Abstract
The bacterial second messenger signaling molecule bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) controls important biological processes such as biofilm formation, virulence response, and motility. This second messenger is sensed by macromolecular targets inside the cell, both protein and RNA, which induce specific phenotypic responses critical for bacterial survival. One class of enzymes responsible for regulating the intracellular concentration of c-di-GMP, and therefore the physiological behavior of the cell, is the EAL domain phosphodiesterases, which degrade the second messenger to its linear form, pGpG. Here, we investigate how base and backbone modifications to c-di-GMP affect the rate of cyclic dinucleotide degradation by an EAL domain protein (CC3396 from Caulobacter crescentus). The doubly substituted thiophosphate analog is highly resistant to hydrolysis by this metabolizing enzyme but can still bind c-di-GMP riboswitch targets. We used these findings to develop a novel ribosyl-phosphate modified derivative of c-di-GMP containing 2′-deoxy and methylphosphonate substitutions that is charge neutral and demonstrate that this analog is also resistant to EAL domain catalyzed degradation. This suggests a general strategy for designing c-di-GMP derivatives with increased enzymatic stability that also possess desirable properties for development as chemical probes of c-di-GMP signaling.
Bacteria have the ability to sense different external signals present in diverse environments and translate these cues into physiological responses that can be critical for their survival1. One mechanism bacteria have evolved to facilitate such behavioral adaptation is through the use of the second messenger signaling molecule bis-(3′-5′)- cyclic dimeric guanosine monophosphate (c-di-GMP) (for recent reviews see2–5). c-di-GMP is ubiquitous within the bacterial domain and is vital for regulating the transition between a sessile, biofilm-forming state and a motile, planktonic existence6. This second messenger also controls the virulence response of pathogenic organisms7 and has been linked to quorum sensing, the process by which bacteria detect and communicate with one another8.
c-di-GMP is synthesized from two molecules of GTP by diguanylate cyclases (DGCs), which contain a GGDEF domain9, and is degraded to the linear 5′-phosphate dinucleotide pGpG by EAL domain phosphodiesterases (PDEs) (Figure 1a)10. A second class of c-di-GMP phosphodiesterases known as the HD-GYP domain proteins have also been identified in some bacterial species, though these enzymes are much less common than the EAL domain proteins11,12. GGDEF and EAL domain proteins are widely distributed throughout the bacterial domain13, suggesting that this second messenger plays an essential biological role in many different species. The opposing activities of these enzymes tightly regulate the concentration of c-di-GMP in the cell, which is directly sensed by downstream targets of the second messenger that act to induce the appropriate phenotypic response2–5.
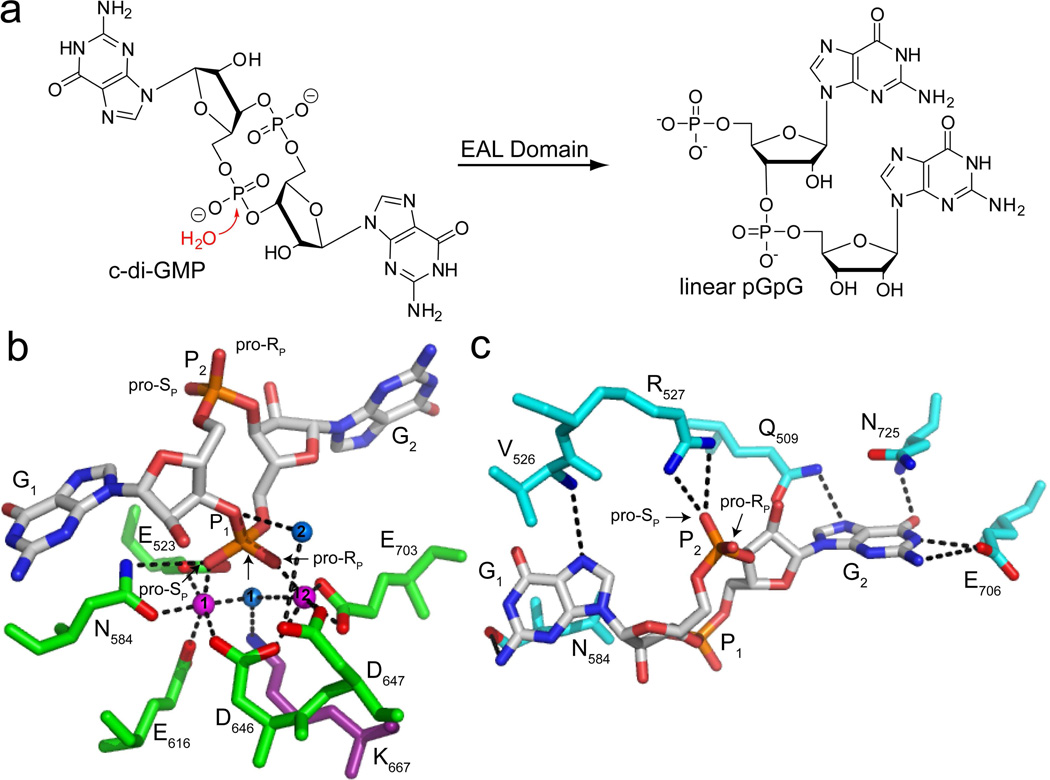
Figure 1.
Recognition and enzymatic hydrolysis of c-di-GMP by EAL domain proteins. (a) Schematic reaction. In the presence of an EAL domain protein, a nucleophilic water molecule attacks one of the c-di-GMP phosphodiester bonds to yield the linear 5′- phosphate product, pGpG. (b) c-di-GMP bound to the active site of the EAL domain protein TBD1265 from Thiobacillus denitrificans (PDB ID 3N3T). c-di-GMP is colored by atom with carbon shown in white, oxygen in red, nitrogen in blue and phosphorous in orange. Water molecules are shown as blue spheres and magnesium ions are shown as magenta spheres. The protein residues in the active site involved in metal coordination are colored in green and oxygens and nitrogens making contacts to c-di-GMP are colored in red and blue, respectively. The highly conserved lysine that stabilizes the negative charge of water 1 and positions it for nucleophilic attack on the scissile phosphate, P1, is shown in purple. Water 2 is predicted to donate a proton to the leaving group. The pro-RP and pro-SP oxygens of P1 and P2 are labeled for sulfur substitution. (c) Recognition of the c-di-GMP bases and the phosphate distal to the site of catalysis, P2 (PDB ID 3N3T). Coloring of c-di-GMP is the same as in (b). Residues in contact with G1 and G2 are colored in cyan and the highly conserved arginine that makes electrostatic contacts to P2 is shown in white. Oxygens and nitrogens of protein side chains are colored as in part (b). Residues involved in G2 recognition are conserved while those involved in G1 recognition are highly variable. The residue corresponding to N725 is most often aromatic (F, W, or Y).
Although c-di-GMP is prevalent in bacteria, both the macromolecular targets that sense this second messenger and the specific mechanisms responsible for the observed behavioral transitions are only beginning to emerge and are highly diverse. Several classes of proteins including PilZ domain proteins14, transcription factors15–17, and degenerate DGCs and PDEs18–20, as well as two structurally distinct classes of riboswitch RNAs21,22, have been identified as c-di-GMP effectors, underscoring the complexity of this key bacterial signaling pathway.
Understanding the metabolism of c-di-GMP is essential to elucidate how different bacterial phenotypes are triggered in response to varying intracellular concentrations of the second messenger. Structural characterization of this class of metabolizing enzymes indicates that c-di-GMP is bound in the extended conformation wherein the guanine bases (termed G1 and G2) are splayed apart from one another (Figure 1b,c)23–25, rather than the eclipsed conformation that is observed for the c-di-GMP binding riboswitches26,27 and many effector proteins15,28,29. Few specific interactions are made to the guanine bases of the second messenger. The most highly conserved residues are involved in coordination of active site metals, the proposed nucleophilic water molecule and the c-di-GMP phosphates (Figures 1b,c). Crystal structures of catalytically active EAL domain proteins reveal two metals in the active site that each contact a different oxygen atom of the scissile phosphate (P1, Figure 1b), suggesting that c-di-GMP hydrolysis is achieved via a two-metal ion mechanism and that both oxygens are critical to catalysis23,25. Several inactive EAL domain proteins that can bind c-di-GMP, but cannot catalyze degradation, have been identified that function as downstream signaling molecules19,30. These proteins contain mutations to the amino acids coordinating the metals, phosphate P1, or the nucleophilic water molecule, highlighting the essential role these residues play in catalysis. The inactivity of such mutants suggests that changes to the c-di-GMP phosphate linkages may also impact reactivity.
Before the EAL domain was identified as the specific enzyme responsible for c-di-GMP degradation, Benziman and coworkers demonstrated that membrane extracts possessing c-di-GMP specific PDE-activity were able to degrade doubly substituted 2′- deoxy and inosine analogs and a singly substituted thiophosphate modified analog31. This suggests that these particular modifications to c-di-GMP are unlikely to yield EAL domain resistant analogs, but these single time point studies did not provide information on how these modifications affect the rate of second messenger degradation or, in some cases, what effect double substitution has on activity. Therefore, we sought to determine which c-di-GMP modifications have the largest effect on catalysis by an EAL domain phosphodiesterase and identify those that render the second messenger resistant to degradation.
Here, we test a series of base, ribose and phosphate modified c-di-GMP derivatives to investigate the specificity of the EAL domain proteins for its cognate ligand. We identified modifications that render the second messenger highly resistant to EAL domain catalyzed degradation and used these findings to develop a charge neutral, EAL domain resistant methylphosphonate c-di-GMP derivative.
MATERIALS AND METHODS
Materials
Radiolabeled c-di-GMP (*c-di-GMP) and unlabeled c-di-GMP were synthesized enzymatically using purified PleD* and tDGC diguanylate cyclase proteins respectively, as previously described32,33. Base and ribose modified analogs were synthesized on solid phase as previously described34. Mono and dithiophosphate analogs were synthesized in solution as previously described35. The absolute stereochemistry of the thiophosphate c-di-GMP analogs was assigned as previously described based on the specificity of P1 and venom phosphodiesterases36. 3′- phosphate CPG beads, guanosine methylphosphonamidite precursors, and Poly-Pak II desalting columns were purchased from Glen Research. 4-(dimethylamino)pyridine (DMAP), tetrahydrofuran (THF), acetic anhydride, anhydrous pyridine, anhydrous acetonitrile (ACN), triethylamine (TEA) and 1-(2-mesitylenesulfonyl)-3-nitro-1H-1,2,4- triazole (MSNT) were purchased from Sigma. The 100 nucleotide class I Vc2 riboswitch RNA from Vibrio cholerae containing 2-aminopurine at position 94 (G94(2AP) RNA) was prepared as previously described34. The wild-type class II aptamer from Clostridium acetobutylicum was cloned and transcribed in vitro using T7 RNA polymerase as previously described27,34.
Expression and purification of the phosphodiesterase protein CC3396 from Caulobacter crescentus
The 6×-histidine tagged EAL domain phosphodiesterase protein CC3396 was purified by affinity chromatography using Ni-NTA agarose (Qiagen) as previously described37. Overnight cultures of E. coli BL21 cells harboring the expression plasmid (pET21) encoding CC3396 were grown at 37°C. The overnight culture was diluted into fresh LB medium and cells were grown to an OD600 of 0.6. Protein expression was then induced for 45 min. at 37°C by adding 0.5 mM isopropyl 1- thio-β-D-galactopyranoside (IPTG). Cells were pelleted and resuspended in lysis buffer (50 mM sodium phosphate pH 7.0, 300 mM NaCl, 5 mM BME) and lysed by passing through a microfluidizer at 15,000 psi. The lysate was cleared by centrifugation (20,000 g) and the cleared lysate was incubated with Ni-NTA agarose for 1 hour at 4°C to allow the protein to bind the column. The column was washed with lysis buffer containing 20 mM imidazole followed by washing with lysis buffer containing 50 mM imidazole. The protein was eluted from the column in 250 mM imidazole and the pure protein fractions were pooled and dialyzed into 25 mM Tris-HCl pH 8.0, 250 mM NaCl, 5 mM BME. Dialyzed protein was concentrated and stored in 10% glycerol.
Phosphodiesterase assays to evaluate degradation rates of c-di-GMP and analogs
Phosphodiesterase activity was evaluated under multiple turnover conditions with saturating concentrations of substrate. Pure CC3396 protein (1 µM) was incubated with 100 µM GTP and 100 µM c-di-GMP (or analog) in 1×PDE buffer (10 mM MgCl2, 25 mM Tris-HCl pH 8.0, 250mM NaCl) at room temperature (22°C). Aliquots were removed at each time point and the reaction was quenched by heating to 95 °C for 5 minutes. Degradation was monitored in the range where the reaction was still linear to measure the initial velocity. Samples were diluted in 50 mM TEAAc buffer (pH 6.0) and compound degradation was analyzed by HPLC on a reverse-phase C8 column using a gradient of 0 to 4% ACN in 50 mM TEAAc pH 6.0 over 15 minutes for most compounds. A steeper gradient was used for the following compounds as indicated: c-(RPRP)-di-Gps 0 to 15% ACN; c-(RPSP)-di-Gps 0 to 10% ACN; c-(RP)-Gps-GMP 0 to 10% ACN; c-didGp(Me)-I and c-di-dGp(Me)-II 0 to 20% ACN. The area under peaks corresponding to linear and cyclic compounds was determined by integration (HPChem Software) and the fraction of linear (FL) product formed was calculated using the following equation:
where AreaL= area of the linear product and AreaC= area of the cyclic compound. FL was multiplied by the substrate concentration to obtain the amount of product formed, which was plotted against time. The data was fit to a line and the slope of the line was used to determine the initial rate (kcat).
Competition phosphodiesterase assays
The degradation of radiolabeled c-di-GMP (*c-di-GMP) to pGpG was monitored by determining the fraction of pGpG formed by separating cyclic and linear products by polyethyleneimine-cellulose thin layer chromatography (PEI-cellulose TLC). For the competition assays, CC3396 (10 nM) was incubated at room temperature for 5 minutes with GTP (100 µM) in 1× PDE buffer before the simultaneous addition of trace amounts of radiolabeled c-di-GMP and an excess of unlabeled competitor analog (100 µM). Aliquots were removed at each timepoint and quenched by the addition of an equal volume of 0.5 M EDTA. Reactions were extracted with a mixture of phenol:chloroform:isoamyl alcohol (25:24:1) to remove the protein before analysis by PEI-cellulose TLC. PEI-cellulose TLC plates were run in a 1:1.5 (v/v) mixture of saturated NH4SO4: 1.5 M KH2PO4, pH 3.6 and the amount of c-di-GMP and pGpG quantified as previously described37.
Chemical synthesis of the methylphosphonate analog
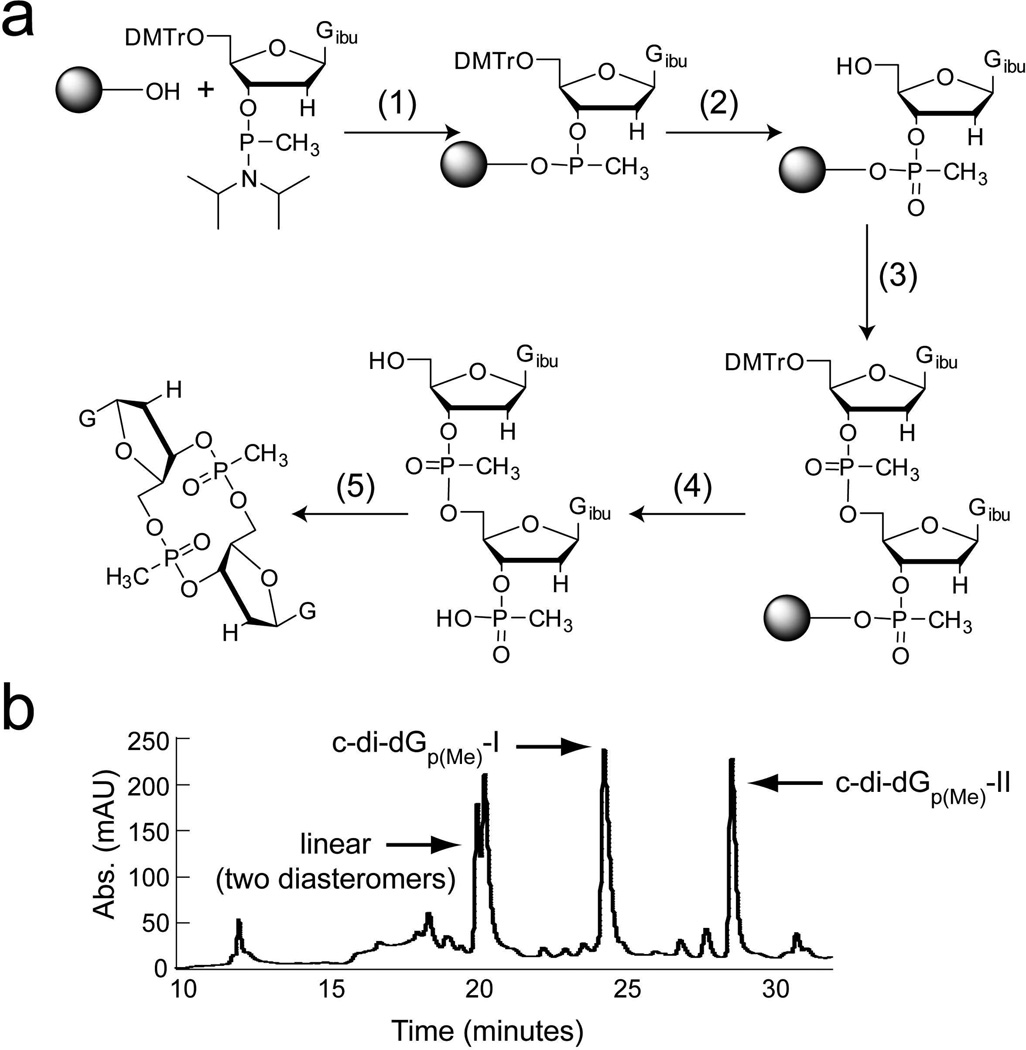
The methylphosphonate c-di-GMP analog was synthesized on solid support (3′-phosphate CPG beads) on a 1 µmole scale as previously described38 with the following adaptations (Figure 2a). The 5′-dimethoxytrityl (DMTr) protected guanosine methylphosphonamidite (50 mM in THF) was coupled to the solid support using 5-benzyl mercaptotetrazole (125 mM in ACN) as the activator. Oxidation was performed using a 0.02 M solution of iodine in THF/pyridine/water (88:10:2) and unreacted sites were capped using a 1:1 mixture of 6.5% DMAP/THF and 10% acetic anhydride/THF. The 5′-DMTr group was removed using 3% dicholoracetic acid in dichloromethane followed by coupling, oxidation, capping and detritylation of the second nucleotide as described above. The linear dinucleotide was cleaved from the solid support by incubating with 10% TEA/ACN and co-evaporated with anhydrous pyridine. The dinucleotide was cyclized in solution using 0.1 M MSNT in pyridine under argon for approximately 96 hours. Global deprotection was afforded by evaporating the cyclization solution and resuspending the dry product in a 45:45:10 mixture of ACN:EtOH:NH4OH. After a 30 min room temperature incubation in the deprotection mixture, an equal volume of ethylenediamine was added and the room temperature incubation was allowed to proceed for an additional 6 hours. The mixture was diluted with water and the pH was adjusted to 7.0 using 6 M HCl. The product was desalted on a Poly-Pak II column (Glen Research) according to the manufacturer’s instructions. The desalted product was lyophilized to dryness and then purified by high performance liquid chromatography (HPLC) on a C18 reverse phase column using a gradient of 0% to 30% acetonitrile in 50 mM TEAAc, pH 6.0 over 30 minutes (Figure 2b). Two of the three possible diastereomers were synthesized and product identity was confirmed by ESI-MS in negative ion and positive ion mode (exact mass= 654.15; observed mass c-di-dGp(Me)-I [M-H]/1= 653.144, [M+H]/1= 655.1473, [M+Na]/1= 677.1346; observed mass c-di-dGp(Me)-II [M-H]/1= 653.1306, [M+H]/1= 655.1335, [M+Na]/1= 677.1207). The major impurity present is the non-cyclized linear intermediate (Figure 2b). Based on the integration of peaks corresponding to the linear and cyclic compounds, the cyclization reaction proceeded with approximately 50% efficiency. The overall yield of cyclic compound was 10% (c-di-dGp(Me)-I = 7% and c-didGp(Me)-II= 3%).
Figure 2.
Synthesis of the double methylphosphonate c-di-GMP analog. (a) Synthetic scheme. The linear dinucleotide was synthesized on solid support and then cyclized in solution. (1) (i) tetrazole/ACN; (2) (i) 3% DCA/DCM (ii) I2/THF/pyridine, (iii) acetic anhydride/DMAP, (iv) 3% DCA/DCM; (3) tetrazole/ACN + guanosine methylphosphonamidite/THF; (4) (i) 3% DCA/DCM, (ii) 10% TEA/ACN; (5) (i) 0.1 M MSNT/pyridine, (ii) ACN/EtOH/NH4OH, (iii) ethylenediamine.
(b) HPLC trace monitored at 254 nm showing purification of the crude reaction synthesis for the methylphosphonate c-di-GMP analog c-di-dGp(Me). Peaks corresponding to the two cyclized diastereomers and the linear, uncyclized molecule are labeled.
Affinity measurements of the methylphosphonate analog for c-di-GMP binding riboswitches
Dissociation constants (Kd’s) of c-di-GMP analogs for the class I and class II riboswitches were determined as previously described by competition gel-shift assay with radiolabeled c-di-GMP34. Radiolabeled c-di-GMP was incubated with riboswitch aptamer RNA (25 nM class I (G94(2AP) variant) or 50 nM class II) and increasing concentrations of analog in folding buffer containing 10 mM MgCl2, 10 mM NaCl, and 10 mM sodium cacodylate pH 6.8 until equilibrium was achieved. Free c-di- GMP and RNA-bound c-di-GMP were separated by native PAGE (100 mM Tris/HEPES pH 7.5, 0.1 mM, 10 mM MgCl2) at 4°C. The amounts of free and bound c-di-GMP were quantified and fit to an equation for competitive binding as previously described to determine the Kd of the competitor analog34.
RESULTS
Monitoring the enzymatic hydrolysis of c-di-GMP and analogs by HPLC
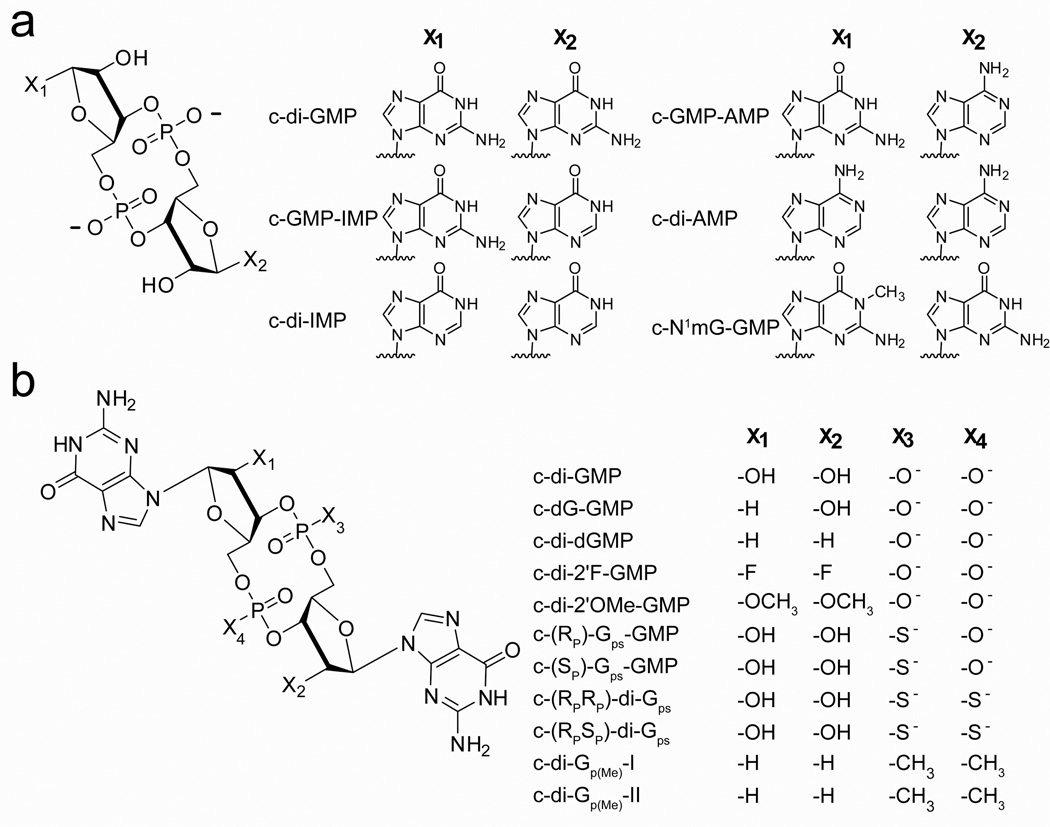
We employed an HPLC-based assay to measure the rate of hydrolysis for c-di-GMP and its analogs by the EAL domain PDE CC3396 from C. crescentus. CC3396 is a prototypical PDE and the activity of this protein has been well characterized in vitro37. It is a GGDEF-EAL protein containing a degenerate GGDEF domain that possesses no diguanylate cyclase activity, but instead acts to allosterically activate the phosphodiesterase by binding to GTP37. Sequence alignment with other structurally and biochemically characterized PDEs indicates that the most highly conserved residues necessary for catalysis are conserved in this protein (supplemental info., Table S1)23,25,39. Using this assay we measured the rate of cyclic dinucleotide degradation for the singly substituted (c-GMP-IMP, c-(RP)-Gps-GMP, c-(SP)-Gps-GMP, c-dG-GMP) and doubly substituted analogs (c-di-IMP, c-di-AMP, c-di-dGMP) previously tested in single time point studies 31. We also tested analogs whose susceptibility to EAL domain catalyzed degradation had not been previously investigated (c-GMP-AMP, c-N1mGGMP, c-di-2′F-GMP, c-di-2′OMe-MP, c-(RPRP)-di-Gps, c-(RPSP)-di-Gps, c-di-dGp(Me)) (Figure 3). With the exception of c-di-IMP, c-di-AMP, and c-di-dGMP, the previous biochemical work only looked at the degradation of singly substituted analogs. Because c-di-GMP is a symmetric molecule, single substitutions are likely to leave the dinucleotide susceptible to degradation by targeting the unsubstituted position.
Figure 3.
Structures of c-di-GMP analogs. (a) Base modified c-di-GMP analogs. Guanine was replaced with inosine, adenosine, or N1-methyl guanosine as indicated by X1 and X2. (b) Ribose and phosphate modified analogs. Modifications made to the ribose rings are indicated by X1 and X2 and those made to the phosphate linkages are indicated by X3 and X4.
The rate of c-di-GMP degradation was measured under saturating conditions of both the second messenger (100 µM, see below) and GTP (100 µM) and the conversion of c-di-GMP to pGpG was detected via HPLC by monitoring the appearance of a peak corresponding to pGpG (retention time = 7.5 minutes) and the disappearance of the peak corresponding to c-di-GMP (retention time= 10 minutes) (Figure 4a). Co-injection of the reaction with a chemically synthesized standard of pGpG confirmed that the peak appearing at 7.5 min. was the correct breakdown product. Under these conditions, the second messenger was completely degraded within 10 min., with the rate of c-di-GMP hydrolysis measured to be 15 mol/(min*mol enzyme) (Figure 4, Table 1). To confirm that we were using saturating concentrations of c-di-GMP, we measured the rate of degradation using 200 µM c-di-GMP and found it to be the same as that measured using 100 µM c-di-GMP (15.3 mol/(min*mol enzyme) at 200 µM c-di-GMP). This indicates that the enzyme is saturated with substrate and that the observed rate corresponds to the kcat. This was also confirmed for the ribosyl-phosphate modified analogs that were susceptible to degradation (Table 1), indicating that all rates measured for these analogs were done under saturating conditions. The rate of c-di-GMP hydrolysis by CC3396 that we measured is in reasonable agreement with that previously reported (115 mol/(min*mol enzyme) at 30°C)37 and the different temperature (37°C vs 30°C) and method used to carry out the two studies likely account for the 8-fold difference in measurements.
Figure 4.
Monitoring the enzymatic degradation of c-di-GMP by HPLC. (a) HPLC traces showing elution of GTP, pGpG, and c-di-GMP. Peaks corresponding to each of these compounds are labeled. After 10 minutes under the experimental conditions used, c-di-GMP is completely degraded. (b) The amount of pGpG formed was measured in the linear range of the reaction to determine the initial velocity of c-di-GMP degradation. Data was collected and analyzed in the same manner for those analogs that were susceptible to degradation.
Table 1.
Rates of enzymatic hydrolysis for c-di-GMP and its nucleotide analogs by the EAL domain phosphodiesterase protein CC3396. All data reported is the average of at least three independent trials.
| Modification | Analog | Rate (kcat) (mol*min−1*mol enzyme−1) |
Fold Change |
|---|---|---|---|
| c-di-GMPa | 15 ± 1.6 | − | |
| Baseb | c-GMP-IMP | 7.0 ± 0.4 | 2.1 |
| c-N1mG-GMP | 4.5 ± 0.7 | 3.3 | |
| c-di-IMP | 0.51 ± 0.06 | 29 | |
| c-GMP-AMP | 0.48 ± 0.04 | 31 | |
| c-di-AMP | n.d.c | − | |
| Ribosea | c-dG-GMP | 6.4 ± 0.2 | 2.3 |
| c-di-dGMP | 1.1 ± 0.6 | 14 | |
| c-di-2′F-GMP | 2.9 ± 0.4 | 5.2 | |
| c-di-2′OMe-GMP | 0.9 ± 0.2 | 17 | |
| Phosphatea | c-(SP)-Gps-GMP | 29 ± 1.8 | 0.5 |
| c-(RP)-Gps-GMP | 8.3 ± 0.6 | 1.8 | |
| c-(RPRP)-di-Gps | n.d. | − | |
| c-(RPSP)-di-Gps | n.d. | − | |
| c-di-dGp(Me)-I | n.d. | − | |
| c-di-dGp(Me)-II | n.d. | − | |
Rates were measured under saturating conditions of substrate and therefore correspond to kcat
Rates measured under multiple turnover conditions with 100 µM substrate and 1µM enzyme.
n.d. indicates no degradation under the conditions tested after 24 hour incubation with CC3396.
Enzymatic hydrolysis of base modified analogs
Previous biochemical studies have demonstrated that mutations to EAL domain residues contacting the c-di-GMP bases do not significantly affect enzymatic activity25,39. This suggests that modifications to the c-di-GMP bases would not be predicted to render the second messenger resistant to enzymatic degradation. We first tested the effects of modifying the c-di-GMP guanine bases with inosine, N1methyl guanine, and adenine substitutions on hydrolysis by the EAL domain protein CC3396 to determine which guanine analogs are most tolerated by the EAL domain (Figure 3a).
With the exception of c-di-AMP, all base modified analogs tested were susceptible to EAL domain catalyzed degradation (Table 1). Second messenger analogs containing either one inosine (c-GMP-IMP) or N1-methyl guanine (c-N1mG-GMP) base in place of a single guanine were both degraded to their corresponding linear products at a rate within 2–3 fold of that for the native second messenger. In contrast, both c-di-IMP and c-GMP-AMP were hydrolyzed approximately 30-fold slower than c-di-GMP (Table 1). However, for the base modified analogs tested, we did not determine if the substrate concentration (100 µM) was saturating and it is likely that the observed decrease in rate for c-di-IMP and c-GMP-AMP is a binding effect rather than a catalytic effect. These data indicate that the EAL domain can accommodate structurally related analogs of guanine in the active site and suggests that specific recognition of only one guanine base is sufficient for second messenger binding and degradation.
In the HPLC profile of c-N1mG-GMP after treatment with CC3396, two product peaks with retention times differing from that of the cyclic dinucleotide were observed (Figure S1a). These peaks likely correspond to the two possible linear products p- N1mG-pG and pGpN1mG. They were formed in unequal amounts, suggesting that CC3396 preferentially binds the modified dinucleotide in one orientation. For c-GMP-IMP, a major product peak and a much smaller minor peak were observed suggesting a preference for binding orientation for that analog as well (Figure S1b). In contrast, only one product peak was observed for c-GMP-AMP (Figure S1c). This suggests there is a preference for binding orientation between the G and the A. These observations are consistent with structural analysis, which predicts that one guanine base is more extensively recognized than the other23,25.
Enzymatic hydrolysis of analogs containing ribose modifications
Based on structural analysis, no specific contacts are made to the 2′-OH’s of the c-di-GMP ribose sugars by the EAL domain (Figure 1b,c)23–25, suggesting that hydroxyl modification would not significantly affect the rate of catalysis. To test this prediction, we first measured the rate of degradation for the 2′-deoxy ligands, c-di-dGMP and c-dG-GMP (Figure 3b). We found no significant effect on degradation for c-dG-GMP (2-fold) however, c-di-dGMP was hydrolyzed approximately 14-fold slower than c-di-GMP (Table 1), suggesting that modification of both ribose rings increases the resistance of the second messenger towards EAL domain catalyzed degradation. To further test this observation, we measured the rate of hydrolysis for 2′-fluoro (c-di-2′F-GMP) and 2′- OMethyl (c-di-2′OMe-GMP) modified analogs (Figure 3). These ribose modifications have been shown to increase the stability of nucleic acids towards nucleases that cleave phosphodiester bonds40. We found that both analogs were hydrolyzed at a slower rate than c-di-GMP, with c-di-2′F-GMP degraded 5-fold slower and c-di-2′-OMe- GMP degraded 17-fold slower (Table 1). The 2′-OMethyl substitutions have a slightly larger effect on hydrolysis than 2′-fluoro substitutions, but both substitutions were degraded with reasonably good efficiency.
Enzymatic hydrolysis of analogs containing phosphate modifications
In the proposed mechanism for c-di-GMP hydrolysis by EAL domain proteins, both nonbridging oxygens of the scissile phosphate are contacted by an active site metal ion (Figure 1b)23,25. Furthermore, it was previously demonstrated that a c-di-GMP analog containing one phosphorothioate linkage is hydrolyzed by membrane extracts possessing c-di-GMP specific PDE activity to only a single linear product, with the modified linkage contained at the internal phosphate of the dinucleotide product31. This suggests that phosphorothioate linkages are resistant to enzymatic hydrolysis by EAL domain proteins and that a second messenger analog with phosphorothioate substitutions in place of both phosphodiester bonds may be resistant to enzymatic degradation. To investigate this hypothesis, we tested mono and dithiophosphate c-di- GMP derivatives for degradation by the EAL protein.
Based on previous reports in the literature, we anticipated that the monothiophosphate c-di-GMP derivatives, c-(RP)-Gps-GMP and c-(SP)-Gps-GMP (Figure 3), would be efficiently degraded31. As expected, both analogs were degraded at a rate within two-fold of that for c-di-GMP (Table 1), confirming that modification to only one of the c-di-GMP phosphates does not significantly affect catalysis. Furthermore, only a single degradation product was detected by HPLC analysis for both the RP and SP derivatives, consistent with the previous observations that incubation of these analogs with membrane extracts containing c-di-GMP specific PDE activity also only yielded a single linear product31.
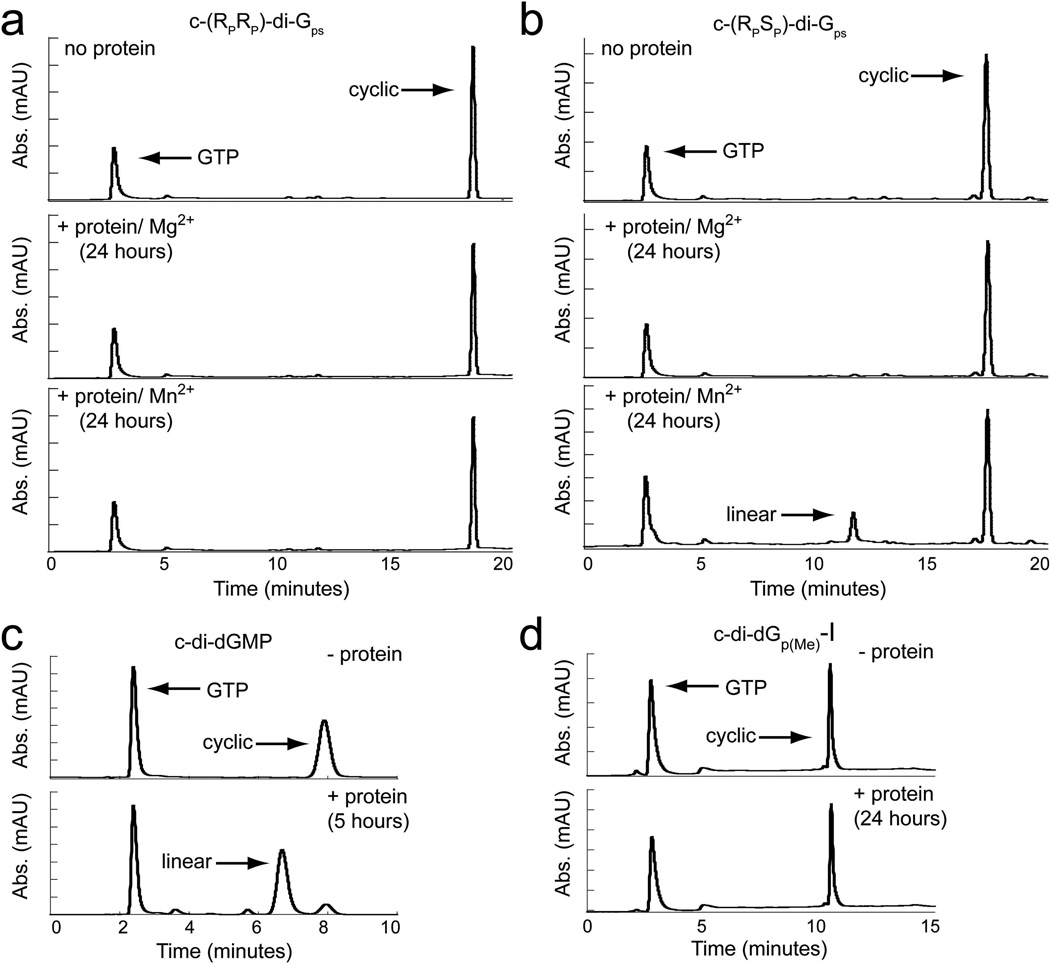
We next tested the ability of the protein to degrade the dithiophosphate analogs, c-(RPRP)-di-Gps and c-(RPSP)-di-Gps (Figure 3). In contrast to the monothiophosphate derivatives, we observed no detectable degradation for either dithiophosphate analog even after a 24-hour incubation with enzyme (Figure 5a,b). Even if 2% of the compound was hydrolyzed after 24 hours, which is the limit of our detection, the rate of hydrolysis would be estimated as 0.001 mol/(min*mol enzyme), which is at least 12,000-fold slower than that of c-di-GMP. This indicates that substitution of either phosphate oxygen of the scissile phosphate for sulfur renders the second messenger highly resistant to EAL domain catalyzed degradation. To ensure that the linear hydrolysis product was not co-eluting with the cyclic starting material, we collected the peak and analyzed it by ESI-MS to determine the mass of the corresponding compound. For both diastereomers, the mass of the cyclic, but not the linear product, was detected (supplemental info, Table S2).
Figure 5.
HPLC traces monitored at 254 nm showing degradation of backbone modified analogs by the EAL domain protein CC3396. Peaks corresponding to GTP, cyclic and linear dinucleotides are labeled. (a) In the absence of protein (top panel) a single peak corresponding to c-(RPRP)-di-Gps is observed. Only a peak corresponding to the cyclic dinucleotide is detected after a 24 hour incubation period with the protein in the presence of either 10 mM MgCl2 (middle panel) or 10 mM MnCl2 (bottom panel). (b) HPLC analysis of c-(RPSP)-di-Gps in the absence (top) and presence of protein (middle and bottom). No degradation product is observed with MgCl2, whereas a peak corresponding to the linear product is observed in the presence of MnCl2.
Approximately 20% of the cyclic product is hydrolyzed after 24 hours. (c) HPLC traces of the c-di-dGMP degradation reaction. The top panel shows the reaction mixture in the absence of protein and the bottom panel is in the presence of protein. After a 5 hour incubation with CC3396, c-di-dGMP is almost completely hydrolyzed (> 85%). (d) HPLC traces of c-di-dGp(Me)-I in the absence (top) and presence (bottom) of CC3396. No hydrolysis products are detectable even after a 24 hour incubation with the protein. The same lack of reactivity was observed for c-di-dGp(Me)-II after treatment with the protein for 24 hours (data not shown).
Because nuclease activity against phosphorothioate containing nucleic acids is often recovered in the presence of manganese41,42, we next tested both dithiophosphate analogs for degradation by the EAL domain PDE with 10 mM manganese in place of 10 mM magnesium in the reaction buffer. Under these conditions, we found that c-(RPSP)- di-Gps was approximately 20% degraded after 24 hours and estimate that the rate of degradation is approximately 1000-fold slower than c-di-GMP (Figure 5b). In contrast, no degradation for c-(RPRP)-di-Gps was observed under these conditions (Figure 5a). This indicates that manganese is able to rescue the activity of the protein, but only for one of the phosphorothioate diastereomers. Taken together, these data demonstrate that replacing both phosphodiester linkages with phosphorothioate linkages renders the second messenger highly resistant to EAL domain catalyzed degradation.
Synthesis and EAL domain catalyzed degradation of a backbone neutral c-di-GMP analog
The enzymatic degradation studies of the dithiophosphate c-di-GMP derivatives suggest that modification of both phosphate linkages renders the second messenger resistant to degradation by EAL domain proteins. To further explore this observation, we synthesized a second backbone modified c-di-GMP derivative containing methylphosphonate linkages in place of both canonical phosphodiester bonds. The methylphosphonate derivative of c-di-GMP, termed c-di-dGp(Me), substitutes a non-bridging phosphate oxygen of each phosphodiester linkage with an isosteric methyl group, resulting in a molecule with a neutral backbone (Figure 3). Because the methylphosphonamidite monomers are only commercially available in the 2′-deoxy version due to the instability of ribo-methylphosphonate linkages43–45, we synthesized the di-2′-deoxy derivative and tested this molecule in comparison to the di-2′-deoxy parental dinucleotide with standard phosphate linkages, c-di-dGMP (Figure 3). We modified the solid phase method for c-di-GMP reported by Kiburu et al to access this cdi- GMP derivative 38. We took advantage of the fact that methyl phosphonamidites starting monomers do not possess any phosphate protecting groups. This allowed for solid-phase dinucleotide synthesis, followed by selective cleavage of the linear dimer from the solid support under mild basic conditions without removing any phosphate protecting groups, or the protecting groups on the bases (Figure 2a). After coupling of both guanosine methyl phosphonamidites to the solid support, the linear dinucleotide was cleaved from the bead via β-elimination under basic conditions, and cyclized in solution.
Replacing one of the non-bridging oxygens on each phosphate renders both phosphates chiral and results in three different possible diastereomers (RPRP, RPSP, or SPSP). The fourth diastereomer (SPRP) is equivalent to the RPSP isomer because of the two fold symmetry within the cyclic dinucleotide. Using solid-phase chemistry as described above, we were able to synthesize and isolate two of the three possible diastereomers after HPLC purification (Figure 2b). This is similar to what was achieved for the synthesis of the phosphorothioate analogs. In that case the cyclization reaction proceeded with stereochemical specificity35,46. Thus, the absence of the third isomer is likely due to the cyclization step producing a linkage of only one chirality (most likely the RP, see discussion). Due to the small amount of product generated from solid phase synthesis, we were not able to make enough material to assign absolute stereochemistry. We have designated these analogs c-di-dGp(Me)-I and c-di-dGp(Me)-II based on their order of elution during HPLC purification (Figure 2b). However, tentative stereochemical assignments are possible based upon their relative affinities to riboswitch targets (binding data suggest isomer I is RPRP and isomer II is RPSP ; see discussion).
To determine if methylphosphonate c-di-GMP derivatives are nuclease resistant, we tested both diastereomers for degradation by the EAL domain protein CC3396. Similar to what we observed for the phosphorothioate derivatives, neither analog was degraded even after a 24 hour incubation period with the protein (Figure 5d). In contrast, the parental dinucleotide, c-di-dGMP, is nearly completely hydrolyzed (> 85%) after 5 hours (Figure 5c). Assuming the maximum possible level of degradation of 2% after 24 hours, the rate of degradation is more than 1000-fold slower than the parental dinucleotide (1.1 mol*min−1*mole enzyme−1, Table 1). Methyl substitution of both phosphodiester bonds results in a c-di-GMP analog that has high resistance to EAL domain catalyzed degradation.
We next sought to determine if the EAL domain resistance of the dithiophosphate and methylphosphonate c-di-GMP analogs results from a weaker affinity for the protein for these analogs or if the protein is unable to perform catalysis on these modified phosphate centers. To differentiate between these two possibilities, we looked at the ability of these analogs to competitively inhibit the degradation of radiolabeled c-di-GMP (*c-di-GMP)47. In the presence of trace amounts of *c-di-GMP and a large excess of an unlabeled competitor analog that is able to bind the EAL domain, *c-di-GMP binding and degradation should effectively be blocked. We found that in the absence of any competitor analog, *c-di-GMP was completely degraded to pGpG after approximately 15 min, whereas in the presence of a large excess of unlabeled c-di-GMP, no degradation was observed as expected (Figure 6a,b). However, in the presence of 100 µM c-di- AMP, which does not bind the EAL domain, we found that *c-di-GMP is degraded at nearly the same rate as in the absence of a competitor (Figure 6c). This establishes that only molecules able to bind the EAL domain can prevent *c-di-GMP degradation under these conditions.
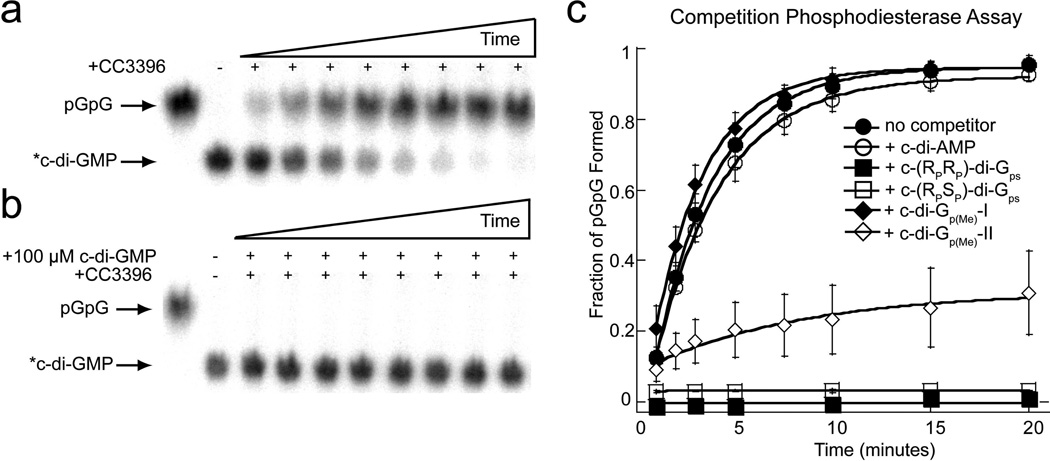
Figure 6.
Degradation of radiolabeled c-di-GMP (*c-di-GMP) in the presence of EAL domain resistant c-di-GMP analogs. (a) PEI-cellulose TLC showing degradation of *c-di-GMP to pGpG by CC3396 over time. (b) Degradation of *c-di-GMP in the presence of unlabeled c-di-GMP. (c) Fraction of pGpG formed from degradation of *c-di-GMP by CC3369 over time in the presence of unlabeled c-di-AMP, dithiophosphate, and methylphosphonate analogs.
To determine if the dithiophosphate analogs bind the EAL domain, we tested their ability to block degradation of *c-di-GMP as described above. In the presence of either 100 µM c-(RPRP)-di-Gps or c-(RPSP)-di-Gps, we found that *c-di-GMP degradation was completely inhibited (Figure 6c). This indicates that the EAL domain can recognize and bind both dithiophosphate diastereomers and suggests that their nuclease resistance results from the inability of the protein to catalyze hydrolysis on these phosphatemodified substrates. We estimated the affinity (Ki) of both the RPRP and RPSP diastereomers for the EAL domain to be within 10-fold of the Km of c-di-GMP (measured as ~ 110 nM), indicating that these analogs may function as effective competitive inhibitors of this enzyme class (c-(RPRP)-di-Gps Ki~ 480 nM; c-(RPSP)-di-Gps Ki~ 820 nM).
For the methylphosphonate derivatives, we found that isomer-I was unable to block second messenger degradation even though the analog concentration was approximately 6-orders of magnitude greater than that of *c-di-GMP. This indicates that c-di-dGp(Me)-I has very little affinity for the EAL protein. For isomer-II, we observed that only 30% of c-di-GMP was degraded to pGpG under the same conditions in which 95% of the substrate was degraded in the absence of any competitor analog (Figure 6c). This indicates that this analog retains some affinity for the EAL domain. Based on the affinity measurements for the dithiophosphate analogs as indicated above along with the competition studies indicating that isomer-II retains some affinity for the EAL domain, we estimate the Ki for isomer-II to be > 10-fold above the Km of c-di-GMP. Taken together, these data suggest that for isomer-I, EAL domain resistance is primarily due to the inability of this compound to bind the protein whereas for isomer-II, resistance stems from a combination of both weaker binding and chemical effects.
Effects of methylphosphonate substitutions on c-di-GMP binding by the class I and class II riboswitches
We previously published a structure-function study of c-di-GMP analog binding to both the class I and class II riboswitches34. With the exception of the novel methylphosphonate derivative developed here, all other analogs tested for EAL domain susceptibility were tested for riboswitch binding, indicating which analogs with increased enzymatic resistance still retain affinity for a known c-di-GMP target. Therefore, we next sought to determine the effects of methylphosphonate substitution of c-di-GMP on the affinity for its second messenger riboswitches.
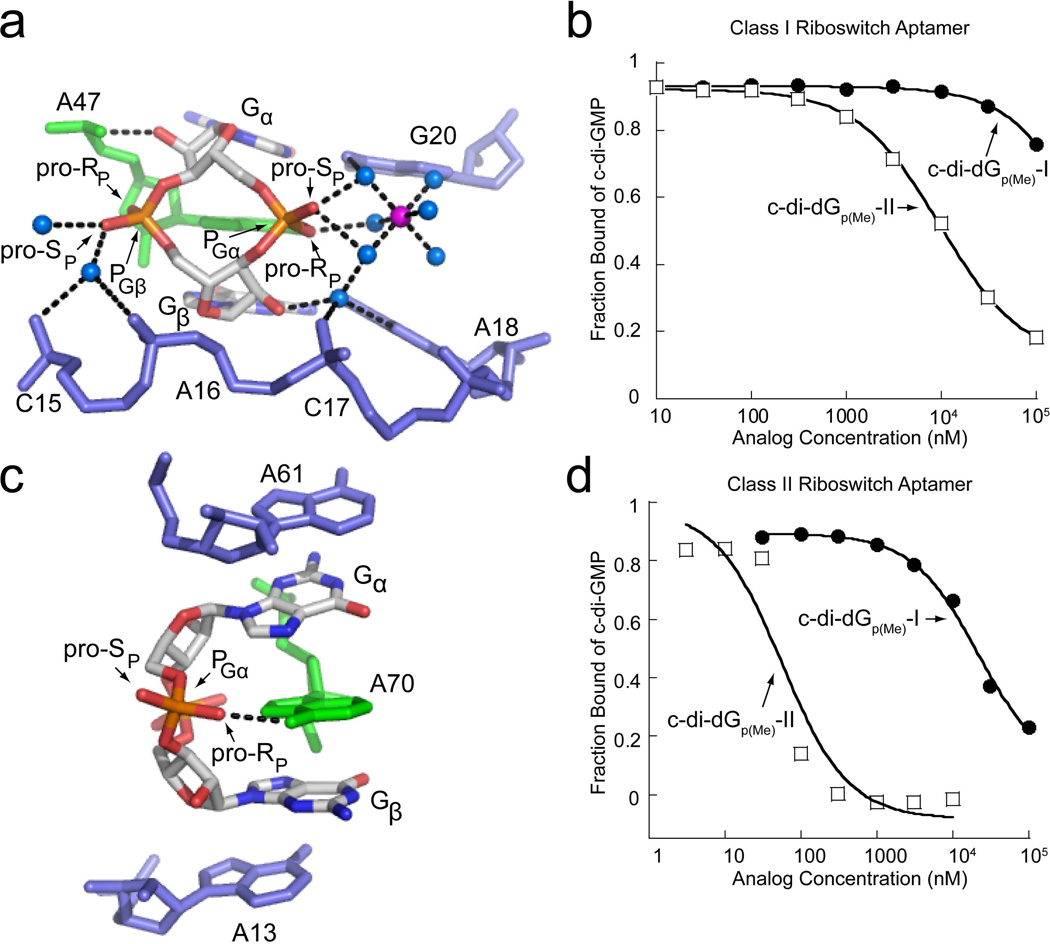
Structural characterization of c-di-GMP binding to the class I and class II riboswitches has revealed that class I more extensively recognizes the ribosylphosphate backbone compared to class II 26,27 (Figure 7a,c). We measured the affinity (Kd) of both methylphosphonate diastereomers for the class I riboswitch (Figure 7b). For c-di-dGp(Me)-I, only slight binding was detectable at high analog concentrations, suggesting that the Kd is > 100 µM (Table 2). In contrast, this RNA bound the second diastereomer, c-di-dGp(Me)-II, with surprisingly good affinity (540 nM). This is 5-fold tighter than the parental dinucleotide c-di-dGMP (Table 2) and indicates that binding of c-di-dGp(Me)-II resulted in improved affinity (−0.9 kcal/mol, Table 1) relative to the dideoxy cyclic dinucleotide analog.
Figure 7.
Backbone recognition of c-di-GMP and binding of methylphosphonate analogs by the class I and class II riboswitch aptamers. (a) c-di-GMP bound to the class I aptamer (PDB ID 3MXH). c-di-GMP is colored by atom with carbon shown in white, nitrogen in blue, oxygen in red, and phosphorous in orange. The highly conserved adenosine that intercalates between the guanine bases of c-di-GMP is colored in green and additional RNA residues are colored in blue. Water molecules are shown as blue spheres and magnesium ions as purple spheres. The pro-RP and pro-SP oxygens are indicated to show the sites of methyl substitutions. Hydrogen bonds are indicated by black dashed lines. (b) Binding curves of methylphosphonate derivatives to the class I riboswitch. The c-di-dGp(Me)-I binding curve is shown as black circles and the c-di-Gp(Me)-II curve is shown as open squares. (c) c-di-GMP bound to the class II aptamer (PDB ID 3Q3Z). Coloring and labeling is the same as in part (a). (d) Binding curves of the methylphosphonate derivatives to the class II riboswitch. Labeling is the same as in part (b).
Table 2.
Kd measurements of methylphosphonate analogs for the class I and class II c-di-GMP binding riboswitches determined by the competition gel-shift assay.
| Class I | Class II | |||||
|---|---|---|---|---|---|---|
| Analog | Kd (nM) | Fold Changea |
ΔΔGbind (kcal/mol)a |
Kd (nM) | Fold Changea |
ΔΔGbind (kcal/mol)a |
| c-di-dGMPb | 2600 ± 88 | − | − | 11 ± 1.2 | − | − |
| c-di-GMPb | 1.4 ± 0.1 | [1900]c | −4.4 | 2.2 ± 0.2 | [5] | −0.9 |
| c-di-dGP(Me)-I | > 100,000 | >38 | >2.2 | 1000 ± 93 | 91 | 2.7 |
| c-di-dGP(Me)-II | 540 ± 57 | [5] | −0.9 | 2.4 ± 0.3 | [5] | −0.9 |
| c-(RPRP)-di-Gps | 150 ± 33 | [17] | −1.7 | 3.6 ± 0.6 | [3] | −0.7 |
| c-(RPSP)-di-Gpsb | 750 ± 63 | [3] | −0.7 | 4.0 ± 0.8 | [3] | −0.6 |
Calculated relative to the parental dinucleotide c-di-dGMP.
Binding data for these compounds was previously reported34.
Brackets indicate fold increase in binding.
Similar studies were performed on the class II riboswitch aptamer (Figure 7d). This RNA bound c-di-dGp(Me)-I significantly weaker than c-di-dGp(Me)-II. The Kd of isomer-I was approximately 1 µM, 90-fold weaker than that of c-di-dGMP (Table 2). As with class I, we found that c-di-dGp(Me)-II analog bound the class II RNA 5-fold tighter than c-didGMP with a Kd nearly identical to that of c-di-GMP (2.4 nM, Table 2). For both riboswitch classes, a gain of 0.9 kcal/mol upon binding isomer-II was observed relative to the parental double deoxy analog (Table 2).
DISCUSSION
The enzymes responsible for the metabolism of c-di-GMP regulate the intracellular concentration of the second messenger, which ultimately controls diverse behaviors in bacteria including biofilm formation, motility and virulence response2,6,9,10. Here, we probed the specificity of the c-di-GMP specific EAL domain PDE CC3396 for its cognate ligand to identify analogs that are resistant to enzymatic hydrolysis. We found that modification of both GMP units of c-di-GMP is necessary to significantly affect the stability of the compound to hydrolysis. Substitution of both phosphates with either phosphorothioate or methylphosphonate linkages yields an EAL domain resistant second messenger analog. In addition, one of the two methylphosphonate diastereomers synthesized and tested (isomer-II) had increased affinity for the c-di- GMP riboswitches that function as downstream effectors in this signaling pathway relative to its parental dinucleotide c-di-dGMP. These results suggest that modifying both c-di-GMP phosphate linkages is a general strategy for designing second messenger analogs that are stable to EAL domain catalyzed degradation yet can still bind known downstream RNA effectors in this signaling pathway.
c-di-GMP has a two-fold axis of symmetry and the second messenger can bind the active site of the EAL domain in two possible orientations, with the modified GMP moiety placed either distal (P2) or proximal (P1) to the site where catalysis is taking place (Figure 1b). As a result, for asymmetric ribose or phosphate modified second messenger analogs in which one GMP moiety remains unmodified, the rate of enzyme catalyzed degradation by the EAL domain PDE was not significantly affected. For example, monothiophosphate and single 2′-deoxy modified ligands are degraded by the PDE at nearly the same rate as the native second messenger whereas the doubly modified ligands with these same substitutions are degraded significantly slower than cdi- GMP.
With the exception of c-di-AMP, modification to the guanine bases does not render the second messenger resistant to EAL domain catalyzed degradation. Substitution of one c-di-GMP base with either inosine or N1methyl guanine, which both are close structural analogs of guanine, had little to no effect on the rate of degradation. An analog with one adenosine substitution, c-GMP-AMP, was still recognized and degraded whereas the doubly substituted analog c-di-AMP was not. This suggests that specific recognition of only one c-di-GMP base is sufficient for binding to the PDE albeit with weaker affinity if the second site is substituted.
Interestingly, c-GMP-AMP has been discovered as a new second messenger in Vibrio cholerae, synthesized by a novel class of dinucleotide cyclases that preferentially produces this dinucleotide over c-di-GMP and c-di-AMP56. The ability of the EAL domain protein studied here to degrade c-GMP-AMP, as discussed above, suggests that specific recognition of only one guanine base is necessary for c-di-GMP recognition and subsequent degradation. However, sequence alignments of EAL domain proteins indicate that guanine base recognition is achieved primarily through highly conserved residues involved in G2 binding, whereas those residues involved in G1 binding are highly variable (Table S1). Therefore, in organisms that utilize both c-di-GMP and potentially c-GMP-AMP, such as Vibrio cholerae, recognition of G1 by endogenous EAL domain phosphodiesterases may be more stringent to specifically discriminate against c-GMP-AMP.
Dithiophosphate c-di-GMP analogs can bind the EAL domain, but what is the molecular basis of their nuclease resistance? Crystal structures of active EAL domain proteins reveal that two metal ions are bound in the active site and that both coordinate the proposed nucleophilic water molecule23,25. In addition, each metal ion coordinates a different non-bridging phosphate oxygen of the c-di-GMP scissile phosphate (P1; Figure 1b). This is different than the classic two metal ion nucleases in which both metal ions coordinate the same phosphate oxygen48 and often show stereochemical specificity for degrading phosphorothioate linkages (RP or SP)49–51. For many systems, making sulfur substitutions in place of oxygen at the site of metal coordination destroys enzymatic activity52. In the case of c-di-GMP, sulfur substitution may simply displace one of the active site metals, significantly affecting the ability of the protein to hydrolyze the scissile phosphate linkage. However, unlike some systems where one oxygen is strongly affected by sulfur substitution and the other is not49–51, both oxygens show strong effects for c-di-GMP degradation. Although dithiophosphate analogs are resistant to enzymatic hydrolysis, these derivatives retain the ability to bind the EAL domain with reasonable affinity and therefore have the potential to function as competitive inhibitors of these c-di-GMP metabolizing enzymes.
Similar to phosphorothioate substitution of c-di-GMP, methylphosphonate substitution also rendered the second messenger resistant to EAL domain catalyzed degradation. However, the methylphosphonate analogs were weaker inhibitors of the EAL domain than the phosphorothioate derivatives. In contrast, both the phosphorothioate and methylphosphonate substituted derivatives of c-di-GMP bound both the class I and class II c-di-GMP riboswitches with good affinity, and in the case of one methylphosphonate diastereomer, with an affinity better than the parental analog. This suggests that these enzymatically stable versions of second messenger analogs may be able to persist longer in the cell and may therefore be useful chemical tools for probing RNA-mediated c-di-GMP signaling in vivo.
While the absolute stereochemistry of the methylphosphonate derivatives are unknown, the data are consistent with the possibility that c-di-dGp(Me)-I is the RPRP diastereomer based on the affinity for riboswitch targets. The interaction between the non-bridging phosphate oxygen of PGα and the adenosine that intercalates between the c-di-GMP bases is the only conserved backbone contact between the otherwise distinct modes of ligand recognition by the two classes of riboswitches27 (Figure 7a,c). This is the only phosphate contact made to c-di-GMP by the class II riboswitch27. Therefore, substitution of the pro-RP oxygen of PGα is expected to have negative effects on binding for both RNAs. Because methyl groups are isosteric with oxygen and the pro-RP oxygen of PGβ is not recognized by either riboswitch, replacing this phosphate oxygen with a methyl group is not expected to significantly impact ligand affinity. The RPSP derivative can bind in two possible orientations, one in which the highly conserved contact to the pro-RP oxygen of PGα is not affected. In contrast, the RPRP diastereomer retains the two-fold symmetry axis and presents a pro-RP oxygen substitution at PGα in both binding orientations. The large loss in affinity for c-di-dGp(Me)-I by both the class I and class II riboswitch suggests that this contact is being disrupted and that isomer-I is the RPRP isomer. Due to the stereochemical specificity of the cyclization step, only two of the three possible diastereomers are produced and each must contain at least one RP linkage. This suggests that c-di-dGp(Me)-II is the RPSP isomer.
While methylphosphonate modification of both c-di-dGMP phosphate linkages had negative effects on binding and catalysis by the EAL domain, positive effects on binding by riboswitch targets of the second messenger were observed. The affinity of one of the methylphosphonate diastereomers (c-di-dGp(Me)-II) for the class II riboswitch was almost identical to that of c-di-GMP and tighter than that of its parental dinucleotide, c-di-dGMP. This suggests that this analog could affect the function of its RNA target molecule to the same extent as the native second messenger, while remaining more stable to degradation by EAL domain PDEs. Although the other methylphosphonate diastereomer (c-di-dGp(Me)-I) bound to the class II riboswitch with weaker affinity, the EAL domain was unable to recognize this compound even at very high analog concentrations. Thus, this EAL domain resistant c-di-GMP derivative shows a strong preference for binding the class II c-di-GMP riboswitch over the EAL domain and could be useful for specifically perturbing RNA mediated signaling processes without affecting the function of phosphodiesterases.
The class I riboswitch aptamer is also able to bind c-di-dGp(Me)-II tighter than the parental dinucleotide c-di-dGMP, despite the significant number of contacts made to the phosphate backbone. Interactions with the 2′-hydroxyl groups make a large contribution to ligand binding by the class I riboswitch34 however, removal of this functional group in the context of the methylphosphonate substitution did not have as large an effect on binding as in the background of the native, charged molecule. This suggests that alleviating electrostatic repulsion between c-di-GMP and the riboswitch RNA increases the affinity of the second messenger for its RNA targets. In addition, previous studies have suggested that a large portion of the binding energy for the c-di-GMP riboswitches results from base stacking interactions34,53. Collectively, these observations indicate that it may be possible to completely replace the ribosyl-phosphate backbone of the second messenger with neutral linkers and maintain affinity for riboswitch targets, provided that the guanine bases are held at the appropriate distance to maintain these high affinity stacking interactions.
Although c-di-dGp(Me)-II binds tighter to c-di-GMP riboswitches than the parental dinucleotide c-di-dGMP, the ribose version of this analog is expected to have even higher affinity, particularly for the class I riboswitch. Although methylphosphonate linkages next to ribose hydroxyls are unstable43–45, it may be feasible to synthesize the ribo-methylphosphonate derivative of c-di-GMP due to the limited conformational flexibility of this cyclic dinucleotide. Wang et al. recently reported the synthesis of a c-di-GMP derivative containing a sulfur substitution in place of a bridging phosphate oxygen that was stable at neutral pH47. When incorporated into linear RNA oligonucleotides, these linkages are highly labile due to in-line attack from the neighboring 2′-hydroxyl. However, when incorporated into c-di-GMP, this linkage proved to be stable because the cyclic backbone of the second messenger constrains the molecule and effectively prevents the 2′-hydroxyl from attacking the scissile phosphate47. Ribo-methylphosphonate linkages incorporated into c-di-GMP may be stable provided that the 2′-OH group is protected until after the cyclization reaction is complete. Improved synthetic methods will be necessary to fully explore the potential of this analog as a chemical tool to study c-di-GMP signaling.
Given that the EAL domain cannot efficiently hydrolyze second messenger analogs with methlyphosphonate or phosphorothioate substitutions, which closely resemble the canonical phosphodiester linkage, it is unlikely that a c-di-GMP derivative with a non-native backbone would be susceptible to enzyme catalyzed degradation. Modifying the phosphate backbone provides not only the opportunity to increase the enzymatic stability of the second messenger but to also increase its cell permeability. Neutral molecules often have increased cell permeability and therefore a greater chance of entering cells via passive diffusion as compared to charged compounds54,55. Because c-di-GMP signaling is used by many pathogenic organisms to control lifestyle changes that often allow the bacteria to infect a host or form biofilms, which are highly resistant to current antibiotic treatments, the ability to target this pathway for therapeutic purposes is desirable. Thus, the degradation resistant phosphorothioate and neutral methyphosphonate derivatives studied here have desirable properties that may make them useful for studying c-di-GMP signaling in vivo and manipulating the biological processes under control of this second messenger.
Supplementary Material
Acknowledgements
We thank all members of the Strobel Lab for advice and helpful discussions; B. Kazmierczak (Yale University) for the CC3396 expression plasmid; U. Jenal (University of Basel) for the gift of the PleD* expression plasmid.
πThis work was supported by National Institutes of Health Grant GM022778 to S.A.S and National Institutes of Health Grant GM79760 to R.A.J.
ABBREVIATIONS
- c-di-GMP
bis-(3′-5′)-cyclic dimeric guanosine monophosphate
Footnotes
Supporting Information. Supplemental results including 2 tables and 2 figures as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 3.Mills E, Pultz IS, Kulasekara HD, Miller SI. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell. Microbiol. 2011;13:1122–1129. doi: 10.1111/j.1462-5822.2011.01619.x. [DOI] [PubMed] [Google Scholar]
- 4.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 5.Sondermann H, Shikuma NJ, Yildiz FH. You’ve come a long way: c-di-GMP signaling. Curr. Opin. Microbiol. 2012;15:140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 7.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1999;1:303–305. [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan RP, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 14.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 15.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRPlike protein (Clp) of Xanthomonas axonopodis pv. citri. J. Bacteriol. 2009;191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duerig A, et al. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newell PD, Monds RD, O’Toole GA. LapD is a bis-(3’,5’)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Y, et al. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 2011;286:2910–2917. doi: 10.1074/jbc.M110.196220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barends TRM, et al. Structure and mechanism of a bacterial lightregulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 24.Minasov G, et al. Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 2009;284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchigvintsev A, et al. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 2010;402:524–538. doi: 10.1016/j.jmb.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith KD, et al. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. Structural basis of differential ligand recognition by two classes of bis-(3’-5’)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7757–7762. doi: 10.1073/pnas.1018857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benach J, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko J, et al. Structure of PP4397 reveals the molecular basis for different cdi-GMP binding modes by Pilz domain proteins. J. Mol. Biol. 2010;398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Navarro MVAS, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross P, et al. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 1990;265:18933–18943. [PubMed] [Google Scholar]
- 32.Paul R, et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao F, et al. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal. Biochem. 2009;389:138–142. doi: 10.1016/j.ab.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 34.Shanahan CA, Gaffney BL, Jones RA, Strobel SA. Differential analogue binding by two classes of c-di-GMP riboswitches. J. Am. Chem. Soc. 2011;133:15578–15592. doi: 10.1021/ja204650q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaffney BL, Veliath E, Zhao J, Jones RA. One-flask syntheses of c-di-GMP and the [Rp,Rp] and [Rp,Sp] thiophosphate analogues. Org. Lett. 12:3269–3271. doi: 10.1021/ol101236b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Veliath E, Kim S, Gaffney BL, Jones RA. Thiophosphate analogs of c-di-GMP: impact on polymorphism. Nucleosides Nucleotides Nucleic Acids. 2009;28:352–378. doi: 10.1080/15257770903044523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 38.Kiburu I, Shurer A, Yan L, Sintim HO. A simple solid-phase synthesis of the ubiquitous bacterial signaling molecule, c-di-GMP and analogues. Mol. Biosyst. 2008;4:518–520. doi: 10.1039/b719423d. [DOI] [PubMed] [Google Scholar]
- 39.Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Basu S, Strobel SA. Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4-P6 domain. RNA. 1999;5:1399–1407. doi: 10.1017/s135583829999115x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narlikar GJ, Herschlag D. Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem. 1997;66:19–59. doi: 10.1146/annurev.biochem.66.1.19. [DOI] [PubMed] [Google Scholar]
- 43.Dertinger D, Uhlenbeck OC. Evaluation of methylphosphonates as analogs for detecting phosphate contacts in RNA-protein complexes. RNA. 2001;7:622–631. doi: 10.1017/s1355838201002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pritchard CE, et al. Methylphosphonate mapping of phosphate contacts critical for RNA recognition by the human immunodeficiency virus tat and rev proteins. Nucleic Acids Res. 1994;22:2592–2600. doi: 10.1093/nar/22.13.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battistini C, Fustinoni S, Brasca MG, Borghi D. Stereoselective synthesis of cyclic dinucloetide phosphorothioates. Tetrahedron. 1993;49:1115–1132. [Google Scholar]
- 47.Wang J, et al. Conservative change to the phosphate moiety of cyclic diguanylic monophosphate remarkably affects its polymorphism and ability to bind DGC, PDE, and PilZ proteins. J. Am. Chem. Soc. 2011;133:9320–9330. doi: 10.1021/ja1112029. [DOI] [PubMed] [Google Scholar]
- 48.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Burgers PM, Eckstein F, Hunneman DH. Stereochemistry of hydrolysis by snake venom phosphodiesterase. J. Biol. Chem. 1979;254:7476–7478. [PubMed] [Google Scholar]
- 50.Connolly BA, Potter BV, Eckstein F, Pingoud A, Grotjahn L. Synthesis and characterization of an octanucleotide containing the EcoRI recognition sequence with a phosphorothioate group at the cleavage site. Biochemistry. 1984;23:3443–3453. doi: 10.1021/bi00310a010. [DOI] [PubMed] [Google Scholar]
- 51.Potter BVL, Connolly BA, Eckstein F. Synthesis and configurational analysis of a dinucleoside phosphate isotopically chiral at phosphorus. Stereochemical course of Penicillium citrinum nuclease P1 reaction. Biochemistry. 1983;22:1369–1377. doi: 10.1021/bi00275a008. [DOI] [PubMed] [Google Scholar]
- 52.Brautigam CA, Steitz TA. Structural principles for the inhibition of the 3’-5’ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 53.Smith KD, Lipchock SV, Strobel SA. Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer. Biochemistry. 2012;51:425–432. doi: 10.1021/bi2016662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varma RS. Synthesis of oligonucleotide analogues with modified backbones. Synlett. 1993;1993:621–637. [Google Scholar]
- 55.Rozners E. Carbohydrate chemistry for RNA interference: Synthesis and properties of RNA analogues modified in sugar-phosphate backbone. Curr. Org. Chem. 2006;10:675–692. [Google Scholar]
- 56.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated Regulation of Accessory Genetic Elements Produces Cyclic Di-Nucleotides for V. cholerae Virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.