Abstract
Geobacter species are important Fe(III) reducers in a diversity of soils and sediments. Mechanisms for Fe(III) oxide reduction have been studied in detail in Geobacter sulfurreducens, but a number of the most thoroughly studied outer surface components of G. sulfurreducens, particularly c-type cytochromes, are not well conserved among Geobacter species. In order to identify cellular components potentially important for Fe(III) oxide reduction in Geobacter metallireducens, gene transcript abundance was compared in cells grown on Fe(III) oxide or soluble Fe(III) citrate with whole-genome microarrays. Outer-surface cytochromes were also identified. Deletion of genes for c-type cytochromes that had higher transcript abundance during growth on Fe(III) oxides and/or were detected in the outer-surface protein fraction identified six c-type cytochrome genes, that when deleted removed the capacity for Fe(III) oxide reduction. Several of the c-type cytochromes which were essential for Fe(III) oxide reduction in G. metallireducens have homologs in G. sulfurreducens that are not important for Fe(III) oxide reduction. Other genes essential for Fe(III) oxide reduction included a gene predicted to encode an NHL (Ncl-1–HT2A–Lin-41) repeat-containing protein and a gene potentially involved in pili glycosylation. Genes associated with flagellum-based motility, chemotaxis, and pili had higher transcript abundance during growth on Fe(III) oxide, consistent with the previously proposed importance of these components in Fe(III) oxide reduction. These results demonstrate that there are similarities in extracellular electron transfer between G. metallireducens and G. sulfurreducens but the outer-surface c-type cytochromes involved in Fe(III) oxide reduction are different.
INTRODUCTION
The mechanisms for electron transfer to Fe(III) oxide in Geobacter species are of interest because Geobacter species play an important role in Fe(III) reduction in a wide diversity of soils, aquatic sediments, and subsurface environments (1). Furthermore, an understanding of the mechanisms for Fe(III) oxide reduction is expected to provide insights into other important types of extracellular electron transfer in Geobacter, such as electron transfer to electrodes (2) and interspecies electron transfer (3, 4). Initial studies on the mechanisms for Fe(III) oxide reduction in Geobacter species were conducted with G. sulfurreducens because it was the first Geobacter species for which a genetic system was developed (5). However, G. metallireducens is a more effective Fe(III) oxide reducer than G. sulfurreducens and has other environmentally significant physiological properties not found in G. sulfurreducens, such as the ability to anaerobically oxidize aromatic hydrocarbons (1), including benzene (6). Therefore, understanding how Fe(III) oxides are reduced in G. metallireducens aids in understanding the physiology of this important model organism and provides the opportunity to find conserved mechanisms for Fe(III) oxide reduction in Geobacter species. A genetic system has recently been developed for G. metallireducens, which now makes such studies feasible (7).
Initial studies suggested that like G. sulfurreducens (8), G.metallireducens requires type IV pili for Fe(III) oxide reduction (7). The pili of G. sulfurreducens possess metal-like conductivity (9), which is distinct from the electron hopping/tunneling associated with other known forms of biological electron transport (10). Measurements of the conductivity of the pili of G. metallireducens have not been reported, but the PilA sequence of G. metallireducens is 76% similar to the G. sulfurreducens PilA sequence. The G. sulfurreducens pili are decorated with the multi-heme c-type cytochrome OmcS (11), which is required for Fe(III) oxide reduction (12). The spacing of OmcS on pili is too great to contribute to conduction of electrons along the length of the pili (11, 13) and multiple lines of additional evidence rule out this possibility (9, 11, 13, 14). Therefore, it has been proposed that the role of OmcS is to facilitate electron transfer from the pili to Fe(III) oxides (15). However, there is no homolog of OmcS in G. metallireducens, in line with the overall poor conservation of outer surface c-type cytochromes in Geobacter species (16).
The only other G. sulfurreducens outer-surface c-type cytochrome known to be essential for Fe(III) oxide reduction is OmcB, which appears to be embedded in the outer membrane (17) and is speculated to facilitate electron transfer from the periplasm to the outer surface (18). There is no homolog to omcB in G. metallireducens, although another c-type cytochrome is found in a syntenous location (19). The only c-type cytochromes known to be involved in Fe(III) reduction that are well conserved between G. sulfurreducens and G. metallireducens are PpcA and MacA, which are located in the periplasm (19–21).
Comparing gene expression during growth on insoluble electron acceptors versus growth on soluble electron acceptors has been a productive strategy for identifying components involved in extracellular electron transfer in G. sulfurreducens (22, 23). Here, we report on components of G. metallireducens likely to be important in electron transfer to Fe(III) oxides identified from gene expression, protein localization, and gene deletion studies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
G. metallireducens (ATCC 53774 and DSM 7210) was routinely cultured under strict anaerobic conditions with 10 mM acetate provided as the sole electron donor as previously described (24). Either Fe(III) citrate (55 mM) or Fe(III) oxide (100 mM) were provided as the sole terminal electron acceptor for Fe(III) reduction studies. Samples of Fe(III) oxide cultures were dissolved in 0.5 N HCl and Fe(II) concentrations were measured using the ferrozine assay as previously described (25).
For genetic manipulations Fe(III) citrate (56 mM) was provided as the electron acceptor along with the addition of ferrous ammonium sulfate (500 μM) and yeast extract (0.1%) to liquid medium and agar plates (7). Genetic manipulations were carried out in an anaerobic chamber containing N2/CO2/H2 (in percent, 83/10/7) atmosphere and at a temperature of 30°C. Escherichia coli was cultivated with Luria-Bertani medium with or without antibiotics (26). All bacterial strains and plasmids are listed in Table S2 in the supplemental material.
SDS-PAGE and protein identification.
The loosely bound surface proteins fraction of G. metallireducens was isolated during mid-exponential growth as previously described (12). Cells grown with Fe(III) oxide provided as the electron acceptor were treated with equal volumes of TPE and oxalate solution prior to cell fractionation. The outer membrane protein fraction was isolated using a previously described method (27). Loosely bound and outer membrane protein fractions were combined, and protein concentration was determined with the bicinchoninic acid (BCA) assay (Pierce, Rockford, IL). c-type cytochromes were identified by separation with SDS-PAGE and stained for heme as previously described (28). Equal amounts of proteins were loaded in each lane. Differentially expressed c-type cytochrome bands from the Tris-Tricine polyacrylamide gel were excised and sent to the Laboratory for Proteomic Mass Spectrometry at the University of Massachusetts Medical School for liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) analysis.
DNA microarrays.
Total RNA for microarray analysis was extracted from quadruplicate cultures of G. metallireducens cells grown with acetate (10 mM)-Fe(III) citrate (55 mM) or acetate (10 mM)-Fe(III) oxide (100 mM) during exponential growth using methods previously described (29). RNA samples were purified with the RNeasy MinElute Clean-Up kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, and treated with the Turbo DNA-free DNase (Ambion, Austin, TX). The RNA samples were tested for genomic DNA contamination by PCR amplification of the 16S rRNA gene (30). The concentration and quality of the RNA samples were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). All RNA samples had A260/A280 ratios of 1.8 to 2.0, indicating high purity (31). cDNA was generated with the TransPlex whole transcriptome amplification kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions.
Whole-genome microarray hybridizations were carried out by Roche NimbleGen, Inc. (Madison, WI). Quadruplicate biological and triplicate technical replicates were conducted for all microarray analyses. Cy3-labeled cDNA was hybridized to oligonucleotide microarrays based on G. metallireducens genome and resident plasmid sequences (accession number NC007515 and NC007517 at GenBank). The microarray results were analyzed with Array 4 Star (DNASTAR, Madison, WI). A gene was considered differentially expressed only if the P value determined by using the Student t test analysis was ≤0.01.
RT-qPCR.
Microarray results were confirmed with reverse transcription-quantitative PCR (RT-qPCR). The Power SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA) and the ABI 7500 Real-Time PCR System were used to amplify and to quantify PCR products. Each reaction consisted of forward and reverse primers at a final concentration of 200 nM, 5 ng of cDNA, and 12.5 μl of Power SYBR green PCR Master Mix (Applied Biosystems). Primer pairs were designed with amplicon size of 100 to 200 bp for the following: Gmet 0557, Gmet 2896, Gmet 0909, Gmet 0930, Gmet 1868, and Gmet 2029. Expression of these genes was normalized with proC expression, a gene shown to be constitutively expressed in Geobacter species (32). The relative levels of expression of the studied genes were calculated by the 2−ΔΔCT method (33). Sequences from all primers used for RT-qPCR are listed in Table S3 in the supplemental material.
Deletion mutant construction and complementation.
The primers used for the construction of mutants are listed in Table S3 in the supplemental material. All mutants were made by replacing the gene of interest with a spectinomycin resistance gene as previously described (7). All restriction digestions were carried out according to the manufacturer's instructions. PCRs were performed with the JumpStart Taq DNA polymerase (Sigma-Aldrich). Briefly, primer pairs were used to amplify by PCR flanking regions of approximately 500 bp downstream and upstream of the target genes using G. metallireducens genomic DNA as a template. PCR products were digested with the AvrII (CCTAGG) (NEB, Beverly, MA) restriction endonuclease, ethanol precipitated, and ligated with the T4 DNA ligase (NEB). The ligation reaction was loaded onto a 1% agarose gel, and a 1-kb band was purified using the Qiaquick gel extraction kit (Qiagen) and cloned into pCR2.1 TOPO cloning vector resulting in pCR2.1up5′+3′dn. Sequences of the cloned products were verified by Sanger sequencing. The spectinomycin resistance cassette was digested with XbaI (TCTAGA) (NEB) from pUC19-SprloxP (7), and the recombinant plasmid pCR2.1up5′+3′dn was digested with AvrII. The spectinomycin resistance cassette was cloned into pCR2.1up5′-′+3′dn to complete the construction of the mutant alleles. Plasmids bearing mutant alleles were linearized by digesting with either KpnI (GGTACC) (NEB) or XhoI (CTCGAG) (NEB) and concentrated by ethanol precipitation. The linearized plasmids were electroporated into G. metallireducens as described previously (7). Replacement of wild-type alleles by mutant alleles in G. metallireducens was verified by PCR. Deletion mutants made in this study were complemented by amplifying the respective genes with their native ribosome binding site (RBS) using G. metallireducens genomic DNA as a template. The resulting PCR products were digested and cloned under the control of a constitutive lac promoter into pCM66 (34).
Microarray data accession number.
Microarray data have been deposited with NCBI GEO under accession number GSE40316.
RESULTS AND DISCUSSION
In order to gain insight into which genes coding for outer cell surface proteins might be important for insoluble Fe(III) oxide reduction in G. metallireducens, gene transcript abundance was compared in cells grown on Fe(III) oxide or Fe(III) citrate (see Table S1 in the supplemental material). The microarray analysis revealed 792 genes differentially expressed at 2-fold change and 95% confidence. A total of 437 of these genes were upregulated with growth on Fe(III) oxide, whereas 355 were downregulated. Additional focus was placed on genes with higher transcript abundance during growth on Fe(III) oxide based on the assumption that genes more highly expressed during growth on Fe(III) oxide are likely to play an important role in this process.
Cytochromes.
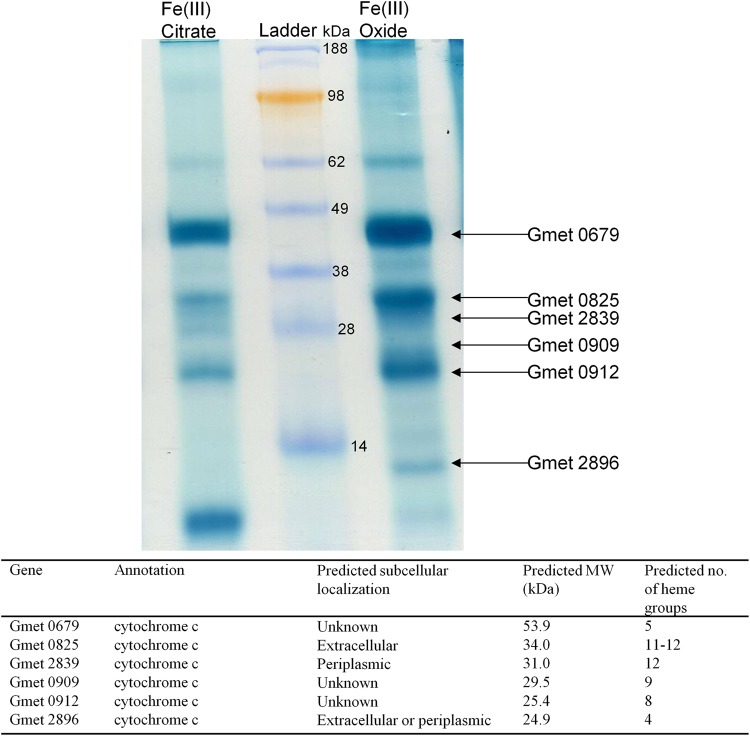
The microarray analysis revealed 23 genes coding for c-type cytochromes that had higher transcript abundance in cells grown on Fe(III) oxide (Table 1). In order to identify additional cytochromes that might have roles in Fe(III) oxide reduction, proteins were isolated from the outer surface protein fraction and stained for heme because cytochromes localized on the outer surface of the cell have the potential to directly interact with extracellular electron acceptors. Heme staining of outer-surface proteins separated by SDS-PAGE revealed numerous protein bands with a stronger signal in preparations of cells grown on Fe(III) oxide versus cells grown on Fe(III) citrate, six of which could be identified via liquid-chromatography/mass spectrometry (Fig. 1). Four of the genes identified (Gmet 0679, Gmet 0825, Gmet 0909, and Gmet 2896) were genes that had higher transcript abundance in cells grown on Fe(III) oxide than in Fe(III) citrate-grown cells (Table 1).
Table 1.
Genes coding for c-type cytochrome upregulated at least 2-fold in G. metallireducens when Fe(III) oxide is the electron acceptor
| Gene | Annotation | Predicted subcellular localizationa | Fold change |
|---|---|---|---|
| Gmet 0909 | Cytochrome c, 9 heme-binding sites | Unknown | 37.8 |
| Gmet 0910 | Lipoprotein cytochrome c, 10 heme-binding sites | Unknown | 32.7 |
| Gmet 0557 | Cytochrome c, 4 heme-binding sites | Extracellular or periplasmic | 18.5 |
| Gmet 0679 | Cytochrome c, 5 heme-binding sites | Unknown | 18.0 |
| Gmet 0558 | Cytochrome c, 27 heme-binding sites | Extracellular or periplasmic | 17.1 |
| Gmet 0571 | Cytochrome c, 26 heme-binding sites | Extracellular or periplasmic | 16.5 |
| Gmet 3091 | Cytochrome c, 2 heme-binding sites | Periplasmic | 13.9 |
| Gmet 1088 | Lipoprotein cytochrome c, 1 heme-binding site | Periplasmic | 11.1 |
| Gmet 0534 | Cytochrome c, 5 heme-binding sites | Unknown | 8.7 |
| Gmet 0155 | Cytochrome c, 1 heme-binding site | Periplasmic | 8.4 |
| Gmet 2470 | Cytochrome c, 27 heme-binding sites | Extracellular or periplasmic | 6.8 |
| Gmet 1868 | Cytochrome c, 4 heme-binding sites | Extracellular | 5.6 |
| Gmet 1866 | Cytochrome c, 3–4 heme-binding sites | Unknown | 5.0 |
| Gmet 0170 | Cytochrome c, 8 heme-binding sites | Periplasmic | 5.0 |
| Gmet 2896 | Cytochrome c, 4 heme-binding sites | Extracellular or periplasmic | 4.4 |
| Gmet 0580 | Lipoprotein cytochrome c, 14 heme-binding sites | Extracellular | 4.2 |
| Gmet 1867 | Cytochrome c, 7–8 heme-binding sites | Unknown | 4.1 |
| Gmet 0142 | Cytochrome c, 8 heme-binding sites | Periplasmic | 3.6 |
| Gmet 1924 | Cytochrome c, 5 heme-binding sites | Unknown | 3.4 |
| Gmet 0913 | Cytochrome c, 9 heme-binding sites | Extracellular | 3.1 |
| Gmet 0581 | Lipoprotein cytochrome c, 27–34 heme-binding sites | Extracellular | 3.0 |
| Gmet 0825 | Cytochrome c, 11–12 heme-binding sites | Extracellular | 2.5 |
| Gmet 0930 | Cytochrome c, 6–8 heme-binding sites | Extracellular or periplasmic | 2.5 |
Subcellular localization predictions were done with PSORTb 3.0 (37).
Fig 1.
c-Type cytochrome content of outer membrane and loosely bound fractions from Fe(III) oxide and Fe(III) citrate cultures. Proteins (2 μg/lane) were separated by SDS-PAGE and stained for heme. Protein ladder was SeeBlue Plus2 Pre-Stained Standard (Invitrogen, Carlsbad, CA). Subcellular localization predictions were done with PSORTb 3.0 (37).
The function of c-type cytochromes whose genes had higher transcript abundance during growth on Fe(III) oxide and/or were identified as outer-surface cytochromes was analyzed by constructing mutant strains in which one of the cytochrome genes was deleted. Six of the c-type cytochrome deletion mutants were unable to grow with Fe(III) oxide as the sole electron acceptor but could grow on Fe(III) citrate (Table 2; see also Fig. S1 in the supplemental material). In each case complementation of the deletion mutants with expression of the appropriate gene in trans partially restored the capacity for Fe(III) oxide reduction (Table 2; see Fig. S1 in the supplemental material).
Table 2.
Fe(III) reduction rate of mutants and complemented strains grown on Fe(III) oxide or Fe(III) citratea
| Strain | Mean Fe(III) reduction rate ± SD |
||
|---|---|---|---|
| Fe(III) oxide [mM Fe(II)/day] | Fe(III) citrate [mM Fe(II)/h] | Complementation Fe(III) oxide [mM Fe(II)/day] | |
| Wild type | 6.1 ± 0.6 | 3.7 ± 0.2 | |
| c-Type cytochrome mutants | |||
| Gmet 2896 | 0.0 ± 0.0 | 3.5 ± 0.2 | 4.0 ± 0.2 |
| Gmet 0930 | 0.0 ± 0.0 | 3.6 ± 0.2 | 3.7 ± 0.4 |
| Gmet 0557 | 0.0 ± 0.0 | 3.6 ± 0.1 | 3.9 ± 0.2 |
| Gmet 0558 | 0.0 ± 0.0 | 3.1 ± 0.2 | 3.6 ± 0.1 |
| Gmet 1867 | 0.0 ± 0.0 | 3.8 ± 0.2 | 3.5 ± 0.1 |
| Gmet 1868 | 0.0 ± 0.0 | 3.9 ± 0.1 | 3.3 ± 0.2 |
| Gmet 0534 | 5.7 ± 0.5 | ||
| Gmet 0571 | 4.9 ± 0.4 | ||
| Gmet 0580 | 5.3 ± 0.5 | ||
| Gmet 0581 | 6.4 ± 0.6 | ||
| Gmet 0679 | 5.7 ± 0.5 | ||
| Gmet 0825 | 4.4 ± 0.4 | ||
| Gmet 0910 | 6.0 ± 0.6 | ||
| Gmet 0912 | 5.5 ± 0.5 | ||
| Gmet 0913 | 5.7 ± 0.4 | ||
| Gmet 1866 | 6.2 ± 0.5 | ||
| Gmet 2470 | 5.5 ± 0.5 | ||
| Gmet 2839 | 6.4 ± 0.5 | ||
| Other mutant | |||
| Gmet 0556 | 0.0 ± 0.0 | 3.9 ± 0.3 | 3.7 ± 0.3 |
| Polysaccharide-associated mutants | |||
| Gmet 2029 | 0.0 ± 0.0 | 3.8 ± 0.3 | 3.5 ± 0.3 |
| Gmet 2030 | 5.9 ± 0.5 | ||
| Gmet 2031 | 6.1 ± 0.5 | ||
| Gmet 2032 | 6.2 ± 0.5 | ||
Each value is the mean and standard deviation of at least three replicates.
Gmet 2896, which was one of the genes required for Fe(III) oxide reduction, had higher transcript abundance during growth on Fe(III) oxide (Table 1) and the Gmet 2896 protein was localized outside the cell (Fig. 1). Gmet 2896 is predicted to encode a tetraheme c-type cytochrome in the same family as OmcE of G. sulfurreducens with 44% amino acid sequence identity (16), which is also found on the outer surface and is predicted to contain four hemes. Gmet 2896 appeared in a band located around 14 kDa (Fig. 1) even though its predicted molecular mass is 24.9 kDa. This suggests that Gmet_2896 might be processed after translation in a manner similar to OmcZ of G. sulfurreducens (23, 27). Long initial lag periods in growth on Fe(III) oxide (12) and current production (22) have suggested a role for OmcE in extracellular electron transfer in G. sulfurreducens, but the ability of this strain to adapt for growth on Fe(III) oxide (12) and current production (9) have demonstrated that OmcE is not essential for these functions.
Like Gmet 2896, Gmet 0930 had higher transcript abundance in Fe(III) oxide-grown cells (Table 1) and is required for Fe(III) oxide reduction (Table 2; see Fig. S1 in the supplemental material). Gmet 0930 is in the same family as the gene for OmcZ (27) of G. sulfurreducens (16). Unlike the Gmet 0930 protein, OmcZ is not required for Fe(III) oxide reduction (23). However, it is also an outer-surface protein (35) and is required for optimal current production (23, 36). Its localization at the interface between anode biofilms and electrodes suggests that it facilitates electron transfer to electrodes in G. sulfurreducens (35).
Gmet 0909 was the most highly upregulated gene coding for a c-type cytochrome in cells grown on Fe(III) oxide (Table 1) and the Gmet 0909 protein was detected outside the cell (Fig. 1). Along with Gmet 0534, Gmet 0909 is the only cytochrome gene with higher transcript abundance during growth on Fe(III) oxide that is conserved across all sequenced Geobacter species (16). Numerous attempts to construct a strain in which Gmet 0909 was deleted failed, suggesting that this gene might also be required for growth on Fe(III) citrate.
The proteins of Gmet 0557 and Gmet 0558 were not detected in the outer surface, but both of these genes, which were predicted to be in the same operon, had much higher transcript levels in cells grown on Fe(III) oxide (Table 1), and are required for Fe(III) oxide reduction (Table 2; see Fig. S1 in the supplemental material). Both proteins are predicted to be localized either in the periplasm or in the extracellular fraction (Table 1). Gmet 0557 is predicted to have four heme binding sites, whereas Gmet 0558 is predicted to have between 23 and 27 heme binding sites. The closest homolog to Gmet 0557 in G. sulfurreducens is OmcP (GSU2913) with 59% amino acid sequence identity. The closest homolog to Gmet 0558 in G. sulfurreducens is OmcO (GSU2912) with 67% amino acid sequence identity. Neither OmcP nor OmcO are essential for Fe(III) oxide reduction by G. sulfurreducens, although transcript levels were higher for both genes during growth on Fe(III) oxide compared to Fe(III) citrate (M. Aklujkar et al., unpublished data).
Gmet 0557 and Gmet 0558 are located in the same operon as Gmet 0556, which is predicted to encode an NHL (Ncl-1–HT2A–Lin-41) repeat-containing protein localized in the extracellular matrix (37) and had higher transcript abundance in cells grown on Fe(III) oxide (Table S1). Gmet 0556 is predicted to contain conserved immunoglobulin-like fold domains, which are considered to play a possible role in cell adhesion in other microorganisms (38). The homolog to Gmet 0556 in G. sulfurreducens, GSU2914, was also more highly expressed with growth on Fe(III) oxide compared to Fe(III) citrate (Aklujkar et al., unpublished). Furthermore, the homologous gene in G. uraniireducens, Gura 3430, was more highly expressed during growth in sediments in which insoluble Fe(III) was expected to be the electron acceptor, compared to cells grown with fumarate as the electron acceptor (39). Deletion of Gmet 0556 produced a strain that could not reduce Fe(III) oxide but was capable of reducing soluble Fe(III) citrate (Table 2; see Fig. S2 in the supplemental material). Expressing Gmet 0556 in trans restored the capacity for Fe(III) oxide reduction (Table 2; see Fig. S2 in the supplemental material). Further functional analysis of this protein seems warranted.
Like Gmet 0557 and Gmet 0558, Gmet 1867 and Gmet 1868 are both located in the same operon, had higher transcript levels in cells grown on Fe(III) oxide (Table 1), and their proteins were not detected in the outer-surface proteins. Both Gmet 1867 and Gmet 1868 are required for Fe(III) oxide reduction (Table 2; see Fig. S1 in the supplemental material). Localization of Gmet 1867 is unclear, whereas Gmet 1868 is predicted to be found in the extracellular matrix (Table 1). Gmet 1867 is predicted to contain 7 to 8 heme-binding sites, and Gmet 1868 is predicted to have 4. The closest homolog to Gmet 1867 and Gmet 1868 in G. sulfurreducens are, respectively, GSU1786 (57% amino acid sequence identity) and GSU1787 (69% amino acid sequence identity). Neither of these genes have been previously reported to be involved with Fe(III) oxide reduction in G. sulfurreducens, although GSU1787 had higher transcript abundance with growth on Fe(III) oxide compared to Fe(III) citrate (Aklujkar et al., unpublished).
Several c-type cytochrome deletion mutants exhibited no phenotype on Fe(III) oxide or Fe(III) citrate (Table 2). Gmet 0910 and Gmet 0913 have no homologs in G. sulfurreducens (16). None of the G. sulfurreducens homologs for Gmet 0534, Gmet 0571, Gmet 0580, Gmet 0581, Gmet 0679, Gmet 0825, Gmet 0912, Gmet 1866, Gmet 2470, and Gmet 2839 have been found to be essential for optimal Fe(III) oxide reduction (Aklujkar et al., unpublished).
Extracellular polysaccharide genes.
A number of genes annotated as contributing to polysaccharide biosynthesis had higher transcript abundance in Fe(III) oxide-grown cells (Table 3). The most highly expressed were Gmet 2028 through Gmet 2032. Genes coding for homologs of Gmet 2028 (GSU1983), Gmet 2029 (GSU1984), Gmet 2030 (GSU1985), Gmet 2031 (GSU1986), and Gmet 2032 (GSU1987) all had higher transcript abundance in G. sulfurreducens when grown on Fe(III) oxide compared to Fe(III) citrate (Aklujkar et al., unpublished). Furthermore, homologs of Gmet 2030 (Gura 1669), Gmet 2029 (Gura 1670), Gmet 2028 (Gura 1670), Gmet 2003 (Gura 2342), and Gmet 0458 (Gura 1672) were upregulated in G. uraniireducens grown on sediments in which insoluble Fe(III) was expected to be the electron acceptor, compared to cells grown with fumarate as the electron acceptor (39), suggesting conserved functions for these proteins in the process of extracellular electron transfer among Geobacter species. It has recently been proposed that another gene (xapD; GSU1501) involved in an extracellular polysaccharide network in G. sulfurreducens contributes to insoluble Fe(III) reduction, biofilm formation, and c-type cytochrome anchoring (40, 41). The homolog of xapD in G. metallireducens (Gmet 1403) was not differentially expressed on Fe(III) oxide compared to ferric citrate.
Table 3.
Polysaccharide biosynthesis-associated genes upregulated at least 2-fold in G. metallireducens when Fe(III) oxide is the electron acceptor (P value cutoff ≤ 0.01)
| Gene | Annotation | Fold change |
|---|---|---|
| Gmet 2030 | Periplasmic polysaccharide biosynthesis/export protein | 20.9 |
| Gmet 2029 | Polysaccharide chain length determinant protein, putative; Wzz family | 14.3 |
| Gmet 2032 | TPR domain lipoprotein | 12.3 |
| Gmet 2028 | Polysaccharide biosynthesis protein, putative | 11.8 |
| Gmet 2031 | Glycosyltransferase domain protein | 7.9 |
| Gmet 2003 | Exopolysaccharide synthesis multitransmembrane protein H (exosortase); EpsH | 4.9 |
| Gmet 2023 | Polysaccharide deacetylase domain protein | 4.7 |
| Gmet 2013 | Polysaccharide deacetylase, putative | 3.8 |
| Gmet 0458 | Polysaccharide biosynthesis protein; CapD | 2.5 |
The potential role of the highly expressed Gmet 2029, Gmet 2030, Gmet 2031, and Gmet 2032 in Fe(III) oxide reduction was investigated by gene deletion. Only the loss of Gmet 2029 resulted in a G. metallireducens strain incapable of reducing Fe(III) oxide (Table 2; see Fig. S3 in the supplemental material).
Gmet 2029 is predicted to encode a polysaccharide chain length determinant protein of the Wzz family. The Wzz proteins of several bacterial species (42–45) are implicated in the length determination of the O polysaccharide, a major component of lipopolysaccharide (LPS). However, members of the Wzz protein family are found throughout the Geobacteraceae family, including G. sulfurreducens (amino acid sequence identity = 39%) which produces a rough LPS without the O polysaccharide (46). The Wzz protein of Pseudomonas aeruginosa also appears to determine the chain length of polysaccharides involved in pilin glycosylation (47, 48). Thus, a potential role of Gmet 2029 is modification of the pili known to play a role in long-range electron transport.
Expression of genes previously identified as important in Fe(III) reduction.
In some instances gene expression patterns were consistent with previous observations of G. metallireducens physiological differences between cells grown on Fe(III) oxide and Fe(III) citrate.
For example, G. metallireducens grown on Fe(III) oxide produces flagella, whereas cells grown on Fe(III) citrate do not (49), and chemotaxis and motility are thought to be important in Fe(III) oxide reduction (7, 49, 50). Genes coding for flagellar or chemotaxis proteins represented a high proportion of the genes with the greatest increase in transcript abundance in Fe(III) oxide-grown cells (Table 4). Gmet 0442, a gene coding for the flagellin protein FliC, was the most highly upregulated gene during growth on Fe(III) oxide.
Table 4.
Motility genes upregulated at least 30-fold in G. metallireducens when Fe(III) oxide is the electron acceptor (P value cutoff ≤ 0.01)
| Gene | Annotation | Fold change |
|---|---|---|
| Gmet 0442 | Flagellin FliC | 94.5 |
| Gmet 0719 | Conserved hypothetical protein | 77.0 |
| Gmet 0438 | Flagellar hook-associated protein FlgK | 56.9 |
| Gmet 0430 | Flagellar basal body rod protein FlgF | 55.8 |
| Gmet 0439 | Flagellar hook-filament junction protein FlgL | 52.7 |
| Gmet 0432 | Flagellar basal body P-ring formation protein FlgA | 49.0 |
| Gmet 0431 | Flagellar basal body rod protein FlgG | 43.8 |
| Gmet 3115 | Flagellar basal body rod protein FlgB | 43.1 |
| Gmet 3112 | Flagellar M-ring mounting plate protein FliF | 42.5 |
| Gmet 3104 | Flagellar operon protein of unknown function DUF3766 | 39.5 |
| Gmet 3098 | Flagellar biogenesis protein FliO | 39.1 |
| Gmet 0427 | Flagellar biogenesis protein FlhF | 37.6 |
| Gmet 0444 | Flagellar filament cap protein FliD | 32.1 |
| Gmet 3101 | Flagellar basal body-associated protein FliL | 31.0 |
The type IV pili of Geobacter sulfurreducens have metallic-like conductivity (9) and are considered to be conduits for electron transfer to Fe(III) oxide (15, 51). G. metallireducens produces type IV pili during growth on Fe(III) oxide, but not on Fe(III) citrate, which was attributed to differences in the expression of the gene for the structural PilA protein (49). Microarray results indicated a slight increase in pilA transcript abundance in Fe(III) oxide-grown cells (1.7-fold; P value = 0.19). The difference in transcript abundance for other genes associated with pili functions was somewhat higher (Table 5). A strain of G. metallireducens in which pilA was deleted was unable to reduce Fe(III) oxide but retained the capacity for Fe(III) citrate reduction, further suggesting the importance of pili in Fe(III) oxide reduction by G. metallireducens (7).
Table 5.
Pilus-associated genes upregulated at least 2-fold in G. metallireducens when Fe(III) oxide is the electron acceptor (P value cutoff ≤ 0.05)
| Gene | Annotation | Fold change |
|---|---|---|
| Gmet 0967 | Type IV pilus tip-associated adhesion PilY1-2 | 4.6 |
| Gmet 1395 | Type IV pilus biogenesis protein PilC | 4.0 |
| Gmet 0974 | Type IV pilus assembly lipoprotein PilP | 3.6 |
| Gmet 3400 | Twitching motility pilus retraction protein; PilT3 | 3.4 |
| Gmet 0975 | Type IV pilus secretin lipoprotein PilQ | 3,2 |
| Gmet 0959 | Type IV prepilin peptidase | 2.0 |
Implications.
These studies demonstrate that although G. metallireducens is closely related to G. sulfurreducens, the outer surface c-type cytochromes that are essential for Fe(III) oxide reduction in these two species are distinct. The identification of a select few outer surface cytochromes from the 90 putative c-type cytochrome genes in the G. metallireducens genome (19) generates a manageable list for future studies on mechanisms for electron transfer to Fe(III) oxide in this organism. Most important will be the development of antibodies or other reagents that will make it possible to determine the localization of the cytochromes as determining whether cytochromes required for Fe(III) oxide reduction are associated with the outer membrane, pili, or in the outer matrix will provide further insights into their functional role.
The identification of an NHL repeat-containing protein and of a Wzz family protein as being essential for Fe(III) oxide reduction demonstrates that there are still unknown components involved in extracellular electron transfer in Geobacter species and that the current understanding of this phenomenon is not complete.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the Office of Science (Biological and Environmental Research), U.S. Department of Energy, award number DE-SC0004114.
Footnotes
Published ahead of print 26 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02954-12.
REFERENCES
- 1. Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru AE, Holmes DE, Franks AE, Orellana R, Risso C, Nevin KP. 2011. Geobacter: the microbe electric's physiology, ecology, and practical applications. Adv. Microb. Physiol. 59:1–100 [DOI] [PubMed] [Google Scholar]
- 2. Lovley DR. 2012. Electromicrobiology. Annu. Rev. Microbiol. 66:391–409 [DOI] [PubMed] [Google Scholar]
- 3. Morita M, Malvankar NS, Franks AE, Summers ZM, Giloteaux L, Rotaru AE, Rotaru C, Lovley DR. 2011. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. mBio 2:e00159–00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, Lovley DR. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415 [DOI] [PubMed] [Google Scholar]
- 5. Coppi MV, Leang C, Sandler SJ, Lovley DR. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang T, Bain TS, Nevin KP, Barlett MA, Lovley DR. 2012. Anaerobic benzene oxidation by Geobacter species. Appl. Environ. Microbiol. 78:8304–8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tremblay P-L, Aklujkar M, Leang C, Nevin KP, Lovley D. 2012. A genetic system for Geobacter metallireducens: role of the flagellin and pilin in the reduction of Fe(III) oxide. Environ. Microbiol. Rep. 4:82–88 [DOI] [PubMed] [Google Scholar]
- 8. Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101 [DOI] [PubMed] [Google Scholar]
- 9. Malvankar NS, Vargas M, Nevin KP, Franks AE, Leang C, Kim BC, Inoue K, Mester T, Covalla SF, Johnson JP, Rotello VM, Tuominen MT, Lovley DR. 2011. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6:573–579 [DOI] [PubMed] [Google Scholar]
- 10. Malvankar NS, Lovley DR. 2012. Microbial nanowires: a new paradigm for biological electron transfer and bioelectronics. Chem. Sus. Chem. 5:1039–1046 [DOI] [PubMed] [Google Scholar]
- 11. Leang C, Qian X, Mester T, Lovley DR. 2010. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl. Environ. Microbiol. 76:4080–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mehta T, Coppi MV, Childers SE, Lovley DR. 2005. Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:8634–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malvankar NS, Mester T, Tuominen MT, Lovley DR. 2012. Supercapacitors based on c-type cytochromes using conductive nanostructured networks of living bacteria. Chem. Phys. Chem. 13:463–468 [DOI] [PubMed] [Google Scholar]
- 14. Malvankar NS, Lau J, Nevin KP, Franks AE, Tuominen MT, Lovley DR. 2012. Electrical conductivity in a mixed-species biofilm. Appl. Environ. Microbiol. 78:5967–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lovley DR. 2011. Live wires: direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination. Energy Environ. Sci. 4:4896–4906 [Google Scholar]
- 16. Butler JE, Young ND, Lovley DR. 2010. Evolution of electron transfer out of the cell: comparative genomics of six Geobacter genomes. BMC Genomics 11:40 doi:10.1186/1471-2164-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qian X, Reguera G, Mester T, Lovley DR. 2007. Evidence that OmcB and OmpB of Geobacter sulfurreducens are outer membrane surface proteins. FEMS Microbiol. Lett. 277:21–27 [DOI] [PubMed] [Google Scholar]
- 18. Lovley DR. Long-range electron transport to Fe(III) oxide via pili with metallic-like conductivity. Biochem. Soc. Trans., in press [DOI] [PubMed] [Google Scholar]
- 19. Aklujkar M, Krushkal J, DiBartolo G, Lapidus A, Land ML, Lovley DR. 2009. The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 9:109 doi:10.1186/1471-2180-9-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Butler JE, Kaufmann F, Coppi MV, Nunez C, Lovley DR. 2004. MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J. Bacteriol. 186:4042–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lloyd JR, Leang C, Hodges Myerson AL, Coppi MV, Cuifo S, Methe B, Sandler SJ, Lovley DR. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holmes DE, Chaudhuri SK, Nevin KP, Mehta T, Methe BA, Liu A, Ward JE, Woodard TL, Webster J, Lovley DR. 2006. Microarray and genetic analysis of electron transfer to electrodes in Geobacter sulfurreducens. Environ. Microbiol. 8:1805–1815 [DOI] [PubMed] [Google Scholar]
- 23. Nevin KP, Kim BC, Glaven RH, Johnson JP, Woodard TL, Methe BA, Didonato RJ, Covalla SF, Franks AE, Liu A, Lovley DR. 2009. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS One 4:e5628 doi:10.1371/journal.pone.0005628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJ, Gorby YA, Goodwin S. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336–344 [DOI] [PubMed] [Google Scholar]
- 25. Anderson RT, Lovley DR. 1999. Naphthalene and benzene degradation under Fe(III)-reducing conditions in petroleum-contaminated aquifers. Bioremediation J. 3:121–135 [Google Scholar]
- 26. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Inoue K, Qian X, Morgado L, Kim BC, Mester T, Izallalen M, Salgueiro CA, Lovley DR. 2010. Purification and characterization of OmcZ, an outer-surface, octaheme c-type cytochrome essential for optimal current production by Geobacter sulfurreducens. Appl. Environ. Microbiol. 76:3999–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas PE, Ryan D, Levin W. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168–176 [DOI] [PubMed] [Google Scholar]
- 29. Holmes DE, Risso C, Smith JA, Lovley DR. 2012. Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. ISME J. 6:146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weisberg WG, Barns SM, Pelletier BA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 32. Holmes DE, Nevin KP, O'Neil RA, Ward JE, Adams LA, Woodard TL, Vrionis HA, Lovley DR. 2005. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl. Environ. Microbiol. 71:6870–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 34. Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065–2075 [DOI] [PubMed] [Google Scholar]
- 35. Inoue K, Leang C, Franks AE, Woodard TL, Nevin KP, Lovley DR. 2011. Specific localization of the c-type cytochrome OmcZ at the anode surface in current-producing biofilms of Geobacter sulfurreducens. Environ. Microbiol. Rep. 3:211–217 [DOI] [PubMed] [Google Scholar]
- 36. Richter H, Nevin KP, Jia H, Lowy DA, Lovley DR, Tender LM. 2009. Cyclic voltammetry of biofilms of wild type and mutant Geobacter sulfurreducens on fuel cell anodes indicates possible roles of OmcB, OmcZ, type IV pili, and protons in extracellular electron transfer. Energy Environ. Sci. 2:506–516 [Google Scholar]
- 37. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bork P, Holm L, Sander C. 1994. The immunoglobulin fold: structural classification, sequence patterns, and common core. J. Mol. Biol. 242:309–320 [DOI] [PubMed] [Google Scholar]
- 39. Holmes DE, O'Neil RA, Chavan MA, N′Guessan LA, Vrionis HA, Perpetua LA, Larrahondo MJ, DiDonato R, Liu A, Lovley DR. 2009. Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J. 3:216–230 [DOI] [PubMed] [Google Scholar]
- 40. Rollefson JB, Levar CE, Bond DR. 2009. Identification of genes involved in biofilm formation and respiration via mini-Himar transposon mutagenesis of Geobacter sulfurreducens. J. Bacteriol. 191:4207–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rollefson JB, Stephen CS, Tien M, Bond DR. 2011. Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J. Bacteriol. 193:1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burrows LL, Charter DF, Lam JS. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481–495 [DOI] [PubMed] [Google Scholar]
- 43. Daniels C, Morona R. 1999. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34:181–194 [DOI] [PubMed] [Google Scholar]
- 44. Guo H, Kong Q, Cheng J, Wang L, Feng L. 2005. Characterization of the Escherichia coli O59 and O155 O-antigen gene clusters: the atypical wzx genes are evolutionary related. FEMS Microbiol. Lett. 248:153–161 [DOI] [PubMed] [Google Scholar]
- 45. Guo H, Yi W, Shao J, Lu Y, Zhang W, Song J, Wang PG. 2005. Molecular analysis of the O-antigen gene cluster of Escherichia coli O86:B7 and characterization of the chain length determinant gene (wzz). Appl. Environ. Microbiol. 71:7995–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vinogradov E, Korenevsky A, Lovley DR, Beveridge TJ. 2004. The structure of the core region of the lipopolysaccharide from Geobacter sulfurreducens. Carbohydr. Res. 339:2901–2904 [DOI] [PubMed] [Google Scholar]
- 47. Faridmoayer A, Fentabil MA, Mills DC, Klassen JS, Feldman MF. 2007. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J. Bacteriol. 189:8088–8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Horzempa J, Dean CR, Goldberg JB, Castric P. 2006. Pseudomonas aeruginosa 1244 pilin glycosylation: glycan substrate recognition. J. Bacteriol. 188:4244–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Childers SE, Ciufo S, Lovley DR. 2002. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416:767–769 [DOI] [PubMed] [Google Scholar]
- 50. Ueki T, Leang C, Inoue K, Lovley DR. 2012. Identification of multicomponent histidine-aspartate phosphorelay system controlling flagellar and motility gene expression in Geobacter species. J. Biol. Chem. 287:10958–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72:7345–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.