Abstract
Dehalococcoides mccartyi strains are obligate organohalide-respiring bacteria harboring multiple distinct reductive dehalogenase (RDase) genes within their genomes. A major challenge is to identify substrates for the enzymes encoded by these RDase genes. We demonstrate an approach that involves blue native polyacrylamide gel electrophoresis (BN-PAGE) followed by enzyme activity assays with gel slices and subsequent identification of proteins in gel slices using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). RDase expression was investigated in cultures of Dehalococcoides mccartyi strain BAV1 and in the KB-1 consortium growing on chlorinated ethenes and 1,2-dichloroethane. In cultures of strain BAV1, BvcA was the only RDase detected, revealing that this enzyme catalyzes the dechlorination not only of vinyl chloride, but also of all dichloroethene isomers and 1,2-dichloroethane. In cultures of consortium KB-1, five distinct Dehalococcoides RDases and one Geobacter RDase were expressed under the conditions tested. Three of the five RDases included orthologs to the previously identified chlorinated ethene-dechlorinating enzymes VcrA, BvcA, and TceA. This study revealed substrate promiscuity for these three enzymes and provides a path forward to further explore the largely unknown RDase protein family.
INTRODUCTION
Chlorinated ethenes and ethanes are widespread groundwater contaminants (1, 2). A viable approach for the remediation of chlorinated solvent contamination is microbial reductive dechlorination (3–5). Phylogenetically diverse bacteria partially dechlorinate tetrachloroethene (PCE) via trichloroethene (TCE) to cis-1,2-dichloroethene (cis-DCE), including Dehalococcoides, Geobacter, Sulfurospirillum, Dehalobacter, and Desulfitobacterium among others (2). Dehalococcoides mccartyi strains are the only organisms known to dechlorinate cis-DCE and vinyl chloride (VC) to nontoxic ethene (6). Some D. mccartyi strains are also capable of catalyzing the reductive dichloroelimination of 1,2-dichloroethane (1,2-DCA) to ethene and 1,2-dichloropropane to propene (7–9).
Reductive dechlorination of these groundwater pollutants is catalyzed by reductive dehalogenases (RDases). The catalytic unit is encoded by the RDase subunit A gene (rdhA). Over 650 rdhA genes have been identified from fully sequenced genomes based on sequence homology, and of these genes, over 100 are from Dehalococcoides (10, 11). Only a few rdhA genes have been functionally characterized because of difficulties inherent to working with slow-growing anaerobes with low biomass yields, the lack of genetic systems for these organisms, and the inability to successfully express functional RDases heterologously. Partial purification of the Dehalococcoides TceA (12), PceA (13), and VcrA (14) RDases enabled the preliminary characterization of their activity and substrate range, but difficulties in obtaining sufficient biomass hampered biochemical studies. Substrates for the BvcA (15) and MbrA (16) RDases were inferred from transcriptional analysis, although biochemical confirmation is still missing. Adrian et al. (17) identified the first chlorobenzene RDase, CbrA, using a combination of clear native polyacrylamide gel electrophoresis (CN-PAGE), enzyme assays, and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) peptide identification. This approach enabled functional attribution without requiring large amounts of biomass. Here we build upon this approach using blue native PAGE (BN-PAGE) (18), which substantially improved recovery of dechlorinating activity after electrophoresis, resulting in higher sensitivity and enabling analysis of a wider range of substrates.
KB-1 is a Dehalococcoides-containing consortium capable of complete PCE hydrogenolysis to ethene via TCE, cis-DCE, and VC as intermediates, as well as 1,2-DCA dichloroelimination to ethene (19, 20). Metagenome sequencing revealed at least 36 rdhA genes in the KB-1 consortium (10), and multiple rdhA genes were transcribed simultaneously when the culture was grown on different chlorinated solvents as electron acceptors (21, 22). D. mccartyi strain BAV1 is capable of hydrogenolysis of all three dichloroethene (DCE) isomers (cis-DCE, trans-DCE, and 1,1-DCE) and VC, as well as the dichloroelimination of 1,2-DCA to ethene (7); 11 rdhA genes were found in its genome (11). Previous gene expression studies implicated bvcA in the reductive dechlorination of VC to ethene (15); however, the RDases that catalyze other dechlorinating reactions are unknown. Therefore, the BN-PAGE approach was applied to identify the RDases expressed and active in strain BAV1 and in the KB-1 consortium.
MATERIALS AND METHODS
Cultures and growth conditions.
The KB-1 consortium, a KB-1 subculture grown on 1,2-DCA (referred to as the 1,2-DCA KB-1 subculture), and pure cultures of D. mccartyi strain BAV1 (7) were used in this study. KB-1 was enriched from sediment from a contaminated site in Ontario (Canada) and contains multiple D. mccartyi strains (23). KB-1 also contains a PCE- and TCE-dechlorinating Geobacter lovleyi strain KB-1 (24). The KB-1 consortium was routinely maintained with TCE as an electron acceptor and methanol and ethanol as electron donors in a defined mineral salts medium (25). The 1,2-DCA KB-1 subculture was maintained for over 4 years with 1,2-DCA as electron acceptor and methanol as an electron donor. D. mccartyi strain BAV1 was isolated from dechlorinating microcosms established with aquifer material collected at the contaminated Bachman Road site in Oscoda, Michigan (7).
Preparation of crude protein extracts.
Prior to preparing cell-free crude extracts, an aliquot of KB-1 culture was separated into two bottles, and the bottles were flushed with N2-CO2 (80/20, vol/vol) to purge residual chlorinated ethenes and ethene. The purged cultures were incubated for 5 days (starvation phase), and the bottles were then flushed with H2-CO2 (80/20, vol/vol) to provide an electron donor and amended with 30 mg/liter (aqueous concentration) of either TCE (230 μM) (TCE-induced cultures) or VC (480 μM) (VC-induced cultures). One day later, cells from 8- to 10-ml culture samples were pelleted by centrifugation for 6 min at 10,000 rpm at 4°C. For the 1,2-DCA KB-1 subculture, 40 ml of culture was collected during the dechlorination of 1,2-DCA to ethene, and cells were collected by centrifugation. Cell pellets were used immediately or stored at −80°C. D. mccartyi strain BAV1 cultures were grown with either cis-DCE or 1,2-DCA as follows. Replicate vessels containing a mineral salts medium (26) supplemented with 5 mM sodium acetate, ∼30 mg/liter (∼300 μM; aqueous concentration) of cis-DCE or 1,2-DCA, and hydrogen (nominal aqueous concentration of 7.5 mM) as the electron donor were inoculated with strain BAV1 (1.3% vol/vol). When at least 50% of the amended cis-DCE had been dechlorinated to VC (with at least traces of ethene produced as well) or at least 50% of the amended 1,2-DCA had been dechlorinated to ethene, 600 ml BAV1 cell suspension was pelleted by centrifugation for 60 min at 4,000 rpm and 15°C. Cell pellets were stored at −80°C.
In an anoxic chamber, fresh or thawed frozen cell pellets were suspended in 200 to 250 μl of anoxic 1× sample buffer provided with the NativePAGE sample prep kit (Invitrogen, Carlsbad, CA) containing 1% (wt/vol) digitonin. During method optimization, four different detergents were compared, including digitonin, dodecyl-β-d-maltoside, taurodeoxycholate, and Triton X-100. Digitonin was found to be the most effective (27). Suspended cell pellets were combined with approximately ∼200 mg of 75-μm-diameter glass beads, sealed in a 1.5-ml screw-top tube, shaken in a bead beater (FastPrep DNA extractor; Savant Instruments, Holbrook, NY) at an intensity of 4.0 for 10 s, and then placed immediately on ice. To separate solubilized proteins from cell debris, the suspensions were centrifuged at 13,000 × g for 10 min at 4°C. For the 1,2-DCA KB-1 subculture only, the cells were lysed by shaking with glass beads on a horizontal vortex mixer (Scientific Industries Inc., Bohemia, NY) at maximum amplitude for three cycles; each cycle consisted of 2 min of shaking and 1 min of incubation in an ice bath. Supernatants (crude protein extracts), containing solubilized proteins, were transferred to new 1.5-ml Eppendorf tubes. Protein concentrations were determined by the Bradford assay (28) with bovine serum albumin as a standard. Before loading the samples on the BN-polyacrylamide gel, crude protein extracts were amended with 5% (wt/vol) G-250 sample additive from the NativePAGE sample prep kit (Invitrogen) to a final concentration of 0.25% (wt/vol).
BN-PAGE gel electrophoresis and staining.
Electrophoresis was performed in an 11°C room using the NativePAGE Novex Bis-Tris gel system (Invitrogen). The anode and cathode buffers were prepared according to the manufacturer's instructions in the NativePAGE running buffer kit (Invitrogen) and were prechilled to 4°C prior to use. A precast gradient Bis-Tris gel (4 to 16% Bis-Tris; 1.0 mm thick; Invitrogen) was placed in the XCell SureLock minicell, and 5 μl of NativeMark unstained protein standard (Invitrogen) was loaded into one lane to serve as the size standard. Volumes of 20 to 25 μl of crude protein extract, corresponding to about 12 to 30 μg of total protein, were loaded into each of the other lanes of the gel. The remaining crude protein extracts were stored on ice to be used as positive controls in subsequent dechlorination assays. Replicate lanes were prepared for (i) staining to visualize protein bands, (ii) excision of gel slices and elution of proteins for SDS-PAGE, and (iii) excision of gel slices for activity assays. The loaded gel was run successively at 150 V for 60 min, then at 250 V for 30 min, and finally at 300 V for 15 min. For the 1,2-DCA KB-1 subculture only, the BN-PAGE electrophoresis was run for 60 min at 150 V, followed by 45 min at 200 V, while the whole chamber was placed in an ice bath. Once electrophoresis was complete, the lane containing the protein ladder and one lane loaded with the crude protein extract were cut from the rest of the gel using a scalpel and silver stained by the method of Nesterenko et al. (29). For the 1,2-DCA KB-1 subculture only, the staining was performed using the “Fast Coomassie G-250 Staining” protocol from Invitrogen (30). The remainder of the gel was stored in anode buffer at 4° or 11°C during the staining procedure. Stained lanes or gel slices were saved at −20° to 4°C in a solution containing 1% (vol/vol) glacial acetic acid for subsequent LC-MS/MS analysis.
Protein quantification in BN-PAGE gels.
The amounts of protein in bands excised from BN-PAGE gel lanes were estimated by comparing the intensity of the stain to those of standards. In this procedure, two gel lanes, one containing a protein ladder and one containing crude protein extract, were first stained following the “Fast Coomassie G-250 Staining” protocol (Invitrogen) (30) and then destained by incubating overnight while gently shaking in 7% (vol/vol) acetic acid to reduce the Coomassie blue background. Then a digital picture (G:BOX Chemi HR16; Syngene) of the two gel lanes was analyzed using ImageJ (http://rsb.info.nih.gov/ij/) comparing gray values of protein bands in the lane with crude protein extracts with those of lanes containing known amounts of protein in the ladder. The amounts of protein in the bands from the ladder had been previously determined in the same way using a series of bovine serum albumin standards. The protein content in BN-PAGE gel slices was not measured in our initial experiments; it was measured only in a later experiment with KB-1 culture extract to investigate enrichment of RDases during BN-PAGE.
Dechlorination activity assays using gel slices.
To determine the locations of proteins in an unstained gel lane, the corresponding lanes with silver-stained proteins were aligned, and gel slices were excised using a scalpel. Individual gel slices were cut into 1-mm square pieces and were transferred to 2 ml crimp-top glass vials. For a positive control, 10 to 25 μl of the crude protein extract from the same original sample was added to an additional glass vial. Dechlorination activity assays were performed essentially as described previously (31). In an anoxic chamber, the samples from TCE-induced and VC-induced KB-1 cultures were tested for TCE and VC dechlorination in 2.0-ml crimp-top vials amended with 1.0 ml assay buffer that contained 100 mM Tris-HCl (pH 7.4), 2 mM titanium citrate, 2 mM methyl viologen, and 30 to 70 mg/liter (aqueous concentration) of chlorinated compounds. The samples from the 1,2-DCA KB-1 subculture were assayed for PCE, TCE, cis-DCE, trans-1,2-dichloroethene (trans-DCE), VC, and 1,2-DCA dechlorination. The samples from the BAV1 culture were assayed for cis-DCE and 1,2-DCA dechlorination in 0.8-ml vials with 0.4 ml assay buffer that contained 100 mM potassium acetate (pH 5.8), 4 mM titanium citrate, 4 mM methyl viologen, and 25 mg/liter (aqueous concentration) of cis-DCE or 1,2-DCA. The crude protein extract from the BAV1 culture grown on cis-DCE was also assayed for PCE, TCE, all three isomers of DCE, VC, and 1,2-DCA. Crimp-top vials were closed with Teflon-coated septa immediately after assay buffer addition, thoroughly mixed, and stored upside down inside the anoxic chamber for 24 to 48 h prior to headspace analysis.
Analysis of dechlorination products.
To determine the concentrations of chlorinated substrates and their dechlorination products following incubation, 250 μl (for KB-1 assays) and 50 μl (for BAV1 assays) headspace samples were removed from activity assay vials and injected directly into a Chrompack CP-3800 gas chromatograph connected to a flame ionization detector (FID) (Varian, Middelburg, the Netherlands) equipped with a 30-m by 0.53-mm GS-Q column (J&W Scientific, Waldbronn, Germany). The following temperature program was used: 100°C for 1 min, temperature increased 50°C/min to 225°C, and hold at 225°C for 2.5 min. The FID was operated at 250°C, with helium as the carrier gas at an input pressure of 680 hPa. For 1,2-DCA KB-1 subcultures only, 300-μl headspace samples were similarly analyzed by gas chromatography as described previously (32).
SDS-PAGE.
While excising gel slices for dechlorination activity assays, parallel gel slices were also excised from a second unstained lane to elute proteins for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). BN-PAGE gel slices were cut into 1-mm square pieces and transferred to 1.5-ml Eppendorf tubes containing 250 μl of SDS elution buffer (100 mM Tris-HCl [pH 7.0] and 0.1% [wt/vol] SDS). Following 12 to 20 h of shaking at 750 rpm, the solution containing eluted proteins was concentrated to 10 to 15 μl using an Amicon ultracentrifugal filter with a 10-kDa cutoff (Millipore, Billerica, MA) following the manufacturer's instructions. The concentrate was then analyzed by SDS-PAGE, and the gels were silver stained by the method of Nesterenko et al. (29).
LC-MS/MS analysis.
The gel slices of interest from the stained lanes of BN-PAGE or SDS-PAGE gels were destained, and the proteins were reduced with 100 mM dithiothreitol, alkylated with 10 mM iodoacetamide, and digested with trypsin as described previously (17). The mass of peptide fragments was determined by liquid chromatography coupled to tandem mass spectrometry via electrospray ionization (LC-ESI-MS/MS) as described previously (33) and by nanoLC-LTQ Orbitrap MS/MS (34). Peptide fragments were identified using the MS/MS ion search in the Mascot server with previously described parameters (34, 35). For the 1,2-DCA KB-1 subculture only, the LC-MS/MS analysis was performed at the Advanced Protein Technology Center of SickKids' Hospital (Toronto, Canada). After reduction, alkylation, and tryptic digestion of proteins (17), the resulting peptides were loaded onto a 150-μm-inner-diameter (i.d.) precolumn (Magic C18 column; Michrom Biosciences) at 4 μl/min and separated over a 75-μm-i.d. analytical column packed into an emitter tip containing the same packing material. The peptides were eluted over 60 min at 300 nl/min using a 0 to 40% acetonitrile gradient in 0.1% formic acid using an EASY n-LC nanochromatography pump (Proxeon Biosystems, Odense, Denmark). The peptides were eluted into a LTQ linear ion trap mass spectrometer (Thermo-Fisher, San Jose, CA) operated in a data-dependent mode. Six MS/MS scans were obtained per MS cycle. The raw data files were searched using X!Tandem (Beavis Informatics) using a parent ion accuracy of 2 Da and a fragment accuracy of 0.5 Da. A fixed modification of carbamidomethyl cysteine and variable modification of oxidized methionine were included in the search.
Reference databases used for LC-MS/MS analysis.
The mass spectra from samples of the BAV1 culture were searched against proteins from the BAV1 genome (NCBI accession no. NC_009455) (11). The genome of BAV1 has 11 rdhA genes, including one transcriptionally identified vinyl chloride reductase gene referred to as bvcA (15). The mass spectra from samples of KB-1 cultures were searched against two reference databases: (i) a database of all predicted protein sequences from the KB-1 metagenome and (ii) a custom RDase database. The KB-1 metagenome was obtained from shotgun sequencing of the KB-1 culture DNA using Sanger sequencing (10). The assembly and annotation of the KB-1 metagenome were performed using the in-house pipelines of the Department of Energy (DoE) Joint Genome Institute (JGI) (Walnut Creek, CA) (10) and can be accessed through the IMG/M platform (http://img.jgi.doe.gov/cgi-bin/m/main.cgi) with the IMG taxon object identification (ID) 2013843002. A clone library of RDase genes generated by Waller et al. (22) identified 15 partial rdhA sequences (KB1_RdhA1 to KB1_RdhA14) in KB-1 cultures. The assembly and annotation of the KB-1 metagenome identified an additional 21 rdhA sequences, 18 of which are complete genes, including a Geobacter rdhA gene (KB1_GeobRD) (24). Because this collection of 36 rdhA sequences from the KB-1 culture was from metagenomic data, it may not cover all rdhA sequences in KB-1 cultures. Therefore, a custom curated protein database containing 182 RdhA sequences (including those from KB-1) was created. These additional RdhA sequences were mined from NCBI and JGI public sequence databases from the genomes of Dehalococcoides and other organisms (see Table S3 in the supplemental material).
RESULTS
RDase expression in BAV1 cultures.
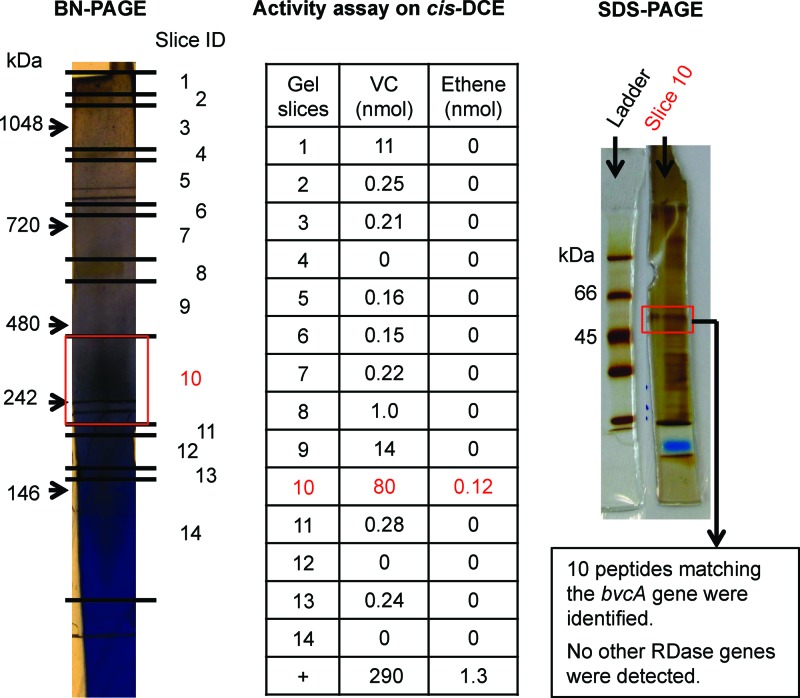
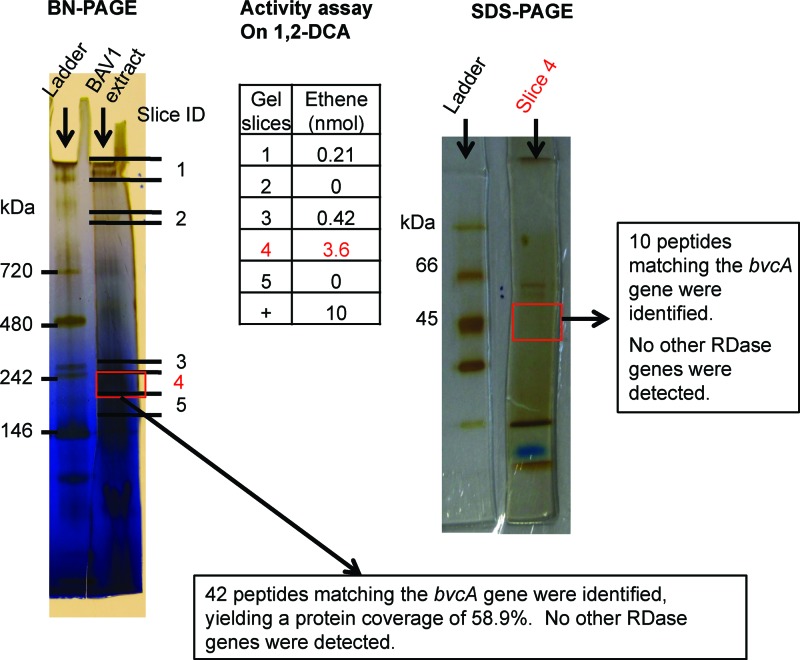
Crude protein extract of Dehalococcoides mccartyi strain BAV1 cultures grown on cis-DCE reductively dechlorinated TCE, cis-DCE, trans-DCE, 1,1-DCE, and VC, but not PCE. 1,2-DCA was transformed by dichloroelimination to ethene (see Table S1 in the supplemental material). The highest dechlorinating activities were observed for the three DCE isomers. These activities are similar to those observed in growing cultures, although the VC dechlorination rate exceeded those of the DCE isomers (7). However, rates observed in growing cultures and in in vitro enzyme assays are not directly comparable because methyl viologen is used as an artificial electron donor in the in vitro assays. After BN-PAGE separation of strain BAV1 crude extracts, cis-DCE dechlorinating activity was mostly constrained to a gel segment around the 242-kDa standard band (Fig. 1). Analysis of proteins eluted from this gel segment by SDS-PAGE revealed a protein band with a molecular mass between 45 and 66 kDa, the molecular mass range of RDases (Fig. 1). LC-MS/MS analysis of the SDS-PAGE gel slice revealed the presence of only one RDase, BvcA (Fig. 1). With crude protein extracts from the BAV1 culture grown on 1,2-DCA instead of cis-DCE, the 1,2-DCA dechlorinating activity was again mostly constrained to a narrow gel slice (3 to 4 mm) around 242 kDa on the BN-PAGE gel (Fig. 2; see Fig. S1 in the supplemental material). Again, LC-MS/MS analysis of the gel slices from both BN-PAGE and SDS-PAGE revealed the presence of only one RDase, BvcA (Fig. 2). Although non-RDase proteins were also detected in the gel slice, BvcA showed the highest coverage (59%) and number of peptide hits (42 hits); BvcA was therefore the most abundant protein in the BN-PAGE gel slice with the highest dechlorinating activity (Fig. 2 and Table 1).
Fig 1.
RDase expression in a culture of the BAV1 strain grown on cis-DCE. Shown are the results from BN-PAGE indicating the positions of gel slices, amounts of dechlorination product(s) obtained with the different gel slices in activity tests, an SDS-polyacrylamide gel of proteins in slice 10, and the mass spectrometric identification of the band at ∼50 kDa. A positive control (+) for dechlorination was assayed with 10 μl crude protein extract, instead of protein from a gel slice.
Fig 2.
RDase expression in a BAV1 culture grown on 1,2-DCA. Shown are the results from BN-PAGE indicating the positions of gel slices, amounts of dechlorination product(s) obtained with the different gel slices in activity tests, an SDS-polyacrylamide gel of proteins in slice 4, and the mass spectrometric identification of the band at ∼50 kDa. A positive control (+) for dechlorination was assayed with 10 μl crude protein extract, instead of protein from a gel slice.
Table 1.
D. mccartyi strain BAV1 proteins identified in the BN-PAGE gel region of enriched dechlorinating activitya
| Protein | NCBI GI no. | No. of peptide hits | Coverage (%) |
|---|---|---|---|
| Reductive dehalogenase, BvcAb | 48995937 | 42 | 59 |
| Nicotinate nucleotide dimethylbenzimidazole phosphoribosyltransferase | 147669271 | 20 | 57 |
| Chaperone protein DnaK | 147669847 | 19 | 33 |
| Chaperonin Cpn10 | 147669874 | 13 | 65 |
| DNA polymerase III, beta subunit | 147669676 | 7 | 21 |
| Transketolase | 147669261 | 7 | 13 |
| Pyruvate ferredoxin oxidoreductase, alpha subunit | 147669303 | 6 | 18 |
| Formate dehydrogenase, alpha subunit | 147668816 | 6 | 9 |
| General substrate transporter | 147669750 | 5 | 11 |
| Chaperonin GroEL | 147669875 | 4 | 9 |
| Hypothetical protein | 147668976 | 3 | 8 |
| Periplasmic binding protein | 147669265 | 2 | 7 |
| GrpE protein | 147669848 | 2 | 14 |
| AIR synthase-like proteinc | 147669977 | 2 | 7 |
| Hypothetical protein (putative S-layer protein) | 147669853 | 2 | 8 |
The BAV1 culture was grown on 1,2-DCA prior to analysis.
The reductive dehalogenase BvcA is shown in boldface type because it is most abundant.
AIR, 5′-aminoimidizole ribonucleotide.
RDase expression in KB-1-derived cultures.
The crude protein extract from KB-1 cultures dechlorinated PCE, TCE, cis-DCE, trans-DCE, VC, and 1,2-DCA in methyl viologen-amended activity tests (see Table S1 in the supplemental material). The crude protein extracts from the 1,2-DCA KB-1 subculture dechlorinated the same substrates (Table S1), despite having been maintained on 1,2-DCA as a growth substrate for 4 years. Dechlorination rates reported in Table S1 were measured over a 24-hour period and were normalized to protein concentration to obtain a rough estimate of the relative specific activity of the enzymes in the crude protein extracts on different substrates. These estimates, ranging from about 4 to 16 nmol · min−1 · mg protein−1 depending on the substrate, were lower than the specific activities determined previously for KB-1, which ranged between 50 and 90 nmol · min−1 · mg protein−1 (36). However, in the previous study, activity was determined after 2 to 4 h (instead of 24 h), and cell extracts were prepared by sonication and without detergents (36).
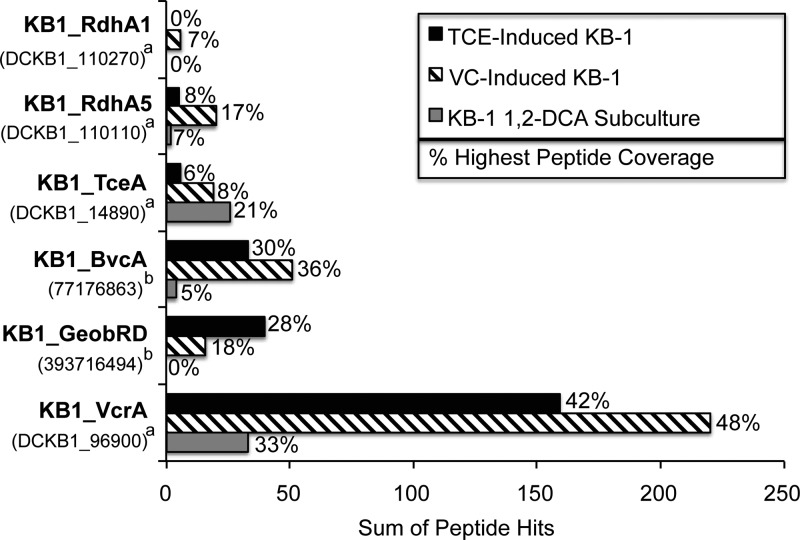
In BN-PAGE gels using crude protein extracts from the three KB-1 cultures (VC-induced KB-1 culture, TCE-induced KB-1 culture, and 1,2-DCA KB-1 subculture), the dechlorinating activity distribution along the gel lanes was essentially identical to that seen with BAV1 cultures. Again, dechlorinating activity was mainly constrained to a narrow region around 242 kDa (Fig. 3; see Fig. S2 and Fig. S3 in the supplemental material). LC-MS/MS analysis was focused on the gel slices with higher activity in order to identify the RDases in the three cultures. In total, only six distinct RDases out of 36 candidate sequences in KB-1 were expressed (Fig. 4). KB1_VcrA, KB1_BvcA, KB1_TceA, and KB1_RdhA5 were expressed in all three cultures (Fig. 4). KB1_VcrA was the most abundant RDase in all three cultures based on the number of peptide hits and coverage (Fig. 4). KB1_RdhA1 was found only in the VC-induced KB-1 culture. KB1_GeobRD, which belongs to Geobacter lovleyi strain KB-1, was found in the TCE-induced culture and to a lesser extent in the VC-induced KB-1 culture, but not in the KB-1 1,2-DCA subculture (Fig. 4).
Fig 3.
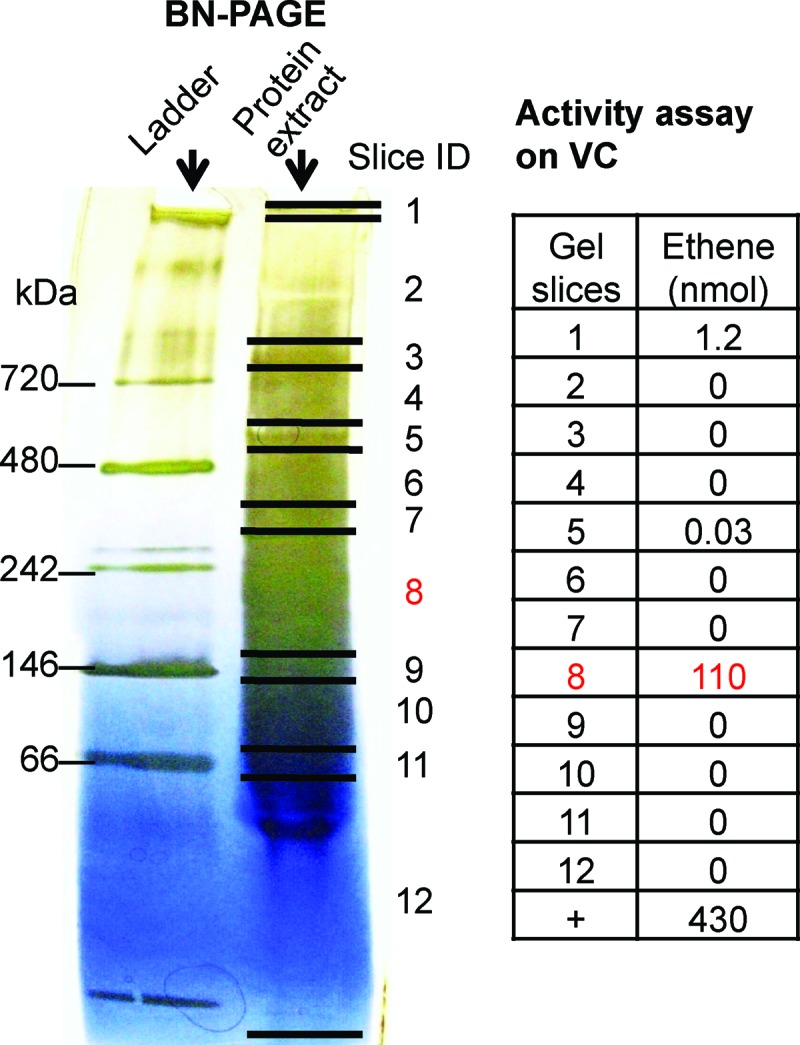
BN-PAGE of protein extracts of the VC-induced KB-1 culture and VC dechlorinating activity in the bands. Shown is the BN-PAGE gel indicating the positions of gel slices. Activity toward VC was measured as nanomoles of ethene produced. A positive control (+) for dechlorination was assayed with 25 μl crude protein extract, instead of protein from a gel slice.
Fig 4.
Peptide hits and coverage of the RDases identified from the three KB-1 related cultures. The MS spectra were searched against a custom RDase database (see Table S4 in the supplemental material). For each culture, three consecutive gel slices covering the active region were subjected to LC-MS/MS separately. Peptide hits were reported by summing up the peptide hits for each protein from the three gel slices. Peptide coverage (shown as a percentage) was reported using the highest coverage seen in the three slices. The values for the number of peptide hits and percent coverage for the three gel slices individually are provided in Table S1 in the supplemental material. The superscript letters a to c indicate the following: a, IMG gene locus tag; b, NCBI GI number.
Investigating RDase enrichment during BN-PAGE.
To determine whether specific activity in the gel slices was higher than that in the crude extract, it was necessary to obtain an estimate of protein content in gel slices. The protein content in gel slices from a KB-1 culture extract was quantified from gel images as described in Materials and Methods. Consistent with all prior experiments, the majority of the activity on TCE, VC, and 1,2-DCA was found in a gel segment around 242 kDa (slice 3 in Fig. S4 in the supplemental material). The total amount of proteins in slice 3 was estimated to be 0.18 μg resulting in specific activities of 77, 90, and 6.9 nmol min−1 mg−1 for VC, TCE, and 1,2-DCA, respectively. The specific activity in slice 3 was 5 to 15 times higher than that in the crude protein extract (Fig. S4), demonstrating enrichment of RDases by BN-PAGE.
Identification of other proteins in active gel slices.
Many non-RDase proteins were identified in selected gel slices of enriched activity when the MS spectra were searched against all proteins and protein fragments from the KB-1 metagenome (see Table S3 in the supplemental material) or proteins from the BAV1 genome (Table 1). In all cultures except the 1,2-DCA KB-1 subculture, the protein with the most peptide hits identified in gel slices showing activity was an RDase. In the 1,2-DCA KB-1 subculture, chaperonin GroEL (Dehalococcoides, IMG gene locus tag DCKB1_11070) and chaperone protein DnaK (Dehalococcoides, IMG gene locus tag DCKB1_174500) were the most abundant proteins, followed by an RDase. GroEL and DnaK were also abundant in all other samples (Table 1 and Table S3). Other non-RDase proteins identified with high peptide hits included the Dehalococcoides α-subunit of pyruvate:ferredoxin oxidoreductase (IMG gene locus tag DCKB1_41710). In samples from the KB-1 cultures, the majority of proteins belonged to Dehalococcoides, consistent with the dominance of Dehalococcoides in the KB-1 consortium. While Geobacter proteins were found in the TCE-induced and VC-induced KB-1 cultures, no Geobacter proteins were detected in the 1,2-DCA KB-1 subculture, consistent with the absence of Geobacter in this subculture.
DISCUSSION
Functional characterization of RDases.
BvcA had previously been associated with VC dechlorination by transcriptional analysis (15). BvcA was the only RDase detected in active BN-PAGE gel segments obtained from electrophoresis of crude protein extracts from the BAV1 culture grown with either cis-DCE or 1,2-DCA (Fig. 1 and 2). It follows that BvcA must then also dechlorinate cis-DCE and 1,2-DCA. Because BN-PAGE does not separate RDases from each other as shown with data for the KB-1 cultures, BvcA, the only RDase detected in the gel segment showing maximum dechlorinating activity, is likely also the only RDase expressed by strain BAV1 under these conditions. BvcA must then be responsible for all dechlorinating activities detected in the crude protein extracts of the BAV1 culture grown on cis-DCE (see Table S1 in the supplemental material). Therefore, in addition to cis-DCE, 1,2-DCA, and VC, the substrates of BvcA include 1,1-DCE, trans-DCE, and TCE, but not PCE. Similar substrate ranges were observed for two other characterized Dehalococcoides RDases, TceA (12) and VcrA (14). These results are consistent with the fact that strain BAV1 grows on all three DCE isomers, VC and 1,2-DCA, but not TCE or PCE. TCE and possibly PCE are however cometabolized in the presence of growth-supporting electron acceptors (7).
In total, only six distinct RDases were detected in the KB-1 cultures (Fig. 4) with three of these orthologous to the characterized Dehalococcoides RDases VcrA, TceA, and BvcA. KB1_VcrA, the most abundant RDase expressed in all KB-1 cultures tested (Fig. 4 and Table 2), shares 97.1% amino acid identity (15 differences/519 amino acids [aa]) with VcrA from D. mccartyi strain VS, which was shown to dechlorinate VC, all three DCE isomers, and TCE, the latter much more slowly (14). KB1_VcrA was also the most abundant in the 1,2-DCA KB-1 subculture, suggesting that it might also dechlorinate 1,2-DCA (Fig. 4 and Table 2). KB1_TceA shares 97.3% amino acid identity (15 differences/560 aa) with TceA from D. mccartyi strain 195; TceA is known to dechlorinate TCE, cis-DCE, 1,1-DCE, trans-DCE (slowly), VC (extremely slowly), and 1,2-DCA, but not PCE (12). KB1_BvcA shares 99.0% amino acid identity (5 differences/516 aa) with BvcA from D. mccartyi strain BAV1; the RDase in this study was found to dechlorinate all three DCE isomers, VC, 1,2-DCA, and TCE, but not PCE. In summary, VcrA, BvcA, and TceA dechlorinate similar substrates with some differences in substrate preferences. However, they are manifestly distinct in pairwise comparisons, sharing 37.2% (TceA and VcrA), 41.2% (TceA and BvcA), and 39.3% (BvcA and VcrA) amino acid identity.
Table 2.
Summary of BN-PAGE analyses for the four different cultures
| Culture | Culture condition before protein extraction | Chlorinated substrate tested on gel slices | Dechlorinating activity detected? | RDases identified in active gel slicesa |
|---|---|---|---|---|
| KB-1 maintained on TCE | Starved for 5 days and then amended with TCE and H2 | TCE | Yes | KB1_VcrA* |
| cis-DCE | Yes | KB1_BvcA* | ||
| trans-DCE | Yes | KB1_GeobRD* | ||
| VC | Yes | KB1_RdhA5 | ||
| 1,2-DCA | Yes | KB1_TceA | ||
| PCE | Yes | |||
| KB-1 maintained on TCE (same culture as above) | Starved for 5 days and then amended with VC and H2 | TCE | Yes | KB1_VcrA* |
| cis-DCE | Yes | KB1_BvcA* | ||
| trans-DCE | Yes | KB1_GeobRD | ||
| VC | Yes | KB1_RdhA5 | ||
| 1,2-DCA | Yes | KB1_TceA | ||
| PCE | Yes | KB1_RdhA1 | ||
| 1,2-DCA KB-1 subculture | Grown exclusively on 1,2-DCA and methanol for >4 years | TCE | Yes | KB1_VcrA* |
| cis-DCE | Yes | KB1_TceA* | ||
| trans-DCE | Yes | KB1_BvcA | ||
| VC | Yes | KB1_RdhA5 | ||
| 1,2-DCA | Yes | |||
| PCE | Yes | |||
| D. mccartyi strain BAV1 | Grown on cis-DCE | TCE | Yes | BvcA |
| cis-DCE | Yes | |||
| trans-DCE | Yes | |||
| 1,1-DCE | Yes | |||
| VC | Yes | |||
| 1,2-DCA | Yes | |||
| PCE | No | |||
| D. mccartyi strain BAV1 | Grown on 1,2-DCA | 1,2-DCA | Yes | BvcA |
The identified RDases are listed in order of decreasing peptide hit counts. Accession numbers are provided in Fig. 4. The dominant RDases are indicated by an asterisk after the RDase name.
KB1_RdhA5 was also detected in all three KB-1 cultures. In a previous transcriptional study (22), the gene encoding KB1_RdhA5 was transcribed with all investigated substrates (TCE, cis-DCE, VC, and 1,2-DCA), corresponding well with the results of this study. In cultures of D. mccartyi strain 195, DET1545, the ortholog of KB1_RdhA5, was the most upregulated at low chlorinated ethene concentrations or respiration rates (37). The low and possibly constitutive expression of this RDase under all conditions complicate a functional assignment, and thus, the substrate(s) of this RDase remains unknown.
KB1_GeobRD shares ∼95% amino acid identity to the two nearly identical RDases (NCBI GI numbers 189425924 and 189425926, differing from each other by only 4/515 aa) of Geobacter lovleyi strain SZ. Both G. lovleyi SZ and G. lovleyi strain KB-1 dechlorinate PCE and TCE to cis-DCE (24) using acetate as the electron donor. Since there are no other RDase sequences in these strains, KB1_GeobRD likely dechlorinates both PCE and TCE, which is consistent with our observations: KB1_GeobRD was relatively more abundant in the TCE-induced KB-1 culture, was less abundant in the VC-induced KB-1 culture, which was normally grown on TCE but amended with VC just prior to analysis, and was not detected in the 1,2-DCA KB-1 subculture (Fig. 4 and Table 2). These results are also consistent with previous microarray data showing the transcription of KB1_GeobRD in TCE- but not in VC-amended KB-1 cultures (21).
Features of BN-PAGE.
Native PAGE is an electrophoresis technique that separates proteins while preserving their native states, enabling subsequent protein identification using activity assays. Previously, Adrian et al. (17) reported the use of clear native PAGE to identify a chlorobenzene RDase (CbrA) from D. mccartyi strain CBDB1. However, when this technique was applied to protein extracts from the KB-1 cultures, no dechlorination activity was recovered from gel slices. BN-PAGE was then investigated and found to significantly increase the activity recovered from the gel. The major difference between CN-PAGE and BN-PAGE is the use of Coomassie blue G-250 to impart negative charges on protein surfaces for greater mobility through the gel (38). Another difference between the BN-PAGE approach described herein and the CN-PAGE used by Adrian et al. (17) is that precast 4 to 16% acrylamide gradient gels (Invitrogen) were used. Another important component of method optimization was the selection of an appropriate nonionic, mild detergent required in the sample buffer to solubilize membrane-associated proteins such as RDases. Of the four different detergents tested—digitonin, dodecyl-β-d-maltoside, taurodeoxycholate, and Triton X-100, digitonin was found to be the most effective.
The BN-PAGE gradient gels (from Invitrogen) separated proteins based on size. Dechlorination activity was highly constrained to regions of electrophoretic mobility corresponding to 242 kDa in all cases studied. The fact that activity was constrained to a particular region suggests enrichment of RDases during electrophoresis. The specific activity in gel slices was found to be 5 to 15 times greater than in crude protein extracts (see Fig. S4 in the supplemental material), further indicating enrichment of RDases by this approach. However, although the BN-PAGE technique can partially separate and enrich RDases from other proteins, it does not separate different RDases from each other. Even when the active gel regions were divided into three consecutive narrow slices (Fig. S2), no significant differences were found in the RDases identified (see Table S2 in the supplemental material). Therefore, further separation of the proteins in each slice is needed to resolve cases where multiple RDases are coexpressed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Benjamin Scheer for assistance in the laboratory and Laura Hug for curation of the RDase database.
Support for this research was provided by the Saxon State Ministry for Science and Art fellowships awarded to both K.E.F. and W.W.M.C. through L.A. K.E.F. acknowledges support through an NSF graduate research fellowship. S.T. received awards from the Government of Ontario through the Ontario Graduate Scholarships in Science and Technology (OGSST) and the Natural Sciences and Engineering Research Council of Canada (NSERC PGS B). L.A. was supported by the European Research Council and the DFG (FOR1530). Support was provided by the Government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405), by the Government of Ontario through the ORF-GL2 program, and by the U.S. Department of Defense through the Strategic Environmental Research and Development Program (SERDP) under contract W912HQ-07-C-0036 (project ER-1586). Metagenome sequencing was provided by the U.S. Department of Energy Joint Genome Institute's Community Sequencing Program (CSP 2010).
Footnotes
Published ahead of print 30 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01873-12.
REFERENCES
- 1. De Wildeman S, Verstraete W. 2003. The quest for microbial reductive dechlorination of C(2) to C(4) chloroalkanes is warranted. Appl. Microbiol. Biotechnol. 61:94–102 [DOI] [PubMed] [Google Scholar]
- 2. Löffler FE, Edwards EA. 2006. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 17:274–284 [DOI] [PubMed] [Google Scholar]
- 3. Lendvay JM, Löffler FE, Dollhopf M, Aiello MR, Daniels G, Fathepure BZ, Gebhard M, Heine R, Helton R, Shi J, Krajmalnik-Brown R, Major CL, Barcelona MJ, Petrovskis E, Hickey R, Tiedje JM, Adriaens P. 2003. Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ. Sci. Technol. 37:1422–1431 [Google Scholar]
- 4. Major DW, McMaster ML, Cox EE, Edwards EA, Dworatzek SM, Hendrickson ER, Starr MG, Payne JA, Buonamici LW. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106–5116 [DOI] [PubMed] [Google Scholar]
- 5. Ward CH, Stroo HF. 2010. In situ remediation of chlorinated solvent plumes. SERDP and ESTCP remediation technology monograph series. Springer, New York, NY [Google Scholar]
- 6. Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. 2012. Dehalococcoides mccartyi gen. nov., sp. nov., obligate organohalide-respiring anaerobic bacteria, relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidetes classis nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. doi:10.1099/ijs.0.034926-0 [Google Scholar]
- 7. He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65 [DOI] [PubMed] [Google Scholar]
- 8. Maymó-Gatell X, Anguish T, Zinder SH. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ritalahti KM, Löffler FE. 2004. Populations implicated in anaerobic reductive dechlorination of 1,2-dichloropropane in highly enriched bacterial communities. Appl. Environ. Microbiol. 70:4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hug L. 2012. A metagenome-based examination of dechlorinating enrichment cultures: Dehalococcoides and the role of the non-dechlorinating organism. Doctoral thesis University of Toronto, Toronto, Canada [Google Scholar]
- 11. McMurdie PJ, Behrens SF, Müller JA, Goke J, Ritalahti KM, Wagner R, Goltsman E, Lapidus A, Holmes S, Löffler FE, Spormann AM. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5:e1000714 doi:10.1371/journal.pgen.1000714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magnuson JK, Romine MF, Burris DR, Kingsley MT. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Müller JA, Rosner BM, von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krajmalnik-Brown R, Hölscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chow WL, Cheng D, Wang S, He J. 2010. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J. 4:1020–1030 [DOI] [PubMed] [Google Scholar]
- 17. Adrian L, Rahnenführer J, Gobom J, Hölscher T. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73:7717–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wittig I, Braun HP, Schägger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]
- 19. Duhamel M, Edwards EA. 2007. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environ. Sci. Technol. 41:2303–2310 [DOI] [PubMed] [Google Scholar]
- 20. Duhamel M, Edwards EA. 2006. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol. Ecol. 58:538–549 [DOI] [PubMed] [Google Scholar]
- 21. Waller AS. 2009. Molecular investigation of chloroethene reductive dehalogenation by the mixed microbial community KB1. Doctoral thesis University of Toronto, Toronto, Canada [Google Scholar]
- 22. Waller AS, Krajmalnik-Brown R, Löffler FE, Edwards EA. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257–8264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duhamel M, Mo K, Edwards EA. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70:5538–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagner DD, Hug LA, Hatt JK, Spitzmiller MR, Padilla-Crespo E, Ritalahti KM, Edwards EA, Konstantinidis KT, Löffler FE. 2012. Genomic determinants of organohalide-respiration in Geobacter lovleyi, an unusual member of the Geobacteraceae. BMC Genomics 13:200 doi:10.1186/1471-2164-13-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edwards EA, Grbić-Galić D. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fletcher KE, Löffler FE, Richnow HH, Nijenhuis I. 2009. Stable carbon isotope fractionation of 1,2-dichloropropane during dichloroelimination by Dehalococcoides populations. Environ. Sci. Technol. 43:6915–6919 [DOI] [PubMed] [Google Scholar]
- 27. Chan WWM. 2009. Characterization of reductive dehalogenases in a chlorinated ethene-degrading bioaugmentation culture. Master's thesis University of Toronto, Toronto, Canada [Google Scholar]
- 28. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 29. Nesterenko MV, Tilley M, Upton SJ. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239–242 [DOI] [PubMed] [Google Scholar]
- 30. Invitrogen 2012. User guide for NativePAGE Novex Bix-Tris gel system: a system for native gel electrophoresis, p 23 Publication 25-0894. MAN0000557 Invitrogen, Carlsbad, CA: http://tools.invitrogen.com/content/sfs/manuals/nativepage_man.pdf [Google Scholar]
- 31. Hölscher T, Görisch H, Adrian L. 2003. Reductive dehalogenation of chlorobenzene congeners in cell extracts of Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 69:2999–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grostern A, Edwards EA. 2006. Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl. Environ. Microbiol. 72:428–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benndorf D, Balcke GU, Harms H, von Bergen M. 2007. Functional metaproteome analysis of protein extracts from contaminated soil and groundwater. ISME J. 1:224–234 [DOI] [PubMed] [Google Scholar]
- 34. Bastida F, Rosell M, Franchini AG, Seifert J, Finsterbusch S, Jehmlich J, Jechalke S, von Bergen M, Richnow HH. 2010. Elucidating MTBE degradation in a mixed consortium using a multidisciplinary approach. FEMS Microbiol. Ecol. 73:370–384 [DOI] [PubMed] [Google Scholar]
- 35. Kellner H, Jehmlich N, Benndorf D, Hoffmann R, Rühl M, Hoegger PJ, Majcherczyk A, Kües U, von Bergen M, Buscot F. 2007. Detection, quantification and identification of fungal extracellular laccases using polyclonal antibody and mass spectrometry. Enzyme Microb. Technol. 41:694–701 [Google Scholar]
- 36. Chan WW, Grostern A, Löffler FE, Edwards EA. 2011. Quantifying the effects of 1,1,1-trichloroethane and 1,1-dichloroethane on chlorinated ethene reductive dehalogenases. Environ. Sci. Technol. 45:9693–9702 [DOI] [PubMed] [Google Scholar]
- 37. Rahm BG, Richardson RE. 2008. Correlation of respiratory gene expression levels and pseudo-steady-state PCE respiration rates in Dehalococcoides ethenogenes. Environ. Sci. Technol. 42:416–421 [DOI] [PubMed] [Google Scholar]
- 38. Wittig I, Schägger H. 2008. Features and applications of blue-native and clear-native electrophoresis. Proteomics 8:3974–3990 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.