Abstract
The reliability of loop-mediated isothermal amplification (LAMP), initially developed for the detection of human herpesvirus 7 (HHV-7), was evaluated in this study. Although a LAMP product was detected in HHV-7 DNA, neither HHV-6 nor human cytomegalovirus DNA produced a product. When agarose gel electrophoresis was used for the detection of LAMP products, the sensitivity of a 30-min HHV-7 LAMP reaction reached 250 copies/tube. The use of turbidity for the detection of the LAMP products gave a sensitivity of 500 and 250 copies/tube for 30- and 60-min reactions, respectively. Following these initial validation studies, clinical samples collected from two patients with primary HHV-7 infections were examined by HHV-7 LAMP. By use of agarose gel electrophoresis, HHV-7 LAMP products could be detected in acute-phase plasma samples but no LAMP product was detectable in convalescent-phase plasma samples from either patient. Since a turbidity assay is less sensitive than agarose gel electrophoresis, no HHV-7 LAMP product could be detected in plasma samples after a 30-min LAMP reaction. After a 60-min LAMP reaction, HHV-7 LAMP product could be detected in acute-phase plasma samples.
Human herpesvirus 7 (HHV-7) was originally isolated in 1990 from CD4+ T lymphocytes obtained from healthy adults (4). The virus, belonging to the Betaherpesvirinae subfamily, is considered to be closely related to HHV-6 based on biological and molecular analyses (2, 19). Soon after the discovery of the virus, HHV-7, in addition to HHV-6, was reported to be a causative agent in exanthem subitum (14, 15). Primary HHV-7 infection usually has a benign and self-limited clinical course but can in rare instances cause several severe complications (16, 18). Furthermore, although the clinical features of HHV-7 reactivation remain obscure, several manifestations, including fatality due to viral reactivation, have been reported for organ transplant recipients (3, 5, 8, 12, 17, 21). It is therefore important to establish a reliable rapid diagnostic procedure for the detection of active HHV-7 infection. Rapid virological diagnosis, however, has proven difficult. Isolation of the virus requires cocultivation with preactivated cord blood mononuclear cells, a difficult procedure to perform in commercial laboratories. In addition, both viral isolation and serological testing require substantial time to obtain final results. While rapid diagnosis by PCR may eventually become practical for the bedside monitoring of active viral infections, it has not yet become common in hospital laboratories due to the requirement for specific equipment (a thermal cycler).
Recently, Notomi et al. (11) reported a novel nucleic acid amplification method, termed loop-mediated isothermal amplification (LAMP), capable of amplifying DNA under isothermal conditions with high specificity, efficiency, and speed. The most significant advantage of LAMP is the ability to amplify specific sequences of DNA under isothermal conditions between 63 and 65°C. This ability allows the method to be performed with only simple and cost-effective reaction equipment amenable to use in hospital laboratories. We sought to establish a LAMP-based HHV-7 DNA amplification method and to examine its reliability in the diagnosis of active HHV-7 infections.
HHV-7 (RK) DNA was used as a positive control to determine appropriate conditions for HHV-7 LAMP. This DNA served to establish LAMP baseline sensitivity and specificity. DNA from HHV-6A (U1102), HHV-6B (Z29), and human cytomegalovirus (HCMV) (AD-169) was used to determine the specificity of HHV-7 LAMP. A plasmid containing the HHV-7 target sequence (pGEMH7S12) was used to determine the limits of assay sensitivity.
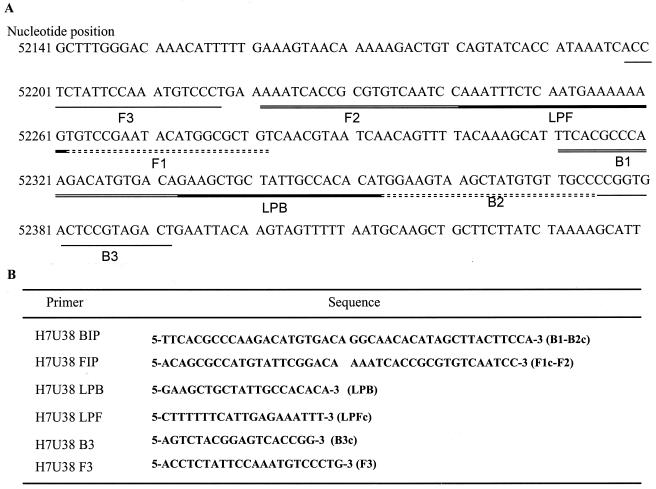
The LAMP reaction was conducted as described by Notomi et al. (11) and Nagamine et al. (10). LAMP requires a set of four specially designed primers (B3, F3, BIP, and FIP) recognizing a total of six distinct sequences (B1 to B3 and F1 to F3) within the target DNA. Primers for HHV-7 LAMP were designed by using Primer Explorer V software to recognize the HHV-7 U38 gene (DNA polymerase gene). The location and sequence of each primer are shown in Fig. 1. BIP, recognizing the U38 gene of HHV-7 (H7U38BIP), consisted of the B1 direct sequence (22 nucleotides [nt]) and the B2 complementary sequence (21 nt). Primer FIP, recognizing the U38 gene of HHV-7 (H7U38FIP), contained the F1 complementary sequence (21 nt) and the F2 direct sequence (20 nt). Primers B3 (H7U38B3) and F3 (H7U38F3), complementary to the U38 gene of HHV-7, were located outside the F2 and B2 regions. Since additional loop primers increase amplification efficiency, we also synthesized loop primers for the HHV-7 U38 gene (H7U38LPB and H7U38LPF). H7U38LPB contained the LPB sequence, while H7U38LPF contained the LPF complementary sequence. LAMP was performed with a 25-μl reaction mixture containing 2.4 μM H7U38FIP and H7U38BIP, each outer primer (H7U38F3 and H7U38B3) at a concentration of 0.4 μM, and each loop primer (H7U38LPF and H7U38LPB) at a concentration of 1.2 μM. The mixture was incubated at 63°C for 30 or 60 min, and then a TERAMECS LA200 (Eiken Chemical, Tokyo, Japan) was used to measure turbidity after 30 or 60 min of LAMP. After the turbidity was measured, LAMP products were subjected to electrophoresis on a 1.5% agarose gel.
FIG. 1.
(A) Locations and names of target sequences used as primers for HHV-7 LAMP within the U38 gene. (B) Name and sequence of each primer used for HHV-7 LAMP. B2c, sequence complementary to B2; F1c, sequence complementary to F1; LPFc, sequence complementary to LPF; B3c, sequence complementary to B3.
Real-time PCR was used to measure the quantity of HHV-7 DNA in each sample. Conditions for real-time PCR to quantify HHV-7 DNA were established by using the U31 gene, encoding the HHV-7 tegument protein, as a target sequence. The forward primer (H7TA3; 5′-AAAGAATGGTTTTGTTCAACTCCAA-3′), the reverse primer (H7TA4; 5′-ACATTCACTTTGCGTGCATTTTC-3′), and the probe (H7TAP2; 5′-TCATCGAGAACATAGGAGAAGCTCCAGCA-3′) recognizing the gene were designed by using Primer Express (PE Applied Biosystems, Foster City, Calif.). PCRs were performed with the TaqMan PCR kit (PE Applied Biosystems) according to the manufacturer's protocol. Standard curves for measuring HHV-7 DNA were constructed by using the CT values obtained from a serially diluted plasmid, pH7TA, which contained HHV-7 U31. The CT value for each sample was plotted on the standard curve, allowing the copy number to be automatically calculated with Sequence Detector v1.6 (PE Applied Biosystems).
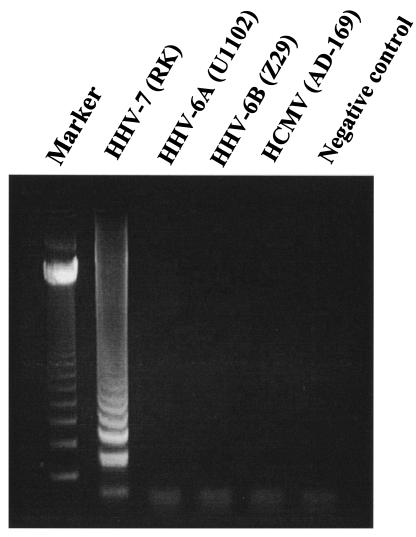
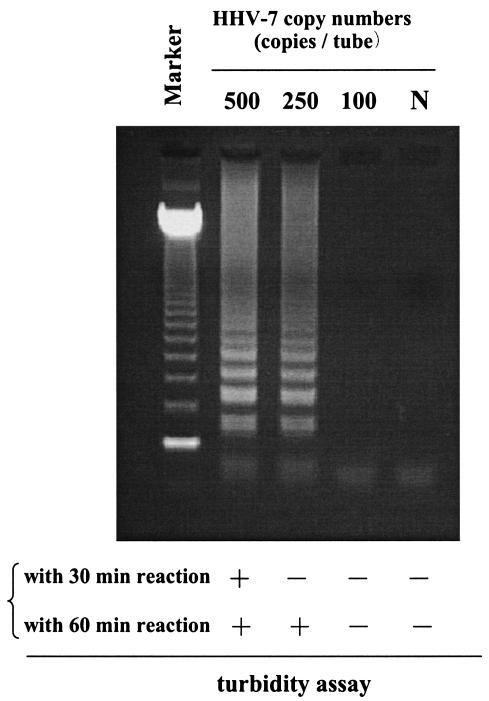
To develop an efficient assay to detect HHV-7 infection, we first evaluated the specificity of the HHV-7 primers. Although amplified HHV-7 DNA demonstrated the typical ladder patterns (Fig. 2), no LAMP products were detected in reactions performed with HHV-6A, HHV-6B, and HCMV DNA. We also determined the sensitivity of this method. Serial dilutions of the pGEMH7S12 plasmid were used to determine assay detection limits. The detection of HHV-7 LAMP products by agarose gel electrophoresis gave a sensitivity of 250 copies/tube for a 30-min reaction (Fig. 3). Detection by the turbidity assay, however, gave sensitivities of 500 and 250 copies/tube for 30- and 60-min LAMP reactions, respectively. As the LAMP-positive samples demonstrated typical time-related changes in turbidity described elsewhere (9), positive and negative samples were easy to discriminate.
FIG. 2.
DNA extracted from Betaherpesvirinae-infected cells was amplified by the HHV-7 LAMP protocol to determine the specificity of the method. Marker, 123-bp DNA ladder marker.
FIG. 3.
Serial dilutions of pGEMH7S12 plasmid DNA were amplified by HHV-7 LAMP to determine the respective sensitivities of each assay. Marker, 123-bp DNA ladder marker; N, no template control.
After initial validation studies, samples collected from two patients with primary HHV-7 infections were analyzed by HHV-7 LAMP. HHV-7 was isolated from peripheral blood collected during the febrile period (day 1 of the illness). Seroconversion of HHV-7 immunoglobulin G antibody titers was later observed in these patients. Results of HHV-7 LAMP and real-time PCR are summarized in Table 1. Agarose gel electrophoresis for the detection of LAMP products detected positive signals in both acute-phase and convalescent-phase blood samples from patient 1 after a 30-min LAMP reaction. For patient 2, a positive signal was detected only in acute-phase blood samples. The HHV-7 LAMP product was detected from plasma only from acute-phase samples; no LAMP products were detected in convalescent-phase plasma samples from either of these two patients. The turbidity assay detected HHV-7 LAMP products in convalescent-phase blood samples from patient 1. Moreover, since the turbidity assay is less sensitive than agarose gel electrophoresis, no HHV-7 LAMP product could be detected in acute-phase plasma samples from either patient by this method. Since the turbidity assay is faster and easier than agarose gel electrophoresis analysis for the detection of LAMP products, it is suitable for bedside monitoring of viral infection. Therefore, we have established a protocol for HHV-7 LAMP with a turbidity assay. After a 60-min LAMP reaction, HHV-7 LAMP product was detectable in only acute-phase plasma samples from the two patients regardless of the detection method used. Copy numbers of HHV-7 DNA in these samples were also measured by using real-time PCR; the results of this assay correlated well with the results of HHV-7 LAMP.
TABLE 1.
Results of HHV-7 LAMP assay for two male patients with primary HHV-7 infections
| Patient | Age (mo) | Sample | Specimen type | Days of sampling | Test result for LAMP reaction timea
|
Copy no. determined by real-time PCR (copies/reaction) | |||
|---|---|---|---|---|---|---|---|---|---|
| 30 min
|
60 min
|
||||||||
| Gel | Turbidity | Gel | Turbidity | ||||||
| 1 | 14 | 1 | Blood | 1 | + | + | ND | ND | 30,649 |
| 2 | Plasma | 1 | + | − | + | + | 205 | ||
| 3 | Blood | 13 | + | + | ND | ND | 785 | ||
| 4 | Plasma | 13 | − | − | − | − | 0 | ||
| 2 | 12 | 5 | Blood | 1 | + | + | ND | ND | 176,831 |
| 6 | Plasma | 1 | + | − | + | + | 762 | ||
| 7 | Blood | 16 | − | − | ND | ND | 26 | ||
| 8 | Plasma | 16 | − | − | − | − | 0 | ||
+, LAMP product was detected by agarose gel electrophoresis (Gel) or by a turbidity assay with a TERAMECS LA200 (Turbidity); −, LAMP product was not detected by agarose gel electrophoresis (Gel) or by a turbidity assay with a TERAMECS LA200 (Turbidity); ND, not done.
HHV-7 LAMP specifically amplified only HHV-7 DNA; no cross-reactivity was observed for other Betaherpesvirinae (HHV-6A, HHV-6B and HCMV) (Fig. 2). The detection limits of HHV-7 LAMP with a 30-min reaction were 250 and 500 copies/tube as determined by agarose gel electrophoresis and by the turbidity assay with the LA200, respectively. Although the turbidity assay is less sensitive than agarose gel electrophoresis, this assay is more appropriate for a bedside monitoring system due to its ease and rapidity. Turbidity gradually increases during LAMP reaction due to increased numbers of the amplified product (9). Therefore, the sensitivity of LAMP is improved when reaction time is increased. The sensitivity of the turbidity assay after 60 min of LAMP reaction increased to a level similar to that for a 30-min reaction with agarose gel electrophoresis analysis being used for detection. These findings demonstrate that HHV-7 LAMP has high specificity and efficiency for the amplification of viral DNA. Although the turbidity assay requires increased time to achieve sensitivity similar to that of agarose gel electrophoresis analysis, the turbidity assay is likely more appropriate for hospital laboratory use.
Since HHV-7 can persist in a latent state in peripheral blood mononuclear cells after primary infection (1, 14), detection of viral DNA in peripheral blood mononuclear cells by sensitive PCR may not be able to differentiate between active and latent infections in organ transplant recipients. In this study, convalescent-phase blood samples collected from patient 1 were positive for HHV-7 LAMP, demonstrating the detection of latent resident viral DNA. This result suggests that HHV-7 LAMP with whole blood as a specimen is too sensitive to be used for the monitoring of active viral infection, as the assay can detect latently infected viral DNA. Detection of viral DNA in plasma, however, appears to correlate well with active HHV-6 infection (13). Similar findings have been reported for several patients with primary HHV-7 infections (6, 7, 20). In this study, two acute-phase plasma samples were positive for HHV-7 after a 30-min LAMP reaction, visualized by agarose gel electrophoresis analysis. An HHV-7 LAMP product was not detected in the convalescent-phase plasma samples, suggesting that plasma should be used as specimens for HHV-7 LAMP testing to distinguish between active and latent infections. The turbidity assay, a more convenient protocol for hospital laboratories, was less sensitive in detecting HHV-7 DNA in acute-phase plasma samples after a 30-min LAMP reaction. A 60-min HHV-7 LAMP reaction, however, was sufficient to detect viral DNA in acute-phase plasma by the turbidity assay. This protocol provides an effective way to monitor active HHV-7 infection in hospital laboratories.
Quantities of HHV-7 DNA in the two patients were also monitored by real-time PCR, currently the most reliable method to measure copy numbers of viral nucleic acids. The results of qualitative HHV-7 LAMP (positive and negative) correspond well to changes in viral load, as determined by real-time PCR. This result also supports the reliability of HHV-7 LAMP as a mechanism to monitor active HHV-7 infection. Since only two patients were evaluated in the present study due to the difficulties in identifying primary HHV-7 infection cases, a larger number of patients with active HHV-7 infections should be examined in future studies to confirm these results.
Acknowledgments
We thank Takeda Chemical Industries, Ltd., Osaka, Japan, for supplying recombinant human interleukin-2, and we gratefully acknowledge Eiken Chemical for their contribution to this work. We also thank Akiko Yoshikawa and Maki Sawamura for technical assistance.
This work was supported in part by Grants-in-Aid for Scientific Research and Open Research Center of Fujita Health University from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a grant from Japan Society for the Promotion of Science (grant JSPS-RFTF97L00703).
REFERENCES
- 1.Asano, Y., S. Suga, T. Yoshikawa, T. Yazaki, and T. Uchikawa. 1995. Clinical features and viral excretion in an infant with primary human herpesvirus 7 infection. Pediatrics 95:187-190. [PubMed] [Google Scholar]
- 2.Berneman, Z. N., D. V. Ablashi, G. Li, M. Eger-Fletcher, M. S. Reitz, Jr., C. L. Hung, I. Brus, A. L. Komaroff, and R. C. Gallo. 1992. Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc. Natl. Acad. Sci. USA 89:10552-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, P. K., K. W. Chik, K. F. To, C. K. Li, M. N. Shing, K. C. Ng, P. M. Yuen, and A. F. Cheng. 2002. Case report: human herpesvirus 7 associated fatal encephalitis in a peripheral blood stem cell transplant recipient. J. Med. Virol. 66:493-496. [DOI] [PubMed] [Google Scholar]
- 4.Frenkel, N., E. C. Schirmer, L. S. Wyatt, G. Katsafanas, E. Roffman, R. M. Danovich, and C. H. June. 1990. Isolation of a new herpesvirus from human CD4+ T cells. Proc. Natl. Acad. Sci. USA 87:748-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths, P. D., M. Ait-Khaled, C. P. Bearcroft, D. A. Clark, A. Quaglia, S. E. Davies, A. K. Burroughs, K. Rolles, I. M. Kidd, S. N. Knight, S. M. Noibi, A. V. Cope, A. N. Phillips, and V. C. Emery. 1999. Human herpesviruses 6 and 7 as potential pathogens after liver transplant: prospective comparison with the effect of cytomegalovirus. J. Med. Virol. 59:496-501. [DOI] [PubMed] [Google Scholar]
- 6.Hara, S., H. Kimura, Y. Hoshino, N. Tanaka, K. Nishikawa, M. Ihira, T. Yoshikawa, and T. Morishima. 2002. Detection of herpesvirus DNA in the serum of immunocompetent children. Microbiol. Immunol. 46:177-180. [DOI] [PubMed] [Google Scholar]
- 7.Ihira, M., T. Yoshikawa, K. Suzuki, M. Ohashi, S. Suga, K. Horibe, N. Tanaka, H. Kimura, S. Kojima, K. Kato, T. Matsuyama, Y. Nishiyama, and Y. Asano. 2002. Monitoring of active HHV-6 infection in bone marrow transplant recipients by real time PCR; comparison to detection of viral DNA in plasma by qualitative PCR. Microbiol. Immunol. 46:701-705. [DOI] [PubMed] [Google Scholar]
- 8.Kidd, I. M., D. A. Clark, C. A. Sabin, D. Andrew, A. F. Hassan-Walker, P. Sweny, P. D. Griffiths, and V. C. Emery. 2000. Prospective study of human betaherpesviruses after renal transplantation: association of human herpesvirus 7 and cytomegalovirus co-infection with cytomegalovirus disease and increased rejection. Transplantation 69:2400-2404. [DOI] [PubMed] [Google Scholar]
- 9.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2002. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 10.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 11.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop mediated isothermal amplification of DNA. Nucleic. Acids Res. 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman, H. K., J. S. Peiris, C. E. Taylor, P. Warwicker, R. F. Jarrett, and C. R. Madeley. 1996. “Cytomegalovirus disease” in renal allograft recipients: is human herpesvirus 7 a co-factor for disease progression? J. Med. Virol. 48:295-301. [DOI] [PubMed] [Google Scholar]
- 13.Secchiero, P., D. R. Carrigan, Y. Asano, L. Benedetti, R. W. Crowley, A. L. Komaroff, R. C. Gallo, and P. Lusso. 1995. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J. Infect. Dis. 171:273-280. [DOI] [PubMed] [Google Scholar]
- 14.Suga, S., T. Yoshikawa, T. Nagai, and Y. Asano. 1997. Clinical features and virological findings in children with primary human herpesvirus 7 infection. Pediatrics 99:E4. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka, K., T. Kondo, S. Torigoe, S. Okada, T. Mukai, and K. Yamanishi. 1994. Human herpesvirus 7: another causal agent for roseola (exanthem subitum). J. Pediatr. 125:1-5. [DOI] [PubMed] [Google Scholar]
- 16.Torigoe, S., W. Koide, M. Yamada, E. Miyashiro, K. Tanaka-Taya, and K. Yamanishi. 1996. Human herpesvirus 7 infection associated with central nervous system manifestations. J. Pediatr. 129:301-305. [DOI] [PubMed] [Google Scholar]
- 17.Ward, K. N., R. P. White, S. Mackinnon, and M. Hanna. 2002. Human herpesvirus-7 infection of the CNS with acute myelitis in an adult bone marrow recipient. Bone Marrow Transplant. 30:983-985. [DOI] [PubMed] [Google Scholar]
- 18.Ward, K. N., P. Kalima, K. M. MacLeod, and T. Riordan. 2002. Neuroinvasion during delayed primary HHV-7 infection in an immunocompetent adult with encephalitis and flaccid paralysis. J. Med. Virol. 67:538-541. [DOI] [PubMed] [Google Scholar]
- 19.Wyatt, L. S., W. J. Rodriguez, N. Balachandran, and N. Frenkel. 1991. Human herpesvirus 7: antigenic properties and prevalence in children and adults. J. Virol. 65:6260-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshikawa, T., M. Ihira, K. Suzuki, S. Suga, K. Iida, Y. Saito, K. Asonuma, K. Tanaka, and Y. Asano. 2000. Human herpesvirus 6 infection after living related liver transplantation. J. Med. Virol. 62:52-59. [PubMed] [Google Scholar]
- 21.Yoshikawa, T., J. Yoshida, M. Hamaguchi, T. Kubota, S. Akimoto, M. Ihira, Y. Nishiyama, and Y. Asano. 2003. Human herpesvirus 7-associated meningitis and optic neuritis in a patient after allogeneic stem cell transplantation. J. Med. Virol. 70:440-443. [DOI] [PubMed] [Google Scholar]