Abstract
Dissemination of Shiga toxin (Stx)-encoding bacteriophages is the most likely mechanism for the spread of Stx-encoding genes and the emergence of new Stx-producing Escherichia coli (STEC). Biofilm has been reported to be a place where horizontal gene transfer by plasmid conjugation and DNA transformation may occur, and in this study, horizontal gene transfer by transduction has been demonstrated. Transfer of Stx-encoding bacteriophages to potentially pathogenic E. coli in biofilm was observed at both 20°C and 37°C. The infection rates were higher at 37°C than at 20°C. To our knowledge, this study is the first to show lateral gene transfer in biofilm mediated by a temperate bacteriophage. The study shows that the biofilm environment can be suitable for transduction events and can thereby be an environment for the emergence of new pathogenic E. coli.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is a food-borne pathogen that may cause diseases ranging from mild diarrhea to hemorrhagic colitis and complications such as the life-threatening hemolytic-uremic syndrome (HUS) (1). An array of virulence characteristics have been described for STEC (reviewed in references 2, 3, and 4), and some of these, such as the eae-encoded adherence factor intimin, are found in other E. coli pathogroups as well. However, the major virulence factor of STEC is the production of Shiga toxins (Stx). This characteristic defines the STEC pathogroup. The stx genes are located within a heterogeneous family of temperate lambdoid bacteriophages. The host range of Stx-encoding bacteriophages is highly variable, and bacteriophage transduction into a wide range of E. coli species and also other related species in the Enterobacteriaceae (e.g., Shigella spp., Citrobacter freundii, and Enterobacter cloacae) has been shown in vitro in planktonic cells (5–7). Moreover, transduction is of importance for the development of emerging pathogenic E. coli, such as, for instance, E. coli O104:H4, which caused a large European outbreak in 2011 (8), and possibly also E. coli O103:H25, which caused an outbreak in Norway in 2006 (9).
Transfer of stx genes by temperate bacteriophages has been reported to take place in the gastrointestinal tracts of various animals (10–12) and in various food matrices at different temperatures (13, 14). Bacterial biofilms are believed to be the natural way of living for the majority of bacterial species. Within bacterial biofilms, vast numbers of bacterial cells live closely together in sessile microbial communities (15). Consequently, the biofilm environment could be an ideal setting for phage-mediated stx gene transfer. Gene transfer by plasmid conjugation and DNA transformation within biofilms has been reported previously (16). The use of lytic bacteriophages on biofilm as an antibacterial strategy has also been described (17, 18), but to the best of our knowledge, incorporation of bacteriophage-carried genes into the bacterial host genome through lysogeny has not previously been shown in biofilms.
The ability to acquire and incorporate foreign DNA through horizontal gene transfer is an important driver of bacterial evolution, including the spread of antibiotic resistance genes and virulence genes (19). Dissemination of Stx-encoding bacteriophages is the most likely mechanism for the emergence of new STEC serotypes. In the present study, we showed that phage-mediated stx2 gene transfer can occur within biofilms.
MATERIALS AND METHODS
Bacteriophage and bacterial strains.
An Stx2-encoding bacteriophage, ϕ731 (Δstx2::cat) (hereafter called ϕ731), in which a chloramphenicol resistance gene (chloramphenicol acetyltransferase; cat) has been inserted into stx2, was used in the experiments. The original bacteriophage was carried by an E. coli O103:H25 isolate from a Norwegian HUS patient (9), but the bacteriophage construct, ϕ731, was carried by E. coli DH5α. As the latter did not produce biofilms in our system, the bacteriophage was transduced into the host E. coli C600, as described below, resulting in E. coli C600:ϕ731, which was then used as the donor strain in the experiments. An eae-positive, stx-negative E. coli O103:H25 strain (2006-22-1199-51-2, an isolate from sheep), hereafter called E. coli O103:H25 1199 (20), was used as the recipient strain in the transduction studies. Characteristics of all the strains are listed in Table 1.
Table 1.
Strain characteristics
| Strain | Description | Serotype | Characteristics | Morphology on CHROMagar O157 | Hemolytic activity | MICa (mg/liter) | Biofilm in microtiter plates (OD595) at 20°C/37°C | Source |
|---|---|---|---|---|---|---|---|---|
| DH5α:ϕ731 | Used for production of donor strain | Δstx2::cat cat+; lacking eae | NDb | ND | ND | 0.004/0.002e | NSVSd | |
| C600 | Host strain | lacking stx, eae, and cat | Blue colonies | Yes | 4 | 1.780/0.101 | NSVS | |
| C600:ϕ731 | Donor strain | Δstx2::cat, lacking eae, cat+ | Blue colonies | Yes | 256 | 2.030/0.237 | This study | |
| 2006-22-1199-51-2 | Recipient strain | O103:H25 | eae+; lacking stx and cat | Purple colonies | No | 2 | 0.097/0.022 | National survey of E. coli in sheep, Norwegian Veterinary Institute |
| 2006-22-1199-51-2:ϕ731 | Transductant | O103:H25 | Δstx2::cat eae+ cat+ | Purple colonies | No | 256c | ND | This study |
MIC for chloramphenicol sensitivity tested by using E-tests (BioMérieux, Marcy I'Etoile, France).
ND, not done.
A selection of transductants were screened for MIC for chloramphenicol sensitivity.
NSVS, Norwegian School of Veterinary Science.
Results for DH5α without bacteriophage.
All strains were stored at −80°C in brain heart infusion broth (BHI; Difco, BD, Franklin Lakes, NJ) supplemented with 15% glycerin (Merck KGaA, Darmstadt, Germany) and recovered on bovine blood agar at 37°C overnight. The bacterial cultures were then transferred to Luria-Bertani broth (LB; Merck KGaA) and incubated statically overnight at 37°C. LB without NaCl (containing Bacto tryptone [10 g/liter] and yeast extract [5 g/liter]) was used as the growth medium in the biofilm assays.
Construction of donor strain E. coli C600:ϕ731.
A stable donor strain, E. coli DH5α:ϕ731 was prepared as previously described (21). First, E. coli DH5α:ϕ731 was grown in 30 ml LB broth with 5 mM CaCl2 to the exponential growth phase (optical density at 600 nm [OD600], 0.3 to 0.5) and then induced with mitomycin C (0.5 μg/μl) (Sigma-Aldrich, St. Louis, MO) and incubated overnight for the production of bacteriophage particles, as described by Muniesa et al. (22). The lysed culture was subsequently centrifuged at 3,000 × g for 10 min and then filtrated through a 0.2-μm Minisart Plus syringe filter (Sartorius Stedim Biotech S.A., Aubagne Cedex, France), giving a filtrate with ϕ731 bacteriophages. The presence of bacteriophages in the filtrate was confirmed by detection of plaques after plating on LB soft agar (5 mM CaCl2) containing E. coli C600.
Ten fold dilutions of the bacteriophage filtrate were prepared, and 100 μl of each dilution was added to 900 μl of a culture of E. coli C600. The bacteria were grown to the exponential phase (OD600, 0.3 to 0.5) in LB broth and incubated at 37°C for 30 min. LB soft agar with 5 mM CaCl2 was mixed with the bacteriophage filtrate and the E. coli C600 culture, plated on LB agar with 5 mM CaCl2 and 25 mg/liter chloramphenicol, and incubated overnight at 37°C. Colonies growng on LB with chloramphenicol (25 mg/liter) were the donor strain containing ϕ731. The inserted construct in E. coli C600:ϕ731 was verified as prophage ϕ731 by the presence of Δstx2::cat and increased chloramphenicol resistance compared to E. coli C600 (Table 1) as described for presumptive transductants below.
Host susceptibility of recipient strain to bacteriophage ϕ731 and transduction experiment with planktonic cells.
Host susceptibility of the recipient strain E. coli O103:H25 1199 to bacteriophage ϕ731 was confirmed by the observation of plaque formation after plating of the bacteriophage filtrate onto LB soft agar (5 mM CaCl2) containing the recipient strain. For the transduction experiment with planktonic cells, overnight cultures of the donor E. coli C600:ϕ731 (1%) and the recipient strain E. coli O103:H25 1199 (1%) were mixed in 30 ml LB without NaCl. Mixed cultures were incubated at 37°C and 20°C for 8 days under static conditions. Every 24 ± 2 h, 10-fold dilutions were plated on CHROMagar O157 (CHROMagar Microbiology, Paris, France) containing chloramphenicol (25 mg/liter) to detect transductants. Colonies that were purple in color were considered presumptive transductants. Two presumptive transductants from each experiment at both temperatures were verified as nonhemolytic E. coli strains and tested serologically with E. coli O103 antiserum for live cultures (Statens Serum Institut, Hillerød, Denmark).
Biofilm formation ability.
Biofilm-forming abilities were tested in microtiter plates (Nunc A/S, Roskilde, Denmark) using a crystal violet binding assay, as previously described by Vestby et al. (23), at 37°C and 20°C with 3 days of incubation. Optical density, indicating the amount of biofilm produced, was measured at 595 nm.
Determination of chloramphenicol concentration for transduction growth medium.
To decide which concentration of chloramphenicol to use in the growth medium during the transduction experiments, the effect of 0 mg, 25 mg, and 50 mg chloramphenicol per liter of growth medium on bacteria in biofilms was tested. Biofilms of the recipient strain were grown on glass slides as described below for the transduction experiment. After 48 ± 2 h, planktonic cells were washed off and the glass slides with the biofilm was transferred to sterile tubes with the growth medium, LB broth without NaCl, containing the different concentrations of chloramphenicol. The biofilms were harvested every 24 ± 2 h up to 6 days at 20°C, as described for the transduction experiment below. The numbers of living cells in the biofilm and the growth medium were determined by plating 10-fold dilutions on bovine blood agar.
Transduction experiments within biofilms. (i) Biofilm formation and addition of donor strain.
An overnight culture of the recipient strain was inoculated (10 μl) into sterile centrifuge tubes (Greiner Bio-One GmbH, Frickenhausen, Germany). An autoclaved glass slide (76 by 26 mm; Gerhard Menzel GmbH, Braunschweig, Germany) was placed in each tube. The glass slides were partially submerged in the broth, as the strains used in the experiments mainly produced biofilms at the liquid-air interface. The tubes were incubated for 48 ± 2 h at 37°C or 20°C to allow the formation of biofilms on the glass slides. After incubation, the glass slides were removed from the broth and planktonic cells were gently washed off using sterile physiological salt water. The slides were then transferred to new tubes containing 10 ml LB broth without NaCl containing chloramphenicol (50 mg/liter). Ten microliters of an overnight culture of the donor strain was added to each tube before further incubation at the chosen temperature for up to 8 days. Initially, the experiments were performed in parallel under both static and shaking (100 rpm) conditions. However, incubation with shaking was found to have no effect on viable biofilm counts, and further experiments were performed under static conditions only.
(ii) Harvesting and enumeration of E. coli within the biofilm.
Biofilm cells were harvested every 24 ± 2 h for the first round of experiments at 20°C and 37°C and then every 48 ± 2 h for the repeats. The glass slides were removed from the tubes, and planktonic cells were gently washed off the slides in sterile physiological salt water. The biofilm was thoroughly scraped off using a sterile cell scraper (BD Falcon, Bedford, MA) and transferred to a reagent tube containing 4 ml sterile saline and about 30 (3-mm-diameter) sterile glass beads. Biofilms were disrupted by vortexing at maximum speed for 40 s. Tenfold dilutions were plated in parallel on CHROMagar O157 medium containing chloramphenicol (25 mg/liter) and bovine blood agar. The CHROMagar O157 medium with chloramphenicol was used to enumerate chloramphenicol-resistant cells of donor E. coli C600:ϕ731 (blue colonies) and transductant E. coli O103:H25 1199:ϕ731 (purple colonies). The bovine blood agar was used for enumeration of cells of E. coli C600 (hemolytic donors) and E. coli O103:H25 (nonhemolytic recipients and transductants) and determination of total bacterial growth. Colonies from the blood agar were tested serologically with E. coli O103 antiserum for live cultures (Statens Serum Institut, Hillerød, Denmark) to confirm that the morphology agreed with what was expected for the serogroup. Each experiment was performed at least three times on separate days, and the results are given as means of all experiments.
Confirmation, stability, and characterization of presumptive transductants.
Five presumptive transductants from each experiment were verified by PCR for the Δstx2::cat construct by using the primers rho and Cm-3 (Table 2), as previously described by Serra-Moreno et al. (24). The donor strain was used as a positive control. The stability of these transductants was verified as follows. The colonies were replated twice on new CHROMagar O157 plates with chloramphenicol (25 mg/liter), followed by PCR to detect the inserted construct. The bacteriophage insertion site was investigated by PCR using the primers Int933W-rev3 and wrbAEDL933-F (Table 2), as previously described by Sekse et al. (9). The insertion site was further confirmed by sequencing of two transductants using the same primers.
Table 2.
Oligonucleotides used in PCR and sequencing analyses
RESULTS
Transduction experiments with planktonic cells.
In transduction experiments where both donor and recipient cells were planktonic, transductants were detected after 24 h at both 37°C and 20°C.
Biofilm-forming abilities.
The biofilm-forming abilities of the recipient and donor strains were investigated on polystyrene in microtiter plates. The OD595 of the recipient strain, E. coli O103:H25 1199, was 0.022 at 37°C and 0.097 at 20°C, indicating a low level of biofilm production (Table 1). The donor strain, E. coli C600:ϕ731, displayed a higher level of biofilm production at both temperatures, i.e., an OD595 of 0.237 at 37°C and an OD595 of 2.030 at 20°C.
Determination of chloramphenicol concentration for transduction growth medium.
With chloramphenicol at a concentration of 25 mg/liter in the biofilm growth medium, planktonic cells of the recipient strain were detected in the medium throughout the experiment. At a chloramphenicol concentration of 50 mg/liter, no planktonic recipient cells were detected in the medium. At this concentration, a small reduction in the number of recipient cells within the biofilm was observed during the first 2 days. After that, the number of recipients within the biofilm remained the same. In addition, this concentration did not affect planktonic growth of the donor cells. Consequently, a chloramphenicol concentration of 50 mg/liter was used in the growth medium during the transduction experiments.
Transduction within biofilm. (i) Transduction within the biofilm at 37°C.
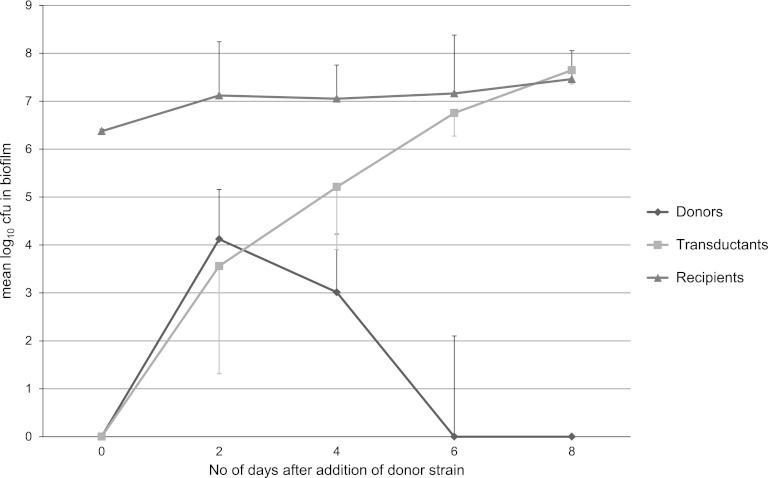
After the initial 2 days of incubation at 37°C, a visible biofilm of the recipient strain E. coli O103:H25 1199 could be seen at the air-liquid interface on both sides of the glass slides. Two days after addition of the donor strain E. coli C600:ϕ731, the amount of donor cells within the biofilm reached a peak before rapidly decreasing. After 6 days, donor cells were no longer detected within the biofilm. Presumptive transductants were found within the biofilm after 2 days. Their number increased steadily throughout the experiment, reaching the same level as the recipient strain at the end of the experiment. Planktonic donor cells and presumptive transductants, but no recipient cells, were detected in the biofilm growth medium. The changes in the components of the biofilm during the experimental period are shown in Fig. 1.
Fig 1.
Donors, recipients, and transductants in the biofilm at 37°C, shown as mean log10 CFU throughout the experiment period. The bars show standard deviations.
(ii) Transduction within the biofilm at 20°C.
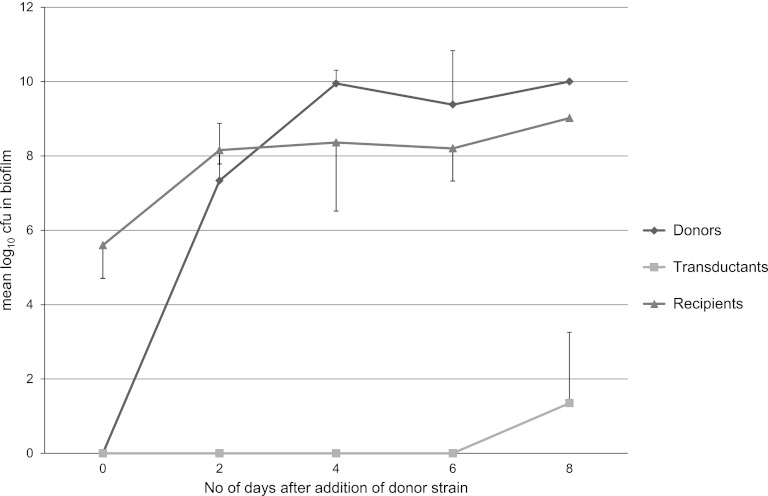
At 20°C, a visible biofilm of the recipient strain, E. coli O103:H25 1199, could be observed after the initial 2 days of incubation. After introduction of the donor strain, the number of donor cells in the biofilm rapidly increased for the first 6 days until it reached approximately 10 times the number of recipient cells. Presumptive transductants were only observed in the biofilm in two of the three experiments, and only on the 8th day of the experiment period. In these two experiments, presumptive transductants were also detected in the biofilm growth medium. In all three experiments, planktonic donor cells, but no recipient cells, were detected in the biofilm growth medium. The changes in the components of the biofilm during the experimental period are shown in Fig. 2.
Fig 2.
Donors, recipients, and transductants in the biofilm at 20°C, shown as mean log10 CFU throughout the experiment period. The bars show standard deviations.
Confirmation, stability, and characterization of presumptive transductants.
Using colony morphology, ability to grow on CHROMagar O157 with chloramphenicol, O103 serotyping, and PCR for the inserted construct, presumptive transductants were confirmed as E. coli O103:H25 1199:ϕ731 in all positive experiments at both temperatures. Furthermore, the stability of the lysogens was confirmed. Bacteriophage ϕ731 used insertion site wrbA in all tested transductants. Two of the isolates were verified by sequencing after screening for a PCR product of the correct size.
DISCUSSION
To the authors' knowledge, this is the first study to show lateral gene transfer in biofilms mediated by a termperate bacteriophage. The experiments were performed using E. coli donor cells containing an Stx2 bacteriophage (Δstx2::cat) and E. coli recipient cells. After the donor was introduced to a biofilm formed by the recipient, we identified transductants with an intact functional bacteriophage within the biofilm at both 20°C and 37°C. The addition of chloramphenicol to the growth medium inhibited survival of planktonic recipient cells, as confirmed in all the experiments by the fact that planktonic cells of the recipient strain were never detected in the growth medium. Thus, the transduction must have taken place within the biofilm.
Differences in transduction events, as well as in the composition of the biofilms at the end of the experiments, could be observed for 20°C and 37°C. At both temperatures, the donor strain rapidly entered the biofilm. However, after 8 days, recipients and transductants dominated the biofilm at 37°C, whereas the donor was the dominant cell type at 20°C. We hypothesize that donors that entered the biofilm at 37°C were lysed, releasing the bacteriophage that infected recipients within the biofilm. In addition, lytic events may also have occurred outside the biofilm, releasing free bacteriophages that entered the biofilm. However, regardless of where the release of bacteriophages took place, infection of recipient cells must have occurred within the biofilm. Furthermore, because transductants were resistant to chloramphenicol, they could move freely between the biofilm and the growth medium and multiply in both places. Some transductants could possibly also act as donors themselves, contributing to increased dissemination of the bacteriophage. All these events would contribute to the high number of transductants detected within the biofilm.
At 20°C, few transductants were observed, and only at the end of the experiment. The number of donors was 10-fold higher than the number of recipients. Although the two strains displayed the same planktonic growth rate in the biofilm medium (results not shown), the donor strain was a much better biofilm producer than the recipient in our biofilm assay at this temperature. This difference could also be observed visually when the two strains produced biofilms on separate glass slides under the same conditions (results not shown). This may be a reason for the high number of donor strain cells within the biofilm at the end of the experiment. Whether the donor/recipient ratio had an impact on the low transduction rate observed in the the biofilm is not known. However, earlier studies indicate that a 10:1 donor/recipient ratio does not influence the transduction rate in other experimental systems (13, 14). It is possible that temperature influenced the rate of transduction. Bacteriophages often show a temperature optimum for both adsorption and replication which reflects the ecological origin of the bacteriophage more closely than the growth optimum of the host bacterium (25). Imamovic et al. (13) reported differing numbers of stx transduction events at 15°C and 22°C. At those temperatures, transduction seemed to be dependent on the bacteriophage, which was not the case at higher temperatures, i.e., 25 to 37°C. Only one bacteriophage was used in the present study, and transduction did occur at 20°C when both donor and recipient cells were planktonic. However, a different result in the biofilm might have been obtained at 20°C with the use of another bacteriophage.
A nationwide outbreak of illness attributable to stx2-positive E. coli O103:H25 occurred in Norway in 2006. The outbreak was caused by a fermented sausage made from contaminated mutton, and stx-negative E. coli O103:H25 was detected both in the fermented sausage and in mutton meat (26). Later, it was shown that stx-negative E. coli O103:H25 is not uncommon among sheep flocks in Norway (5.8% [20]), but stx-positive E. coli O103:H25 has not been detected. Even though the possibility that a small undetected reservoir of stx-positive E. coli O103:H25 may exist in Norwegian sheep cannot be excluded, the Stx2 bacteriophage may also have been introduced somewhere along the food chain. In the present study, the method used for investigating transduction in biofilms was therefore optimized for E. coli O103:H25 as the recipient strain. The method can easily be modified to suit other strains as long as they have some biochemical differences that will identify them in the plate counting step. Here we used both hemolytic activity on bovine blood agar and different colors of the strains on the chromogenic medium, CHROMagar O157. Furthermore, we used chloramphenicol to make sure that growth of the recipients occurred within the biofilm only. McGannon et al. (27) found that subinhibitory levels of antibiotics targeting transcription, translation, or the cell wall did not increase Stx production, and thereby not the level of bacteriophage induction either. This contrasts with the situation when using antibiotics targeting DNA synthesis. As chloramphenicol inhibits protein synthesis, it is not among the antibiotics that increase toxin production and bacteriophage induction. The use of chloramphenicol should therefore not have an effect on the experimental outcome. We have also performed experiments with an E. coli O103:H2 strain as the recipient, but the growth of this strain on CHROMagar O157 was so similar to the donor that it was difficult to quantify transductants. However, we did observe E. coli O103:H2 transductants in the biofilm (results not shown), indicating that transduction events in the biofilm, as demonstrated here, may also occur for other E. coli serotypes.
In conclusion, this study demonstrated phage-mediated stx2 gene transfer within a biofilm of potentially pathogenic E. coli. This indicates that the biofilm can be an environment for the emergence of new pathogenic E. coli strains.
ACKNOWLEDGMENTS
This study was supported by grant no. 178161/I10 from the Research Council of Norway.
The E. coli strain DH5α:ϕ731 (Δstx2::cat) was kindly provided by the Norwegian School of Veterinary Science. We are grateful to Hannah Joan Jøgensen for language proofing and valuable scientific contributions.
Footnotes
Published ahead of print 26 November 2012
REFERENCES
- 1. Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caprioli A, Morabito S, Brugereb H, Oswald E. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289–311 [DOI] [PubMed] [Google Scholar]
- 3. Law D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microbiol. 88:729–745 [DOI] [PubMed] [Google Scholar]
- 4. Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80:1420–1438 [DOI] [PubMed] [Google Scholar]
- 5. Herold S, Karch H, Schmidt H. 2004. Shiga toxin-encoding bacteriophages—genomes in motion. Int. J. Med. Microbiol. 294:115–121 [DOI] [PubMed] [Google Scholar]
- 6. James CE, Stanley KN, Allison HE, Flint HJ, Stewart CS, Sharp RJ, Saunders JR, McCarthy AJ. 2001. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 67:4335–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmidt H, Bielaszewska M, Karch H. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage phi3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sekse C, Muniesa M, Wasteson Y. 2008. Conserved Stx2 phages from Escherichia coli O103:H25 isolated from patients suffering from hemolytic uremic syndrome. Foodborne Pathog. Dis. 5:801–810 [DOI] [PubMed] [Google Scholar]
- 10. Acheson DW, Reidl J, Zhang X, Keusch GT, Mekalanos JJ, Waldor MK. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sekse C, Solheim H, Urdahl AM, Wasteson Y. 2008. Is lack of susceptible recipients in the intestinal environment the limiting factor for transduction of Shiga toxin-encoding phages? J. Appl. Microbiol. 105:1114–1120 [DOI] [PubMed] [Google Scholar]
- 12. Toth I, Schmidt H, Dow M, Malik A, Oswald E, Nagy B. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69:7242–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imamovic L, Jofre J, Schmidt H, Serra-Moreno R, Muniesa M. 2009. Phage-mediated Shiga toxin 2 gene transfer in food and water. Appl. Environ. Microbiol. 75:1764–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Picozzi C, Volponi G, Vigentini I, Grassi S, Foschino R. 2012. Assessment of transduction of Escherichia coli Stx2-encoding phage in dairy process conditions. Int. J. Food Microbiol. 153:388–394 [DOI] [PubMed] [Google Scholar]
- 15. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 16. Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255–261 [DOI] [PubMed] [Google Scholar]
- 17. Doolittle MM, Cooney JJ, Caldwell DE. 1995. Lytic infection of Escherichia coli biofilms by bacteriophage T4. Can. J. Microbiol. 41:12–18 [DOI] [PubMed] [Google Scholar]
- 18. Kay MK, Erwin TC, McLean RJ, Aron GM. 2011. Bacteriophage ecology in Escherichia coli and Pseudomonas aeruginosa mixed-biofilm communities. Appl. Environ. Microbiol. 77:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelly BG, Vespermann A, Bolton DJ. 2009. Horizontal gene transfer of virulence determinants in selected bacterial foodborne pathogens. Food Chem. Toxicol. 47:969–977 [DOI] [PubMed] [Google Scholar]
- 20. Urdahl AM, Sunde M, Bruheim T, Cudjoe K, Hopp PK. 2009. Kartlegging av Ecoli hos sau—resultater fra prøver samlet inn i 2007. Veterinærinstituttets rapportserie 02-2009. Norwegian Veterinary Institute, Oslo, Norway [Google Scholar]
- 21. Muniesa M, de Simon M, Prats G, Ferrer D, Pañella H, Jofre J. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71:4554–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muniesa M, Serra-Moreno R, Jofre J. 2004. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx genes appeared conserved. Environ. Microbiol. 6:176–725 [DOI] [PubMed] [Google Scholar]
- 23. Vestby LK, Moretro T, Langsrud S, Heir E, Nesse LL. 2009. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet. Res. 5:20 doi:10.1186/1746-6148-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Serra-Moreno R, Acosta S, Hernalsteens JP, Jofre J, Muniesa M. 2006. Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol. Biol. 7:31 doi:10.1186/1471-2199-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fry JC, Day MJ. 1992. Release of genetically engineered and other micro-organisms. Cambridge University, Cambridge, England [Google Scholar]
- 26. Sekse C, O'Sullivan K, Granum PE, Rorvik LM, Wasteson Y, Jorgensen HJ. 2009. An outbreak of Escherichia coli O103:H25—bacteriological investigations and genotyping of isolates from food. Int. J. Food. Microbiol. 133:259–264 [DOI] [PubMed] [Google Scholar]
- 27. McGannon CM, Fuller CA, Weiss AA. 2010. Different classes of antibiotics differentially influence Shiga toxin production. Antimicrob. Agents Chemother. 54:3790–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]