrolidonyl arylamidase activity; +, positive; −, negative; (w), weak reaction; S, susceptible; R, resistant.
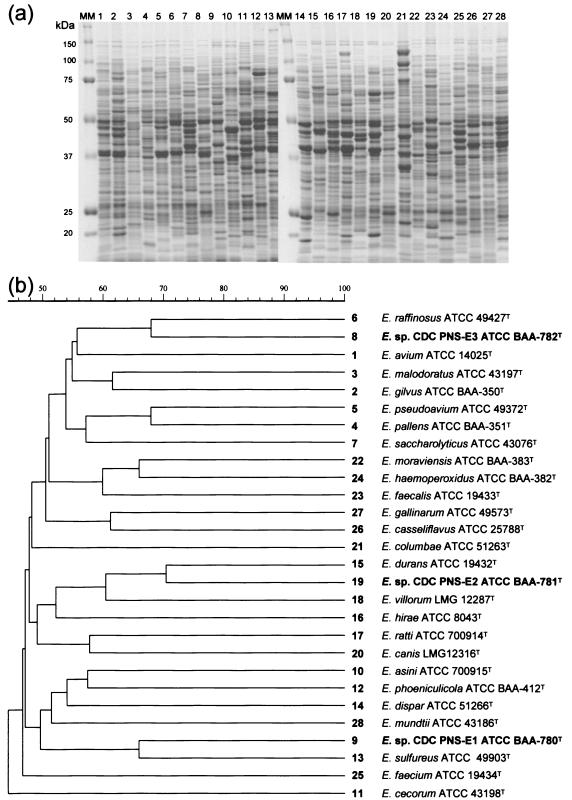
To clarify the identification, electrophoretic analysis of the whole-cell protein profiles was performed. The profiles generated are shown in Fig. 1. The profiles were unique for each strain and were not similar to the profiles of any previously described species of Enterococcus.
FIG. 1.
(a) SDS-PAGE profiles of whole-cell protein extracts of Enterococcus sp. nov. CDC PNS-E1, Enterococcus sp. nov. CDC PNS-E2, Enterococcus sp. nov. CDC PNS-E3, and other enterococcal species. MM, molecular mass marker. (b) Dendrogram resulting from a computer-assisted analysis of the protein profiles in panel a. Numbers on the right correspond to the lanes in panel a. The scale represents average percentages of similarity.
The results of DNA-DNA relatedness experiments were consistent with the results of the whole-cell protein profile analysis (Table 2), since percentages of homology with all 25 known species of Enterococcus were lower than 70% and met the criteria for species definition (15, 19).
TABLE 2.
Levels of DNA relatedness of the three new species of Enterococcus and other enterococcal species
| Source of unlabeled DNA | RBRa of labeled DNA from:
|
||
|---|---|---|---|
| SS-1728T | SS-1729T | SS-1730T | |
| Enterococcus sp. nov. CDC PNS-E1 SS-1728T | 100 | 12 | 11 |
| Enterococcus sp. nov. CDC PNS-E2 SS-1729T | 18 | 100 | 12 |
| Enterococcus sp. nov. CDC PNS-E3 SS-1730T | 9 | 12 | 100 |
| E. asini ATCC 700915T | 15 | 11 | 13 |
| E. avium ATCC 14025T | 13 | 10 | 41 |
| E. canis LMG 12316T | 8 | 11 | 9 |
| E. casseliflavus ATCC 25788T | 11 | 9 | 6 |
| E. cecorum ATCC 43198T | 13 | 6 | 9 |
| E. columbae ATCC 51263T | 9 | 8 | 10 |
| E. dispar ATCC 51266T | 9 | 15 | 10 |
| E. durans ATCC 19432T | 14 | 21 | 11 |
| E. faecalis ATCC 19433T | 11 | 22 | 11 |
| E. faecium ATCC 19434T | 15 | 28 | 15 |
| E. gallinarum ATCC 49573T | 10 | 15 | 11 |
| E. gilvus ATCC BAA-350T | 11 | 10 | 32 |
| E. haemoperoxidus ATCC BAA-382T | 6 | 8 | 6 |
| E. hirae ATCC 8043T | 11 | 22 | 11 |
| E. malodoratus ATCC 43197T | 11 | 14 | 32 |
| E. moraviensis ATCC BAA-383T | 6 | 8 | 8 |
| E. mundtii ATCC 43186T | 14 | 17 | 13 |
| E. pallens ATCC BAA-351T | 11 | 14 | 21 |
| E. phoeniculicola ATCC BAA-412T | 9 | 15 | 10 |
| E. pseudoavium ATCC 49372T | 11 | 14 | 35 |
| E. raffinosus ATCC 49427T | 9 | 11 | 32 |
| E. ratti ATCC 700914T | 8 | 16 | 8 |
| E. saccharolyticus ATCC 43076T | 16 | 14 | 12 |
| E. sulfureus ATCC 49903T | 17 | 13 | 14 |
| E. villorum LMG 12287T | 12 | 17 | 9 |
RBR, relative binding ratio at 55°C.
The G+C content of strain SS-1728T was 40.3 mol%, that of strain SS-1729T was 38.1 mol%, and that of strain SS-1730T was 39.0 mol%. These values are similar to published values for both streptococci and enterococci (7, 14).
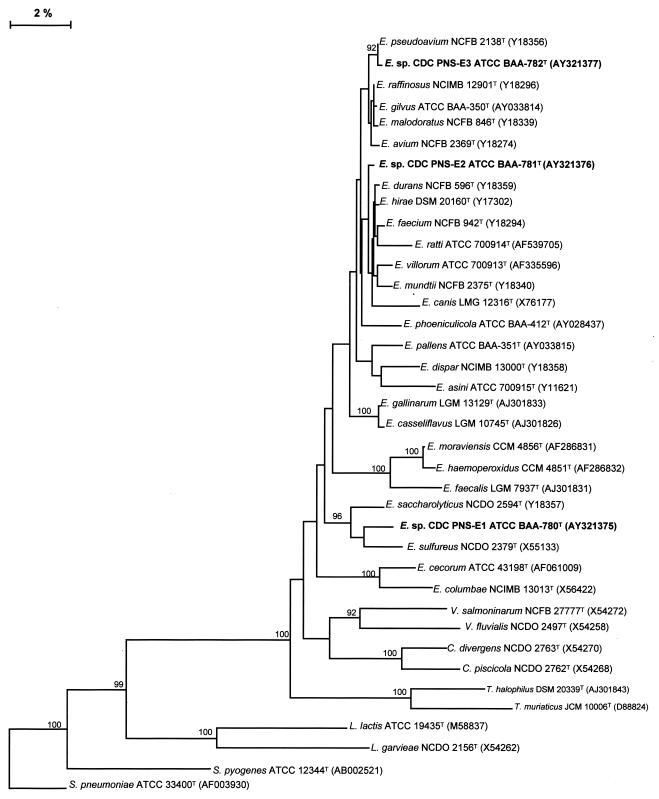
The relationship among the almost-complete 16S rDNA sequences of the strains was confirmed by a pairwise comparison: isolate SS-1728T (Enterococcus sp. nov. CDC PNS-E1) (1,479 bp) was most closely related to E. sulfureus, isolate SS-1729T (Enterococcus sp. nov. CDC PNS-E2) (1,483 bp) was most closely related to E. durans and isolate SS-1730T, and Enterococcus sp. nov. CDC PNS-E3 (1,482 bp) was most closely related to E. pseudoavium. A multiple-sequence alignment analysis, trimmed to consensus, comparing those three 16S rRNA gene sequences with sequences of other species of Enterococcus and related genera in GenBank is shown in Fig. 2.
FIG. 2.
Phylogenetic tree of enterococci based on comparative analysis of the 16S rDNA sequences, showing the relationship among Enterococcus sp. nov. CDC PNS-E1, Enterococcus sp. nov. CDC PNS-E2, Enterococcus sp. nov. CDC PNS-E3, other enterococcal species, and species of related genera. The limit criterion to show the bootstrap numbers was above 85%.
On the basis of our findings, we suggest that the three new species be included in the conventional test scheme, proposed by Teixeira and Facklam (16), used to identify the species of Enterococcus and some related genera, as shown in Table 3. These biochemical tests are used to identify enterococcal cultures to the species level. According to the results of such testing, isolate SS-1728T can be easily differentiated from the other members in group IV by the combination of results for arabinose, sorbitol, raffinose, pigment, pyruvate, and methyl-α-d-glucopyranoside (MGP). Isolate SS-1729T can be differentiated in group II by the results for sorbitol, raffinose, tellurite, motility, pigment, pyruvate, and MGP, and isolate SS-1730T can be differentiated from members of group I by the results for arabinose, raffinose, pigment, sucrose pyruvate, and MGP.
TABLE 3.
Phenotypic characteristics used for identification of Enterococcus species, including Enterococcus sp. nov. CDC PNS-E1, Enterococcus sp. nov. CDC PNS-E2, and Enterococcus sp. nov. CDC PNS-E3, and some physiologically related species of other gram-positive coccia
| Group and species | Phenotypic characteristicsb
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAN | SOR | ARG | ARA | SBL | RAF | TEL | MOT | PIG | SUC | PYU | MGP | |
| I | ||||||||||||
| E. avium | + | + | − | + | + | − | − | − | − | + | + | V |
| E. gilvus | + | + | − | − | + | + | − | − | + | + | + | − |
| E. malodoratus | + | + | − | − | + | + | − | − | − | + | + | V |
| E. pallens | + | + | − | − | + | + | − | − | + | + | − | + |
| E. pseudoavium | + | + | − | − | + | − | − | − | − | + | + | + |
| E. raffinosus | + | + | − | + | + | + | − | − | − | + | + | V |
| E. saccharolyticusc | + | + | − | − | + | + | − | − | − | + | − | + |
| Enterococcus sp. nov. CDC PNS-E3 | + | + | − | − | + | − | − | − | − | − | + | − |
| II | ||||||||||||
| E. faecalis | +* | − | +* | − | + | − | + | − | − | +* | + | − |
| E. haemoperoxidusc,d | + | − | + | − | − | − | − | − | − | + | − | + |
| Lactococcus sp. | + | − | + | − | − | − | − | − | − | + | − | − |
| E. faecium | +* | − | + | + | V | V | − | − | − | +* | − | − |
| E. casseliflavus | + | − | +* | + | V | + | −* | +* | +* | + | V | + |
| E. gallinarum | + | − | +* | + | − | + | − | +* | − | + | − | + |
| E. mundtii | + | − | + | + | V | + | − | − | + | + | − | − |
| Enterococcus sp. nov. CDC PNS-E2 | + | − | + | − | − | − | − | − | − | + | − | − |
| III | ||||||||||||
| E. dispar | − | − | + | − | − | + | − | − | − | + | + | + |
| E. durans | − | − | + | − | − | − | − | − | − | − | − | − |
| E. hirae | − | − | + | − | − | + | − | − | − | + | − | − |
| E. ratti | − | − | + | − | − | − | − | − | − | − | − | − |
| E. villorum | − | − | + | − | − | − | − | − | − | − | − | − |
| IV | ||||||||||||
| E. asinic | − | − | − | − | − | − | − | − | − | + | − | − |
| E. cecorumc | − | − | − | − | + | + | − | − | − | + | + | − |
| E. phoeniculicolac | − | − | − | + | − | + | − | − | − | + | − | + |
| E. sulfureus | − | − | − | − | − | + | − | − | + | + | − | + |
| Enterococcus sp. nov. CDC PNS-E1 | − | − | − | − | − | − | − | − | − | + | + | + |
| V | ||||||||||||
| E. canisc | + | − | − | + | − | − | − | − | − | + | + | + |
| E. columbaec | + | − | − | + | + | + | − | − | − | + | + | − |
| E. moraviensisc | + | − | − | + | − | − | − | − | − | + | + | + |
| Vagococcus fluvialis | + | − | − | − | + | − | − | + | − | + | − | + |
Adapted from reference 16 with permission.
Abbreviations and symbols: MAN, mannitol; SOR, sorbose; ARG, arginine; ARA, arabinose; SBL, sorbitol; RAF, raffinose; TEL, 0.04% tellurite; MOT, motility; PIG, pigment; SUC, sucrose; PYU, pyruvate; MGP, methyl-α-d-glucopyranoside; +, 90% or more of the strains are positive; −, 10% or fewer of the strains are positive; V, variable (11 to 89% of the strains are positive); *, occasional exceptions occur (<3% of strains show aberrant reactions).
Phenotypic characteristics based on data from type strains.
Late mannitol positive (5 days of incubation).
A particular finding has drawn our attention to strain SS-1729T: this isolate formed no zone of growth inhibition around the vancomycin disk used in the testing scheme for presumptive identification at the genus level. The MICs of vancomycin and teicoplanin for that strain were >256 and ≥16 μg/ml, respectively, and the presence of the vanA gene was detected. Those are important resistance characteristics that are becoming more common among the two enterococcal species, E. faecalis and E. faecium, that are frequently found as agents of nosocomial outbreaks (2), but they have only sporadically been found in other species of enterococci. Additionally, that strain also showed resistance to high levels of gentamicin (MIC, >500 μg/ml) and streptomycin (MIC, >1,000 μg/ml), resistance to rifampin (MIC, >4 μg/ml), and susceptibility to penicillin, ampicillin, chloramphenicol, doxycycline, and linezolid. The recovery from a clinically significant human source of a previously unknown bacterial species (strain SS-1729T) carrying such resistance markers raises special concern about its potential for playing a role as a new human pathogen and about the dissemination of antimicrobial resistance genes. Strain SS-1730T showed resistance to rifampin (MIC, >4 μg/ml), intermediate resistance to linezolid (MIC, 4 μg/ml), and susceptibility to vancomycin, gentamicin, streptomycin, penicillin, ampicillin, chloramphenicol, and doxycycline. Strain SS-1728T was susceptible to all antimicrobial agents mentioned above.
This report provides data on the phenotypic and genotypic characterization of three new enterococcal species that may represent new etiological agents of human infections and calls attention to the fact that one of them (Enterococcus sp. nov. CDC PNS-E2) harbors the vancomycin resistance gene vanA. These new species can easily be differentiated from other related enterococcal species by a few physiological tests or by electrophoretic whole-cell protein profile analysis, which can be recommended as a reliable and relatively simple method, since protein profiles correlate with the results of DNA relatedness experiments and 16S rDNA sequencing.
Description of Enterococcus sp. nov. CDC PNS-E1 (Centers for Disease Control and Prevention Proposed New Species of Enterococcus 1).
Cells are gram-positive cocci occurring as short chains, in pairs, and singly. Nonpigmented, alpha-hemolytic, small colonies of up to 0.5 mm in diameter are formed on sheep blood agar at 37°C and are unaffected by the absence or presence of 5% CO2. It is catalase negative, nonmotile, and susceptible to vancomycin. Growth occurs at 10°C, at 45°C, and in broth containing 6.5% NaCl. It is positive for pyrrolidonyl arylamidase activity, leucine aminopeptidase activity, hydrolysis of esculin in the presence of bile, pyruvate utilization, and Voges-Proskauer test. Acid is produced from lactose, maltose, MGP, and sucrose. Acid is not produced from arabinose, glycerol, inulin, mannitol, melibiose, raffinose, ribose, sorbitol, sorbose, and trehalose. It is negative for production of gas in MRS broth, hydrolysis of arginine, hippurate, and tellurite. It reacts with the AccuProbe Enterococcus genetic probe, and Lancefield extracts react with CDC group D antiserum. The type strain is SS-1728T (= ATCC BAA-780T = CCUG 47860T), isolated from blood of a patient in Evanston, Ill.
Description of Enterococcus sp. nov. CDC PNS-E2 (Centers for Disease Control and Prevention Proposed New Species of Enterococcus 2).
Cells are gram-positive cocci occurring as short chains, in pairs, and singly. Nonpigmented, alpha-hemolytic, small colonies of up to 0.5 mm in diameter are formed in sheep blood agar at 37°C and are unaffected by the absence or presence of 5% CO2. It is catalase-negative, nonmotile, and resistant to vancomycin (MIC, >256 μg/ml; harbors the vanA gene). Growth occurs at 10°C, at 45°C, and in broth containing 6.5% NaCl. It is positive for pyrrolidonyl arylamidase activity, leucine aminopeptidase activity, hydrolysis of esculin in the presence of bile, hydrolysis of arginine, and Voges-Proskauer test. Acid is produced from glycerol, lactose, maltose, mannitol, ribose, sucrose, and trehalose. Acid is not produced from arabinose, inulin, melibiose, MGP, raffinose, sorbitol and sorbose. It is negative for production of gas in MRS broth, hydrolysis of hippurate, pyruvate utilization, and tellurite. It reacts with the AccuProbe Enterococcus genetic probe, and Lancefield extracts react with CDC group D antiserum. The type strain is SS-1729T (= ATCC BAA-781T = CCUG 47861T), isolated from blood of a patient in Los Angeles, Calif.
Description of Enterococcus sp. nov. CDC PNS-E3 (Centers for Disease Control and Prevention Proposed New Species of Enterococcus 3).
Cells are gram-positive cocci occurring as short chains, in pairs, and singly. Nonpigmented, alpha-hemolytic, small colonies of up to 0.5 mm in diameter are formed in sheep blood agar at 37°C and are unaffected by the absence or presence of 5% CO2. It is catalase negative, nonmotile, and susceptible to vancomycin. Growth occurs at 10°C and in broth containing 6.5% NaCl but not at 45°C. It is positive for pyruvate utilization and has a weakly positive leucine aminopeptidase activity. Acid is produced from glycerol, maltose, mannitol, ribose, sorbitol, sorbose, and trehalose. Acid is not produced from arabinose, inulin, lactose, melibiose, MPG, raffinose, and sucrose. It is negative for pyrrolidonyl arylamidase activity, hydrolysis of esculin in the presence of bile, production of gas in MRS broth, Voges-Proskauer test, hydrolysis of arginine, and hippurate. It reacts with the AccuProbe Enterococcus genetic probe, and Lancefield extracts fail to react with CDC group D antiserum. The type strain is SS-1730T (= ATCC BAA-782T = CCUG 47862T), isolated from brain tissue of an 11-month-old patient from Honolulu, Hawaii.
Acknowledgments
We are grateful to Roberta B. Carey, Janet Hindler, and the Hawaii State Department of Health for providing strains to the Streptococcus Laboratory at the Centers for Disease Control and Prevention.
This study was supported in part by Conselho Nacional de Desenvolvimento Científico e Técnologico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Financiadora de Estudos e Projetos (FINEP), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and Ministério Da Ciência e Tecnologia (MCT/PRONEX), Brazil. M. G. S. Carvalho was supported by a CNPq postdoctoral fellowship.
REFERENCES
- 1.Brenner, D. J., A. C. McWhorter, J. K. Leete Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbial. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, N. C., R. C. Cooksey, B. C. Hill, J. M. Swenson, and F. C. Tenover. 1993. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob. Agents Chemother. 37:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Graef, E. M., L. A. Devriese, M. Vancanneyt, M. Baele, M. D. Collins, K. Lefebvre, J. Swings, and F. Haesebrouck. 2003. Description of Enterococcus canis sp. nov. from dogs and reclassification of Enterococcus porcinus Teixeira et al. 2001 as a junior synonym of Enterococcus villorum Vancanneyt et al. 2001. Int. J. Syst. Evol. Microbiol. 53:1069-1074. [DOI] [PubMed] [Google Scholar]
- 5.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devriese, L. A., M. Vancanneyt, P. Descheemaeker, M. Baele, H. W. Van Landuyt, B. Gordts, P. Butaye, J. Swings, and F. Haesebrouck. 2002. Differentiation and identification of Enterococcus durans. E. hirae and E. villorum. J. Appl. Microbiol. 92:821-827. [DOI] [PubMed] [Google Scholar]
- 7.Devriese, L. A., G. N. Dutta, J. A. E. Farrow, A. Van De Kerchkhove, and B. A. Philips. 1983. Streptococcus cecorum, a new species isolated from chickens. Int. J. Syst. Bacteriol. 33:772-776. [Google Scholar]
- 8.Facklam, R. R., M. D. G. S. Carvalho, and L. M. Teixeira. 2002. History, taxonomy, biochemical characteristics, and antibiotic susceptibility testing of enterococci, p. 1-54. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. American Society for Microbiology, Washington, D.C.
- 9.Law-Brown, J., and P. R. Meyers. 2003. Enterococcus phoeniculicola sp. nov., a novel member of the enterococci isolated from the uropygial gland of the red-billed woodhoopoe, Phoeniculus purpureus. Int. J. Syst. Evol. Microbiol. 53:683-685. [DOI] [PubMed] [Google Scholar]
- 10.Mandel, M., L. Igambi, J. Bergendahl, M. L. Dodson, Jr., and E. Scheltgen. 1970. Correlation of melting temperature and cesium chloride buoyant density of bacterial deoxyribonucleic acid. J. Bacteriol. 101:333-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merquior, V. L. C., J. M. Peralta, R. R. Facklam, and L. M. Teixeira. 1994. Analysis of electrophoretic whole-cell protein profiles as a tool for characterization of Enterococcus species. Curr. Microbiol. 28:149-153. [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing (M100-S11). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Segonds, C., T. Heulin, N. Marty, and G. Chabanon. 1999. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 37:2201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schleifer, K. H., and R. Kilpper-Balz. 1984. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int. J. Syst. Bacteriol. 34:31-34. [Google Scholar]
- 15.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossella-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 16.Teixeira, L. M., and R. R. Facklam. 2003. Enterococcus, p. 422-433. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 17.Teixeira, L. M., R. R. Facklam, A. G. Steigerwalt, N. E. Pigott, V. L. C. Merquior, and D. J. Brenner. 1995. Correlation between phenotypic characteristics and DNA relatedness within Enterococcus faecium strains. J. Clin. Microbiol. 33:1520-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 20.Whiley, R. A., and M. Kilian. 2003. International Committee on Systematics of Prokaryotes subcommittee on the taxonomy of staphylococci and streptococci: minutes of the closed meeting, 31 July 2002, Paris, France. Int. J. Syst. Evol. Microbiol. 53:915-917. [Google Scholar]