Abstract
Mangotoxin production was first described in Pseudomonas syringae pv. syringae strains. A phenotypic characterization of 94 P. syringae strains was carried out to determine the genetic evolution of the mangotoxin biosynthetic operon (mbo). We designed a PCR primer pair specific for the mbo operon to examine its distribution within the P. syringae complex. These primers amplified a 692-bp DNA fragment from 52 mangotoxin-producing strains and from 7 non-mangotoxin-producing strains that harbor the mbo operon, whereas 35 non-mangotoxin-producing strains did not yield any amplification. This, together with the analysis of draft genomes, allowed the identification of the mbo operon in five pathovars (pathovars aptata, avellanae, japonica, pisi, and syringae), all of which belong to genomospecies 1, suggesting a limited distribution of the mbo genes in the P. syringae complex. Phylogenetic analyses using partial sequences from housekeeping genes differentiated three groups within genomospecies 1. All of the strains containing the mbo operon clustered in groups I and II, whereas those lacking the operon clustered in group III; however, the relative branching order of these three groups is dependent on the genes used to construct the phylogeny. The mbo operon maintains synteny and is inserted in the same genomic location, with high sequence conservation around the insertion point, for all the strains in groups I and II. These data support the idea that the mbo operon was acquired horizontally and only once by the ancestor of groups I and II from genomospecies 1 within the P. syringae complex.
INTRODUCTION
The bacterial plant pathogen Pseudomonas syringae is ubiquitous in nature and often causes economically important plant diseases. This bacterial species establishes an epiphytic population in association with a plant host's surfaces prior to infection. There are at least 50 pathovars, many of which cause a wide range of plant diseases that exhibit diverse symptoms, such as leaf or fruit lesions, cankers, blasts, and galls (1–7).
P. syringae produces several type III effectors and virulence factors, but the role of its toxins is particularly significant during symptom development (1, 8–10). The current study is focused on mangotoxin, which inhibits the enzyme ornithine N-acetyltransferase (11, 12). Mangotoxin was initially detected in strains from P. syringae pv. syringae (11); however, its production was recently observed in strains of pathovar avellanae (13). Recent studies have reported the involvement of the mgo (8, 12, 14) and mbo (15) operons in mangotoxin production and P. syringae pv. syringae virulence. The mbo operon is composed of six genes, and all of them are directly involved in mangotoxin production.
The development of specific methods for pathogen detection in planta is very important. In this sense, different PCR methods have been developed and improved to detect different P. syringae pathovars. For example, one PCR method used primers that were based on the 16S-23S ribosomal genes to identify strains of P. syringae pv. actinidae in which the PCR resulted in a specific amplicon of this pathovar (16). A multiplex PCR assay has been developed for the identification and differentiation of P. fragi, P. lundensis, and P. putida on the basis of the coamplification of different carbamoyl phosphate synthase small-subunit gene fragments (17). Another example is the amplification of the gene iaaL, yielding a 454-bp fragment that allows the identification and detection of P. syringae pv. savastanoi strains (18). In the current study, we have developed a sensitive and specific method for the detection of mangotoxin-producing strains by a PCR that is based on the amplification of an mbo operon region in P. syringae. The specificity of the mbo operon within mangotoxin biosynthesis was essential for the planning and development of this method (15).
Additionally, the study of specific genes for the biosynthesis and production of virulence factors, as in the case of the mbo operon, could also be used in phylogenetic studies. Sawada et al. (19) revealed a remarkable degree of congruence between two housekeeping genes (gyrB and rpoD) and two components of the pathogenesis-associated type III secretion system (hrpS and hrpL), leading to the conclusion that the type III secretion system was acquired prior to the diversification of the P. syringae pathovars. The diversity of P. syringae strains has been further explored by physical mapping of the ribosomal gene cluster, revealing that the size and structure of P. syringae genomes vary greatly by pathovar and that large-scale genomic rearrangements are common (20). In this study, a phylogenetic analysis using housekeeping genes has enabled us to establish a basis for the comparison and study of the mbo operon across a group of strains belonging to different pathovars of P. syringae. Characterizing the allocation and internal organization of the mbo operon has also been realized to obtain information about the evolutionary history of mbo genes in the P. syringae complex. Moreover, sequencing of the mbo operon in four strains belonging to pathovar syringae was carried out to perform a comparison.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this work are described in Table 1. Escherichia coli CECT831 (Colección Española de Cultivos Tipo, Spain) was used as the indicator strain for antimetabolite toxin production and was cultured at 37°C using Luria-Bertani medium (41). P. syringae was routinely propagated at 28°C using King's medium B (42).
Table 1.
Production of antimetabolite toxins and possession of the mbo operon by a collection of P. syringae strains
| P. syringae pathovar and strain | Host and place of isolation | Source or referencea | Antimetabolite toxin producedb |
mbo operonc |
|
|---|---|---|---|---|---|
| Detection | Insertion site | ||||
| Cit7d* | Orange, USA | Baltrus et al. (21) | ND | + | + |
| aceris MAFF302273* | Maple, USA | Baltrus et al. (21) | ND | − | − |
| actinidae | |||||
| NCPPB3738 | Kiwi, Japan | Murillo et al. (13) | Phaseolotoxin | − | − |
| NCPPB3739 | Kiwi, Japan | Murillo et al. (13) | Phaseolotoxin | − | − |
| NCPPB3740 | Kiwi, Japan | Murillo et al. (13) | Phaseolotoxin | − | − |
| NCPPB3871 | Kiwi, Italy | Murillo et al. (13) | Phaseolotoxin | − | − |
| MAFF302091* | Kiwi, Japan | Baltrus et al. (21) | ND | − | − |
| aesculi | |||||
| 2250* | Chestnut, UK | Green et al. (22) | ND | − | − |
| NCPPB3681* | Chestnut, India | Green et al. (22) | ND | − | − |
| aptata | |||||
| DSM50252 | Sugar beet, USA | Baltrus et al. (21) | − | + | + |
| LMG5059 | Sugar beet, USA | BCCM/LMG | − | + | + |
| LMG5532 | Sugar beet, Italy | BCCM/LMG | − | + | + |
| LMG5646 | Sugar beet, New Zealand | BCCM/LMG | − | + | + |
| avellanae | |||||
| ISPaVe011 | Hazelnut, Italy | Murillo et al. (13) | Mangotoxin | + | + |
| ISPaVe2056 | Hazelnut, Italy | Murillo et al. (13) | Mangotoxin | + | + |
| coronafaciens | |||||
| CECT4389 | Oat, Canada | Arrebola et al. (11) | Tabtoxin | − | − |
| ICMP3113* | Oat, UK | Yamamoto et al. (21) | ND | − | − |
| glycinea | |||||
| A29-2* | Soybean, | Baltrus et al. (21) | ND | − | − |
| NCPPB2411* | Soybean, New Zealand | Yamamoto et al. (23) | ND | − | − |
| japonica MAFF301072* | Barley, Japan | Baltrus et al. (21) | ND | + | + |
| lachrymans | |||||
| MAFF301315* | Cucumber, Japan | Baltrus et al. (21) | ND | − | − |
| MAFF302278* | Cucumber, USA | Baltrus et al. (21) | ND | − | − |
| maculicola ICMP 3935* | Broccoli, New Zealand | Yamamoto et al. (23) | ND | − | − |
| mori MAFF301020* | Mulberry, Japan | Baltrus et al. (21) | ND | − | − |
| morsprunorum MAFF302280* | Plum, Japan | Baltrus et al. (21) | ND | − | − |
| oryzae 1_6* | Rice, Japan | Reinhardt et al. (24) | ND | − | − |
| phaseolicola | |||||
| 1448A | Bean, Ethiopia | Teverson (25) | Phaseolotoxin | − | − |
| CYL314 | Bean, Spain | Rico et al. (26) | − | − | − |
| pisi | |||||
| 1704B | Pea, France | Baltrus et al. (21) | Mangotoxin | + | + |
| HRI203 | Pea, New Zealand | Yamamoto et al. (23) | Mangotoxin | + | + |
| NCPPB1365 | Pea, Canada | NCPPB | Mangotoxin | + | + |
| savastanoi | |||||
| NCPPB3335 | Olive, France | Pérez-Martínez et al. (27) | − | − | − |
| NCPPB639* | Olive, Yugoslavia | Yamamoto et al. (23) | ND | − | − |
| syringae | |||||
| 7A7 | Ornamental pear, USA | Sundin and Bender (28) | − | − | − |
| 7C6 | Ornamental pear, USA | G. W. Sundin | Mangotoxin | + | + |
| 7B12 | Ornamental pear, USA | G. W. Sundin | − | − | − |
| 7B40 | Ornamental pear, USA | Sundin and Bender (28) | Mangotoxin | + | + |
| 7D46 | Ornamental pear, USA | G. W. Sundin | − | − | − |
| 7F29 | Ornamental pear, USA | Sundin et al. (29) | Mangotoxin | + | + |
| 8B48 | Ornamental pear, USA | Sundin et al. (29) | − | − | − |
| 8C32 | Ornamental pear, USA | Sundin et al. (29) | Mangotoxin | + | + |
| 8C43 | Ornamental pear, USA | Sundin et al. (29) | − | − | − |
| 8F21 | Ornamental pear, USA | Sundin et al. (29) | Mangotoxin | + | + |
| 1444-5 | Laurel, Spain | Arrebola et al. (11) | − | + | + |
| 1507-7 | Hawthorn, Spain | Arrebola et al. (11) | − | − | − |
| 1559-9 | Mango, Spain | Arrebola et al. (11) | Mangotoxin | + | + |
| B728a | Bean, USA | Feil et al. (30) | − | − | − |
| CECT127 | Lilac, UK | CECT | Mangotoxin | + | + |
| CECT4429 | Lilac, UK | CECT | Mangotoxin | + | + |
| CFBP3388 | Vetch, France | Tourte and Manceau (31) | Mangotox./phaseolotox. | + | + |
| CRD 09-87 | Chickling pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| DAR77787 | Mango, Australia | Young (33) | Mangotoxin | + | + |
| DAR77789 | Mango, Australia | Young (33) | Mangotoxin | + | + |
| EPSMV3 | Pear, Spain | Arrebola et al. (11) | − | − | − |
| EPS17A | Pear, Spain | Arrebola et al. (11) | Mangotoxin | + | + |
| FF5 | Ornamental pear, USA | Sohn et al. (34) | − | + | + |
| ITACyL 488 | Vetch, Spain | Martín-Sanz et al. (35) | Unknown | − | − |
| ITACyL 522 | Chickling pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| ITACyL 523 | Chickling pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| ITACyL 524 | Chickling pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| ITACyL 525 | Chickling pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| ITACyL 526 | Grass pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| ITACyL 527 | Grass pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| ITACyL 528 | Grass pea, Spain | Martín-Sanz et al. (32) | − | − | − |
| ITACyL 529 | Vetch, Spain | Martín-Sanz et al. (32) | − | − | − |
| NCPPB1239 | Bean, Kenya | NCPPB | Mangotoxin | + | + |
| Ps-5 | Mango, Israel | UMA-LC | Mangotoxin | + | + |
| Ps-6 | Mango, Israel | Arrebola et al. (11) | Mangotoxin | + | + |
| Ps-10 | Mango, Israel | Arrebola et al. (11) | Mangotoxin | + | + |
| Ps-35 | Mango, Israel | Arrebola et al. (11) | Mangotoxin | + | + |
| UMAF0049 | Mango, Spain | Cazorla et al. (36) | Mangotoxin | + | + |
| UMAF0081 | Mango, Spain | Cazorla et al. (36) | Mangotoxin | + | + |
| UMAF0158 | Mango, Spain | Cazorla et al. (36) | Mangotoxin | + | + |
| UMAF0167 | Mango, Spain | Cazorla et al. (36) | − | + | + |
| UMAF0170 | Mango, Spain | Cazorla et al. (36) | Mangotoxin | + | + |
| UMAF0176 | Mango, Spain | Arrebola et al. (11) | Mangotoxin | + | + |
| UMAF0209 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0214 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0217 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0220 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0221 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0222 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0223 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0225 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF0226 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF1003 | Mango, Spain | Arrebola et al. (11) | Mangotoxin | + | + |
| UMAF1060 | Mango, Spain | Gutiérrez-Barranquero et al. (37) | Mangotoxin | + | + |
| UMAF2007 | Mango, Portugal | Arrebola et al. (11) | Mangotoxin | + | + |
| UMAF2008 | Mango, Portugal | UMA-LC | Mangotoxin | + | + |
| UMAF2025 | Mango, Portugal | Cazorla et al. (36) | Mangotoxin | + | + |
| UMAF2026 | Mango, Portugal | Cazorla et al. (36) | Mangotoxin | + | + |
| UMAF2676 | Bean, South Africa | Arrebola et al. (11) | − | − | − |
| UMAF2700 | Mango, Italy | UMA-LC | Mangotoxin | + | + |
| UMAF2702 | Mango, Italy | Gutiérrez-Barranquero et al. (37) | Mangotoxin | + | + |
| UMAF2801 | Mango, Spain | Gutiérrez-Barranquero et al. (37) | Mangotoxin | + | + |
| UMAF2802 | Mango, Spain | Gutiérrez-Barranquero et al. (37) | Mangotoxin | + | + |
| UMAF2805 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF2808 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF2811 | Mango, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF3028 | Mango, Spain | Arrebola et al. (11) | Mangotoxin | + | + |
| UMAF4002 | Tomato, Spain | Arrebola et al. (11) | Mangotoxin | + | + |
| UMAF6024 | Purple phlomis, Spain | UMA-LC | Mangotoxin | + | + |
| UMAF6016 | Chestnut, Spain | Arrebola et al. (11) | − | − | − |
| UMAF6582 | Peach, Spain | Arrebola et al. (11) | − | − | − |
| tabaci ATCC 11528 | Tobacco, USA | ATCC | Tabtoxin | − | − |
| tagetis CECT4430 | Marigold, Zimbabwe | CECT | − | − | − |
| tomato | |||||
| DC3000 | Tomato, UK | Moore et al. (38) | − | − | − |
| DCT6D1 | Tomato, Canada | Moore et al. (38) | Unnamed | − | − |
| K40* | Tomato, USA | Cai et al. (39) | ND | − | − |
| NCPPB1108* | Tomato, UK | Cai et al. (39) | ND | − | − |
| T1* | Tomato, Canada | Almeida et al. (40) | ND | − | − |
| UMAF4007 | Tomato, Spain | Arrebola et al. (11) | Unnamed | − | − |
| UMAF6018 | Tomato, Spain | Arrebola et al. (11) | Unnamed | − | − |
ATCC, American Type Culture Collection; CECT, Colección Española de Cultivos Tipo, Spain; CFBP, Collection Française de Bactéries Associées aux Plantes, France; DSM, German Collection of Microorganisms and Cell Cultures, Germany; BCCM/LMG, Belgian Coordinated Collections of Microorganisms, Belgium; NCPPB, National Collection of Plant Pathogenic Bacteria, United Kingdom; ICMP, International Collection of Microorganisms from Plants, New Zealand; MAFF, Ministry of Agriculture, Forestry and Fisheries, Japan; UMA-LC, IHSM-UMA-CSIC, University of Málaga, Microbiology and Plant Pathology Laboratory Collection.
ND, not determined; −, antimetabolite toxins not detected; Mangotox./phaseolotox., mangotoxin/phaseolotoxin.
The presence or absence (+ and −, respectively) of the mbo operon was determined by specific PCR amplification or from bioinformatic analyses of the published genomes from the NCBI (strains marked *); the operon was in all cases inserted in the same genomic location as in strain P. syringae pv. syringae UMAF0158.
No pathovar designation.
Detection of P. syringae antimetabolite toxin production.
Antimetabolite toxin production was assayed using the previously described indicator technique (43, 44) that evaluates growth inhibition of E. coli on Pseudomonas minimal medium (PMS). Briefly, P. syringae strains to be assayed were stabbed onto a double layer of the E. coli strain CECT831 indicator bacterium and incubated at 22°C for 24 h, followed by an additional 24 h of incubation at 37°C. The production of an antimetabolite toxin was deduced from the appearance of inhibition haloes around the P. syringae strains, and the identity of the biochemical step that was inhibited by the antimetabolite toxin was confirmed by reversion of the haloes in separate PMS plates containing 100 μl of a 100 mM solution of the corresponding amino acids, l-glutamate, N-acetyl-l-glutamate, N-acetyl-ornithine, l-citrulline, and l-arginine (Fig. 1).
Fig 1.
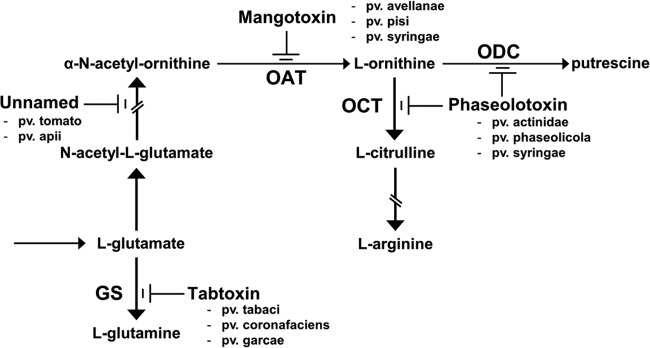
Schematic representation of the arginine-glutamine and polyamine biosynthesis pathways. The enzymatic targets that are inhibited by antimetabolite toxins mangotoxin, phaseolotoxin, tabtoxin, and an unnamed phytotoxin that was produced by different P. syringae pathovars are shown. Target enzymes have the following abbreviations: GS, glutamine synthetase; OAT, ornithine N-acetyltransferase; OCT, ornithine carbamoyltransferase; and ODC, ornithine decarboxylase. The pathovars of P. syringae producing each toxin are indicated.
General molecular techniques.
Standard molecular biology techniques were used for experimentation (41). Genomic DNA was prepared using a Jet-Flex genomic DNA purification kit (Genomed) following the manufacturer's instructions. DNA concentrations and quality were determined with a NanoDrop ND-1000 (NanoDrop Technologies) spectrophotometer and by agarose gel electrophoresis.
DNA sequence analysis and primer design.
Multiple-sequence alignments were constructed with the program ALIGN X of the Vector NTI suite (version 9.0; InforMax). Primers for conserved regions of the selected target genes were designed using Primer3 software (45).
PCR amplification was done using GoTaq DNA polymerase (Promega); amplification conditions consisted of an initial denaturation step at 94°C for 1 min, followed by 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min and a final extension step at 72°C for 7 min in a Techne TC-412 thermal cycler. Amplification products were analyzed by agarose gel electrophoresis and stained with ethidium bromide.
The conservation of the mbo operon insertion point in different P. syringae strains was examined by PCR using primers (mboIS-For/mboIS-Rev; see Table S1 in the supplemental material) that overlapped the right operon border. In the same way, the mbo operon synteny was determined using the three primer pairs mboAB-For/mboC-Rev, mboC-For/mboDE-Rev, and mboDE-For/mboF-Rev (see Table S1 in the supplemental material), which amplified overlapping DNA fragments that cover the whole mbo operon (see Fig. 7).
Fig 7.
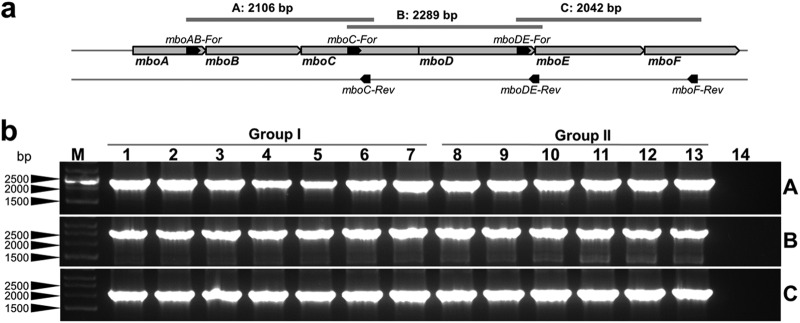
Internal organization of the mbo operon. (a) Schematic diagrams of the overlapping primers used for determining the organization of the mbo operon. The size of each amplicon is also shown. (b) Results of amplification in the selected strains belonging to groups I and II. P. syringae pv. syringae B728a (a non-mangotoxin producer) was included as a negative control. Lanes correspond to the following: 1, P. syringae pv. syringae 7C6; 2, P. syringae pv. syringae DAR77787; 3, P. syringae pv. syringae EPS17A; 4, P. syringae pv. syringae NCPPB1239; 5, P. syringae pv. syringae UMAF4002; 6, P. syringae pv. syringae UMAF0167; 7, P. syringae pv. syringae UMAF0158; 8, P. syringae pv. aptata DSM50252; 9, P. syringae pv. avellanae ISPaVe2056; 10, P. syringae pv. pisi NCPPB1365; 11, P. syringae pv. syringae 1444-5; 12, P. syringae pv. syringae FF5; 13, P. syringae pv. syringae CFBP3388; 14, P. syringae pv. syringae B728a; M, molecular size marker (HyperLadder I, Bioline).
Phylogenetic analysis.
A phylogenetic analysis based on partial rpoD and gyrB sequences was done using sequences of Pseudomonas spp. that were available in the NCBI database and generated by us from the P. syringae strains described in Table 1. Primers rpoDFor2/rpoDRev2 and gyrBFor2/gyrBRev2 (13) or rpoDFor3/rpoDRev3 and gyrBFor3/gyrBRev3 (see Table S1 in the supplemental material) were used for rpoD and gyrB amplification, and purified amplicons were sequenced by Macrogen. Partial sequences of rpoD (807 bp) and gyrB (890 bp) were concatenated, resulting in an alignment of 1,697 sites.
Other phylogenetic analyses were done using concatenated partial or complete sequences available in the databases of the housekeeping genes rpoD and gyrB (4,032 sites) and gltA, pgi, recA, and rpoD (5,871 sites); the partial sequence of the mgoA gene (2,582 sites), which encodes a nonribosomal peptide synthetase involved in mangotoxin production (12); or the partial sequences of the mbo genes, which are involved in mangotoxin biosynthesis (15). Searches for sequence similarity in the NCBI databases were carried out using BLAST algorithms (46). Genome and nucleotide sequences were visualized and manipulated using the Artemis genome browser (47) and compared using the Artemis comparison tool (ACT) (48) in combination with WebACT (49). Concatenated sequences from the same strain were treated as a single sequence; multiple-sequence alignments were done using the Muscle program, and determination of the optimal nucleotide substitution model and phylogenetic tree construction were done using MEGA5 software (50). P. fluorescens Pf-5 or P. syringae pv. oryzae 1_6 was used as the outgroup. Neighbor-joining, maximum-parsimony, and maximum-likelihood phylogenetic trees of the individual gene sequences and concatenated data sets were generated using MEGA5 with the optimal model parameters and the option of complete deletion to eliminate positions containing gaps. Confidence levels for the branching points were determined using 1,000 bootstrap replicates.
Nucleotide sequence accession numbers.
All gltA, gyrB, pgi, recA, and rpoD gene sequences were deposited in GenBank under accession numbers JX867777 to JX867781, JX867783 to JX867860, JX867862 to JX867932, and JX878889 to JX878894. Sequences of the mbo operons and mgoA genes were deposited in GenBank under accession numbers JX878400 to JX878403.
RESULTS
Genes for mangotoxin production are restricted to some pathovars of genomospecies 1.
Antimetabolite toxin production by the 94 P. syringae strains representing 20 pathovars and 6 genomospecies listed in Table 1 was evaluated with an E. coli in vitro inhibition assay (43, 44). Mangotoxin production was detected in diverse strains of P. syringae pv. avellanae, pisi, and syringae (Table 1), confirming the earlier descriptions of pathovars syringae (11) and avellanae (13) as mangotoxin producers. Three pathovar tomato strains produced an unnamed antimetabolite toxin (Fig. 1) that inhibits the conversion of N-acetyl-l-glutamate to N-acetyl-ornithine (11, 51). Additionally, strain P. syringae pv. syringae ITACyL 488, which was isolated from vetch, produced inhibition haloes in the E. coli inhibition test that could not be reverted with any of the tested amino acids. Therefore, this strain might be producing a new toxin with characteristics that remain to be determined.
The phenotypic identification of mangotoxin-producing strains by the E. coli inhibition assay is cumbersome and time-consuming. We therefore developed a PCR method to detect strains that harbored the mbo operon, which is essential for mangotoxin production (15). Among 20 primer pairs assayed that covered single or adjacent genes of the mbo operon, we selected pair mbo24-For/mbo24-Rev (see Table S1 in the supplemental material) to amplify a 692-bp fragment overlapping genes mboA and mboB because it produced a strong and specific band, with no spurious banding (data not shown). We then detected the presence or absence of the mbo operon using primer pair mbo24-For/mbo24-Rev in 94 strains belonging to 20 P. syringae pathovars and 6 genomospecies. We obtained a 692-bp specific amplicon for 59 of the analyzed strains, which belonged to 5 pathovars; all of them were part of genomospecies 1 (Fig. 2 and 3; Table 1). However, only 52 of these strains produced mangotoxin in the E. coli inhibition test: 4 P. syringae pv. aptata strains and 3 P. syringae pv. syringae strains (UMAF0167, 1444-5, and FF5) yielded a strong amplification band (Fig. 2) but did not produce mangotoxin.
Fig 2.
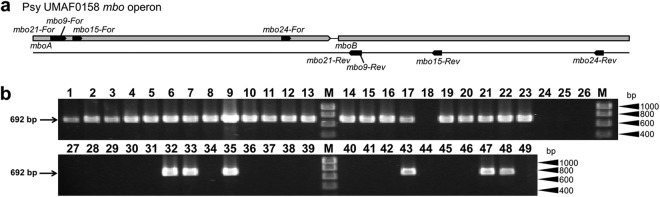
Detection by PCR of a specific mangotoxin biosynthesis operon sequence in different P. syringae genomospecies 1 strains. (a) Schematic representation of the four primer pairs designed for mbo operon detection in P. syringae strains. (b) Gel electrophoresis showing the specific 692-bp product generated with primers mbo24-For and mbo24-Rev from strains harboring the mbo operon. Lanes correspond to the following: 1, P. syringae pv. syringae 7C67; 2, P. syringae pv. syringae 7F29; 3, P. syringae pv. syringae 8F21; 4, P. syringae pv. syringae 1559-9; 5, P. syringae pv. syringae CECT127; 6, P. syringae pv. syringae CECT4429; 7, P. syringae pv. syringae CFBP3388; 8, P. syringae pv. syringae DAR77787; 9, P. syringae pv. syringae DAR77789; 10, P. syringae pv. syringae EPS17A; 11, P. syringae pv. syringae NCPPB1239; 12, P. syringae pv. syringae Ps-5; 13, P. syringae pv. syringae UMAF0081; 14, P. syringae pv. syringae UMAF0214; 15, P. syringae pv. syringae UMAF0225; 16, P. syringae pv. syringae UMAF1060; 17, P. syringae pv. syringae UMAF6024; 18, P. syringae pv. syringae UMAF6016; 19, P. syringae pv. pisi 1704B; 20, P. syringae pv. pisi HRI203; 21, P. syringae pv. pisi NCPPB1365; 22, P. syringae pv. avellanae ISPaVe011; 23, P. syringae pv. avellanae ISPaVe2056; 24, P. syringae pv. syringae 7D46; 25, 8B48; 26, P. syringae pv. syringae 8C43; 27, P. syringae pv. syringae ITACyL 488; 28, P. syringae pv. syringae ITACyL 521; 29, P. syringae pv. syringae ITACyL 523; 30, P. syringae pv. syringae ITACyL 526; 31, P. syringae pv. syringae ITACyL 529; 32, P. syringae pv. syringae 1444-5; 33, P. syringae pv. syringae FF5; 34, P. syringae pv. syringae EPSMV3; 35, P. syringae pv. syringae UMAF0167; 36, P. syringae pv. tomato DC3000; 37, P. syringae pv. tomato DCT6D1; 38, P. syringae pv. tomato UMAF4007; 39, P. syringae pv. tomato UMAF6018; 40, P. syringae pv. coronafaciens CECT4389; 41, P. syringae pv. phaseolicola 1448A; 42, P. syringae pv. phaseolicola CYL314; 43, P. syringae pv. aptata DSM50252; 44, P. syringae pv. syringae 1507-7; 45, P. syringae pv. syringae B728a; 46, P. syringae pv. syringae CECT4430; 47, P. syringae pv. syringae 0049; 48, P. syringae pv. syringae UMAF0158; 49, P. syringae pv. tabaci ATCC 11528; M, molecular size marker (HyperLadder I; Bioline).
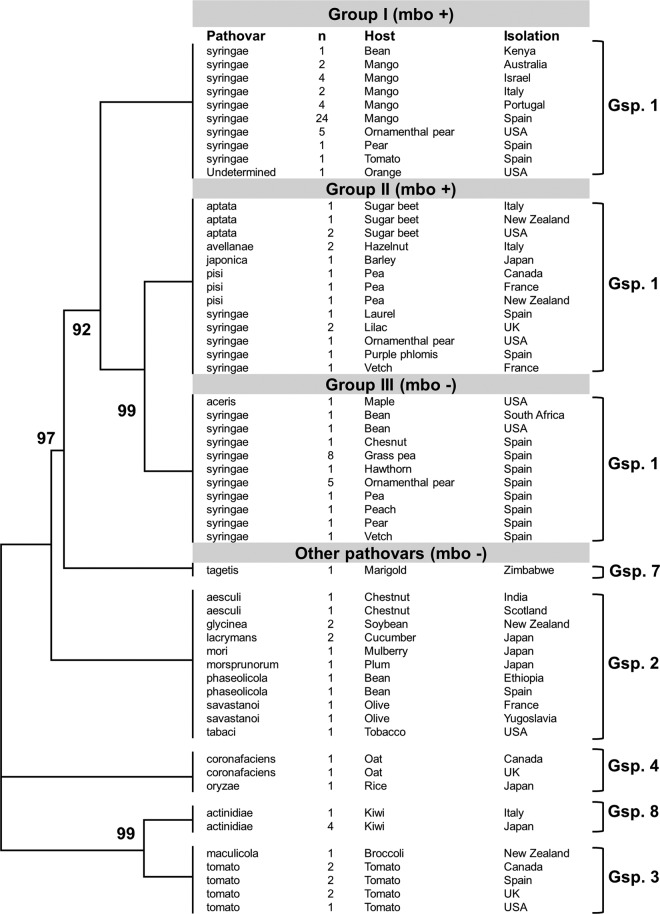
Fig 3.
Schematic neighbor-joining tree of P. syringae strains belonging to different pathovars and harboring or not the mbo genes (mbo + and mbo −, respectively) for mangotoxin production. The neighbor-joining tree was constructed with MEGA5 using the concatenated partial sequences of rpoD and gyrB. The Tamura-Nei substitution model with gamma correction was used for tree construction. Bootstrap values (1,000 repetitions) are shown on each branch. One hundred thirteen strains of different P. syringae pathovars were analyzed. The pathovar, host, and country of isolation are presented on the right, together with the number of strains (indicated by n) and the genomospecies (Gsp.) to which they belong, according to Gardan et al. (52). Evolutionary distances are given in units of nucleotide substitutions per site. The topology was similar among trees produced by the maximum-parsimony and maximum-likelihood methods. Sequences of rpoD and gyrB were extracted from published genome sequences or were sequenced for this work. For more detailed information, see Fig. S1 in the supplemental material.
Additionally, we did not detect a specific amplicon or mangotoxin production in 35 strains (Fig. 2b; Table 1) belonging to the pathovars actinidae (n = 4), coronafaciens (n = 1), phaseolicola (n = 2), savastanoi (n = 1), syringae (n = 21), tabaci (n = 1), tagetis (n = 1), and tomato (n = 4). Furthermore, a BLAST search of sequenced P. syringae genomes belonging to 17 pathovars (http://www.pseudomonas-syringae.org/) (21) revealed the presence of the mbo operon in strains Cit7 (without pathovar assignation) and MAFF301072 (P. syringae pv. japonica).
In summary, the characterization of a large collection of P. syringae strains belonging to 20 pathovars (Table 1) by phenotypic assays, PCR amplification, and/or analysis of their sequenced genomes disclosed the presence of the mbo operon for mangotoxin biosynthesis in strains from five pathovars (pathovars aptata, avellanae, japonica, pisi, and syringae), all of which belong to genomospecies 1 (52).
Elucidation of the mbo operon evolutionary history.
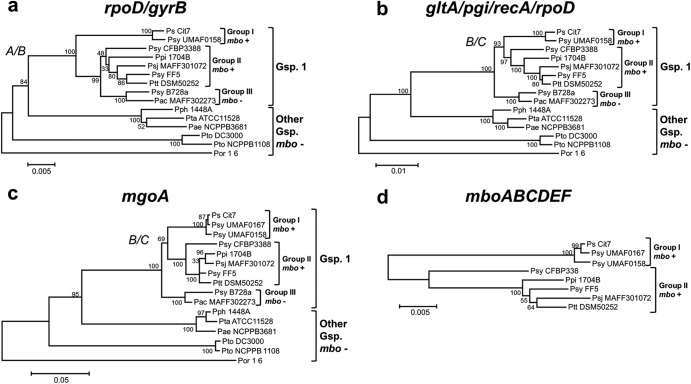
To study the mbo operon evolutionary history within the P. syringae complex, we examined the phylogeny of diverse strains using concatenated sequences of internal fragments of housekeeping genes rpoD and gyrB (total, 1,697 sites). Trees obtained by diverse clustering methods showed the same topology (Fig. 3 and 4a; see Fig. S1 in the supplemental material), and the strains of genomospecies 1 were clustered in three groups within all trees, two of which contained all of the strains harboring the mbo operon. Among the mangotoxin-producing strains, group I contained 45 strains of P. syringae pv. syringae, which were isolated from woody hosts, mainly mangoes, whereas group II contained 16 strains from the pathovars aptata, avellanae, japonica, pisi, and syringae, which were isolated from diverse woody and herbaceous hosts. Group III clustered all the strains from genomospecies 1 that did not contain the mbo operon, which included 1 pathovar aceris strain plus 21 pathovar syringae strains. Finally, the remaining strains comprised 30 non-mangotoxin producers from different genomospecies that clustered separately. In this phylogenetic analysis, group III diverged after the separation of groups I and II, suggesting that the mbo operon was acquired once by the common ancestor of groups I, II, and III and later lost in group III. An alternative explanation is that the mbo operon was acquired independently by the putative ancestors of groups I and II (Fig. 3 and 4a; see Fig. S1 in the supplemental material). To confirm this result, we performed further phylogenetic analyses using additional housekeeping genes (Fig. 4b to d). For these analyses, we used only those strains with an available genome sequence plus two pathovar syringae strains representative of group I (UMAF0158) and group II (CFBP3388).
Fig 4.
Phylogeny of toxigenic and nontoxigenic P. syringae pv. syringae strains using diverse nucleotide sequences. Neighbor-joining trees were constructed with MEGA5 using the concatenated complete sequences of rpoD and gyrB (a), concatenated partial sequences of gltA, pgi, recA, and rpoD (b), a partial sequence of gene mgoA (c), and concatenated partial sequences of the mboABCDEF genes (d); the substitution models Tamura 3-parameter (mgoA and mbo genes) and Tamura-Nei (other genes) with gamma correction, as indicated by the model analysis module of MEGA5, were employed. Bootstrap values (1,000 repetitions) are shown on branches. The mbo operon acquisition alternatives are marked as follows: A, mbo operon acquisition by the common ancestor of groups I, II, and III and loss in group III; B, mbo operon acquisition twice independently by groups I and II; C, mbo operon acquisition by the common ancestor of groups I and II. P. syringae pathovars are abbreviated as follows: Ps, P. syringae (no pathovar assigned); Pac, P. syringae pv. aceris; Pae, P. syringae pv. aesculi; Ptt, P. syringae pv. aptata; Psj, P. syringae pv. japonica; Ppi, P. syringae pv. pisi; Pph, P. syringae pv. phaseolicola; Psy, P. syringae pv. syringae; Pta, P. syringae pv. tabaci; Pto, P. syringae pv. tomato; Por, P. syringae pv. oryzae. The trees were rooted with P. syringae pv. oryzae 1_6. Evolutionary distances are given in units of number of nucleotide substitutions per site. The topology was similar for trees produced by the maximum-parsimony and maximum-likelihood methods. Sequences from some strains were extracted from published genome sequences.
In the different trees that were built using concatenated sequences of the gltA, pgi, recA, and rpoD genes (5,871 sites), a partial sequence of the mgoA gene (2,582 sites), or the mbo genes (5,114 sites), all strains clustered as in the gyrB-rpoD phylogenetic tree. However, and in contrast to the gyrB-rpoD tree, group III diverged before the separation of groups I and II. Because this topology has strong bootstrap support, the multilocus sequence typing and mgoA trees suggest that the mbo operon was acquired by groups I and II in either one or two separate acquisition events and after their separation from group III.
The mbo operon is highly conserved among the two phylogroups.
Phylogenetic analyses clearly indicated that the strains harboring the mbo operon cluster in two well-separated phylogroups, groups I and II (Fig. 3 and 4; see Fig. S1 in the supplemental material). We therefore examined whether the operon is conserved in organization, sequence, and physical location in both phylogroups in order to gain support for one of the possible scenarios explaining the evolutionary history of the mbo operon.
To further evaluate the conservation of the mbo operon in the two phylogroups, we obtained the complete sequence of this operon from P. syringae pv. syringae strains UMAF0158, UMAF0167 (both group I), and CFBP3388 (group II) (see Table S2 in the supplemental material) and compared them with the sequence of the operon from the draft genome sequence of strains P. syringae Cit7 (group I), and P. syringae pv. aptata DSM50252, pisi 1704B, and syringae FF5 (the last three strains are from group II) (see Table S2 in the supplemental material). Sequence analysis of the seven strains reveals that the mbo operon maintains synteny and is generally well conserved, ranging in size from 6,769 to 6,801 bp. The operon harbors six open reading frames (ORFs) and shows a 55 to 56% G+C content, which is lower than the average G+C content of the P. syringae pv. syringae B728a chromosome (59.23%) (30, 53). The mbo operon is highly conserved among strains of group I, with nucleotide identity values being higher than 99%; however, the nucleotide identity among strains of group II varies between 96.5 and 98.5%, which is not surprising, given that this group includes strains from different pathovars. Nevertheless, the mbo operon is well conserved when both groups are compared, with levels of nucleotide identity being higher than 94.2% in pair comparisons (see Table S2 in the supplemental material).
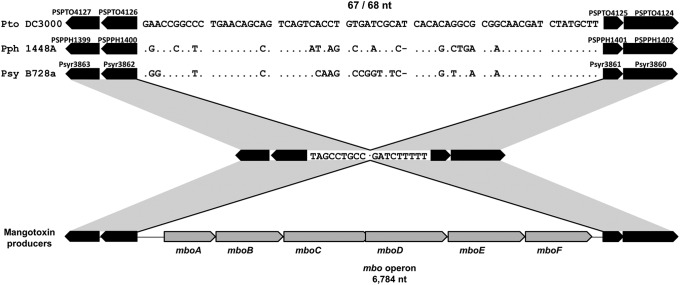
We examined the mbo operon point of insertion by PCR using a primer pair that overlaps the right border of the operon (Fig. 5). The amplification of DNA from 19 strains from group I and 15 strains from group II produced an amplicon of the same size for all strains (692 bp), whereas no amplification was observed for strains belonging to group III (Fig. 5). Additionally, the comparison of the mbo operon sequence from the seven strains indicated above (see Table S2 in the supplemental material) showed that the operon was inserted in the same location in strains from both groups I and II. A comparison of the nucleotide sequences from mangotoxin-producing and non-mangotoxin-producing strains indicates that the insertion of the mbo operon is correlated with the loss of a highly conserved 67- or 68-bp sequence (Fig. 6). This sequence is present in the closed genome sequences of P. syringae pv. phaseolicola 1448A, P. syringae pv. syringae B728a, and P. syringae pv. tomato DC3000 (Fig. 6) and is also conserved in sequence and position within the genomes of other P. syringae and P. fluorescens strains that do not produce mangotoxin (data not shown), indicating that this small fragment is ancestral to P. syringae and has been lost during the acquisition of the mbo operon. This evidence suggests that the mbo operon has never been acquired by strains from group III, which contain the 67-bp fragment.
Fig 5.
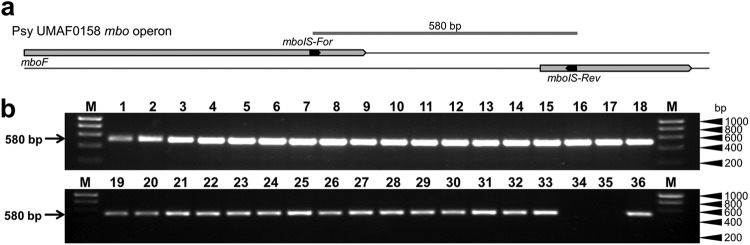
Mapping of the mbo operon insertion point in P. syringae strains. (a) Schematic representation of the PCR amplification strategy for the 3′ end of the mbo operon. Primers mboIS-For/mboIS-Rev (amplicon of 580-bp) were designed between the 3′ end of the mboF gene and the next ORF downstream of the mbo operon. (b) Representative results of amplification using 34 strains that harbor the mbo operon and two non-mangotoxin-producing strains that were used as negative controls. Lanes correspond to the following: 1, P. syringae pv. aptata DSM50252; 2, P. syringae pv. aptata LMG5059; 3, P. syringae pv. aptata LMG5532; 4, P. syringae pv. aptata LMG5646; 5, P. syringae pv. avellanae ISPaVe011; 6, P. syringae pv. avellanae ISPaVe2056; 7, P. syringae pv. pisi HRI203; 8, P. syringae pv. pisi NCPPB1365; 9, P. syringae pv. pisi 1704B; 10, P. syringae pv. syringae 7C6; 11, P. syringae pv. syringae 7B40; 12, P. syringae pv. syringae 7F29; 13, P. syringae pv. syringae 1444-5; 14, P. syringae pv. syringae 1559-9; 15, P. syringae pv. syringae CECT127; 16, P. syringae pv. syringae CECT4429; 17, P. syringae pv. syringae CFBP3388; 18, P. syringae pv. syringae DAR77787; 19, P. syringae pv. syringae EPS17A; 20, P. syringae pv. syringae FF5; 21, P. syringae pv. syringae NCPPB1239; 22, P. syringae pv. syringae Ps-10; 23, P. syringae pv. syringae UMAF0049; 24, P. syringae pv. syringae UMAF0081; 25, P. syringae pv. syringae UMAF0167; 26, P. syringae pv. syringae UMAF0209; 27, P. syringae pv. syringae UMAF0221; 28, P. syringae pv. syringae UMAF1060; 29, P. syringae pv. syringae UMAF2008; 30, P. syringae pv. syringae UMAF2700; 31, P. syringae pv. syringae UMAF2802; 32, P. syringae pv. syringae UMAF4002; 33, P. syringae pv. syringae UMAF6024; 34, P. syringae pv. phaseolicola 1448A; 35, P. syringae pv. syringae B728a; 36, P. syringae pv. syringae UMAF0158; M, molecular size marker (HyperLadder I, Bioline).
Fig 6.
mbo operon insertion site analysis. Alignment of the nucleotide sequences bordering the mbo operon with the corresponding sequences in non-mangotoxin-producing strains (P. syringae pv. phaseolicola 1448A, P. syringae pv. syringae B728a, and P. syringae pv. tomato DC3000). The nucleotide differences with respect to strain DC3000 are indicated, while dots indicate identical nucleotides and a hyphen shows an indel. The CDSs flanking the point of insertion of the mbo operon are shown in black, with indication of their locus tag numbers in the genomes of P. syringae pv. tomato DC3000, P. syringae pv. phaseolicola 1448A, and P. syringae pv. syringae B728a. nt, nucleotides.
Finally, the internal organization of the mbo operon was determined by PCR using three pairs of primers that produced overlapping amplicons. These primers yielded identical amplification products for all the analyzed strains from groups I and II (Fig. 7), indicating a widely conserved synteny. All of these data showed that the mbo operon was acquired only once during the evolutionary history of P. syringae, by a common ancestor of groups I and II.
DISCUSSION
In previous studies by our group, the production of mangotoxin was shown to be associated with P. syringae pv. syringae and to contribute to its virulence in tomato plants (8, 11). Recently, Murillo et al. (13) reported the production of mangotoxin by strains of pathovar avellanae, as well as the simultaneous production of two antimetabolite toxins, mangotoxin and phaseolotoxin, by strain P. syringae pv. syringae CFBP3388. In the current study, we evaluated the production of mangotoxin and other antimetabolite toxins in 94 strains belonging to different pathovars of the P. syringae complex. Besides pathovars avellanae and syringae, we detected production of mangotoxin in three strains of pathovar pisi, which is in contrast to previous analyses that did not find toxin-producing strains in the last pathovar (44, 51). This discrepancy could be due to several reasons, including differential analysis conditions that could impact toxin production; an inherent inability of previously analyzed strains to produce mangotoxin, in spite of having the biosynthesis genes, as we observed with strain FF5; or, since we examined here strains from only one of the two P. syringae pv. pisi lineages (54), the possibility that the previously analyzed strains belong to a phylogenetic lineage that does not produce toxins.
We designed a primer pair specific for the mbo operon that was used to screen a collection of 94 strains from 20 P. syringae pathovars. The primers were highly specific, yielding amplification only in strains that produced mangotoxin in an in vivo inhibition test or in strains that contained the mbo operon, confirming the usefulness of this primer pair. All of the strains isolated from lesions of apical necrosis of mango so far have been shown to produce mangotoxin during the inhibition bioassay (11, 55), supporting the idea that this primer pair could potentially be used for the detection of P. syringae pv. syringae in mango plants and for broader disease diagnosis. The PCR and the inhibition analyses, combined with the search for mangotoxin biosynthesis genes in the P. syringae genomes from the databases, allowed us to determine that the mangotoxin biosynthesis operon is present only in strains from P. syringae pv. aptata, avellanae, japonica, pisi, and syringae, all of which belong to genomospecies 1. The amplification of overlapping DNA fragments as well as the sequence comparisons showed that the composition, structure, and sequence of the mbo operon are highly conserved in all these pathovars. Although P. syringae pv. syringae FF5, UMAF0167, and UMAF1444-5 and the four P. syringae pv. aptata strains yielded the expected amplification products for the mbo operon or contained the mbo operon in their genome sequence, we did not detect production of mangotoxin in the in vivo inhibition test for any of the strains. This is in contrast to a previous report showing that three strains of pathovar aptata produced an antimetabolite toxin inhibiting the arginine/ornithine biosynthesis pathway (51). However, the enzymatic step(s) affected could not be identified because the reversion of inhibition with purified amino acids followed an atypical pattern, suggesting the production of more than one antimetabolite toxin or of a toxin inhibiting different enzymatic steps. Additionally, transformants of strains FF5 and UMAF0167 that contained the mbo operon were able to produce mangotoxin in the inhibition assay (15), suggesting that they do not contain any mutation outside the mbo operon that could prevent the biosynthesis or functionality of this toxin. Although we did not find premature stop codons in any of the coding DNA sequences of the mbo operon from the genomes of strains FF5 and DSM50252, it is still possible that these operons contain specific mutations that prevent the biosynthesis of an active toxin with respect to the operon in strain UMAF0158. This is not unlikely, because instances of nontoxigenic isolates containing a toxin biosynthesis cluster have already been described for P. syringae pathovars that produce phaseolotoxin or tabtoxin (2, 8, 26, 56).
To study the evolutionary history of the mbo operon, we performed independent phylogenetic analyses using diverse housekeeping genes, as well as the mgoA operon, which is essential for mangotoxin biosynthesis but is present in all examined P. syringae strains. All of these analyses were highly congruent and revealed the same major clades, which were in agreement with previous results of studies that used different combinations of housekeeping genes (19, 21, 57–59). In particular, strains of genomospecies 1 containing the mbo operon clustered in two well-defined groups that mirror the relative degree of host specialization found in P. syringae pv. syringae (60). From these, group I contains P. syringae pv. syringae strains that were mainly isolated from woody hosts, particularly mango, whereas group II is enriched in strains from herbaceous hosts belonging to diverse pathovars. An important inconsistency among the different phylogenetic trees was identified with respect to the branching order of groups I, II, and III (Fig. 4), which results in some uncertainty over the number of times that the mbo operon has been acquired by the strains of genomospecies 1. This type of discrepancy is commonly found in phylogenetic analyses of P. syringae when using different housekeeping genes or even different genes from an operon from the hrp cluster, particularly for very closely related taxa, and has been interpreted to be a consequence of genetic exchange and recombination (21, 59, 61–63). Examination of the mbo operon insertion site offers further significant clues about its evolutionary history, in that the insertion site was the same for all the strains and in all cases was associated with the loss of a continuous 67-bp fragment that is ancestral to P. syringae. The possibility that the mbo operon was acquired by an ancestor of genomospecies 1 and later lost by the ancestor of group III is difficult to maintain because this loss implies the concomitant reacquisition of the 67-bp fragment. Therefore, the simplest explanation implies that the mbo operon was acquired by a putative ancestor of groups I and II and has since evolved separately within both groups. Nevertheless, we cannot discount the less likely possibility that the operon was acquired by the ancestor of one of these two groups and then horizontally transferred to the ancestor of the other group, where the operon evolved independently. However, we could predict a very limited lateral transfer of the mbo operon because it is not linked to integrases, transposon sequences, or any other obvious repeated sequences; in contrast, the phaseolotoxin biosynthesis cluster from P. syringae pv. phaseolicola and P. syringae pv. actinidae is associated with integrases, which probably mediated its horizontal exchange between these two pathovars (13, 20, 64). This would suggest that the mbo operon was acquired by illegitimate recombination, a process that was shown to operate during the diversification of effector genes in P. syringae (65) but is predicted to occur with a very low frequency (66), further supporting the single-acquisition-event hypothesis.
In conclusion, the specificity of the mbo operon has allowed the design of specific primers and a PCR protocol that permits a fast and efficient method for identifying potential mangotoxin-producing strains. With this PCR method, we have detected the presence of the mbo operon in five pathovars of P. syringae from genomospecies 1, whereas mangotoxin production has been reported for the first time in P. syringae pv. pisi. The high conservation of the structure, sequence, and genomic location of the mbo operon, together with the phylogenetic analysis showing that the mbo operon is present in only two groups from genomospecies 1 of the P. syringae complex, strongly suggests that the mbo operon has been horizontally acquired only once during the evolution of this bacterial species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the regional government of Andalucía (Spain), grants from CICE-Junta de Andalucía, Ayudas Grupo PAIDI AGR-169, and Proyecto de Excelencia (P07-AGR-02471) and Plan Nacional I+D+I grant AGL2008-55311-CO2-01 (Ministerio de Ciencia e Innovación), cofinanced by FEDER (European Union). Plan Propio of the University of Málaga funded a stay by V.J.C. at the Universidad Pública de Navarra, Spain. V.J.C. was supported with a fellowship from the Junta de Andalucía, Spain, and E.A. was supported with a JAEDoc grant from the CSIC, which was cofinanced by ESF.
We thank George W. Sundin, Alberto Martín-Sanz, and Constantino Caminero for providing isolates of P. syringae. We also thank Menno van der Voort for critically reading the manuscript and the research group of Cayo Ramos for their help with some aspects of this research.
Footnotes
Published ahead of print 9 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03007-12.
REFERENCES
- 1. Bender CL, Alarcón-Chaidez F, Gross DC. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gutiérrez-Barranquero JA, Arrebola E, Bonilla N, Sarmiento D, Cazorla FM, de Vicente A. 2012. Environmentally friendly treatment alternatives to Bordeaux mixture for controlling bacterial apical necrosis (BAN) of mango. Plant Pathol. 61:665–676 [Google Scholar]
- 3. Kennelly MM, Cazorla FM, de Vicente A, Ramos C, Sundin GW. 2007. Pseudomonas syringae diseases of fruit trees: progress toward understanding and control. Plant Dis. 91:4–17 [DOI] [PubMed] [Google Scholar]
- 4. Krieg NR, Holt JG. 1984. Bergey's manual of systematic bacteriology, vol 1 Williams & Wilkins, Baltimore, MD [Google Scholar]
- 5. Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13:614–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mansfield JW. 2009. From bacterial avirulence genes to effector functions via the hrp delivery system: an overview of 25 years of progress in our understanding of plant innate immunity. Mol. Plant Pathol. 10:721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramos C, Matas IM, Bardaji L, Aragón IM, Murillo J. 2012. Pseudomonas savastanoi pv. savastanoi: some like it knot. Mol. Plant Pathol. 13:998–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arrebola E, Cazorla FM, Codina JC, Gutiérrez-Barranquero JA, Pérez-García A, de Vicente A. 2009. Contribution of mangotoxin to the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Int. Microbiol. 12:87–95 [PubMed] [Google Scholar]
- 9. Arrebola E, Cazorla FM, Pérez-García A, de Vicente A. 2011. Chemical and metabolic aspects of antimetabolite toxins produced by Pseudomonas syringae pathovars. Toxins 3:1089–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arrebola E, Cazorla FM, Pérez-García A, de Vicente A. 2011. Genes involved in the production of antimetabolite toxins by Pseudomonas syringae pathovars. Genes 2:640–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arrebola E, Cazorla FM, Durán VE, Rivera E, Olea F, Codina JC, Pérez-García A, de Vicente A. 2003. Mangotoxin: a novel antimetabolite toxin produced by Pseudomonas syringae inhibiting ornithine/arginine biosynthesis. Physiol. Mol. Plant Pathol. 63:117–127 [Google Scholar]
- 12. Arrebola E, Cazorla FM, Romero D, Pérez-García A, de Vicente A. 2007. A nonribosomal peptide synthetase gene (mgoA) of Pseudomonas syringae pv. syringae is involved in mangotoxin biosynthesis and is required for full virulence. Mol. Plant-Microbe Interact. 20:500–509 [DOI] [PubMed] [Google Scholar]
- 13. Murillo J, Bardaji L, Navarro de la Fuente L, Führer ME, Aguilera S, Álvarez-Morales A. 2011. Variation in conservation of the cluster for biosynthesis of the phytotoxin phaseolotoxin in Pseudomonas syringae suggests at least two events of horizontal acquisition. Res. Microbiol. 162:253–261 [DOI] [PubMed] [Google Scholar]
- 14. Arrebola E, Carrión VJ, Cazorla FM, Pérez-García A, Murillo J, de Vicente A. 2012. Characterisation of the mgo operon in Pseudomonas syringae pv. syringae UMAF0158 that is required for mangotoxin production. BMC Microbiol. 12:10 doi:10.1186/1471-2180-12-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carrión VJ, Arrebola E, Cazorla FM, Murillo J, de Vicente A. 2012. The mbo operon is specific and essential for biosynthesis of mangotoxin in Pseudomonas syringae. PLoS One 7:e36709 doi:10.1371/journal.pone.0036709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rees-George J, Vanneste JL, Cornish DA, Pushparajah IPS, Yu J, Templeton MD, Everett KR. 2010. Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S-23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathol. 59:453–464 [Google Scholar]
- 17. Ercolani GL. 1985. The relation between dosage, bacterial growth and time for disease response during infection of bean leaves by Pseudomonas syringae pv. phaseolicola. J. Appl. Microbiol. 58:63–75 [Google Scholar]
- 18. Penyalver R, García A, Ferrer A, Bertolini E, López MM. 2000. Detection of Pseudomonas savastanoi pv. savastanoi in olive plants by enrichment and PCR. Appl. Environ. Microbiol. 66:2673–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sawada H, Suzuki F, Matsuda I, Saitou N. 1999. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49:627–644 [DOI] [PubMed] [Google Scholar]
- 20. Sawada H, Kanaya S, Tsuda M, Suzuki F, Azegami K, Saitou N. 2002. A phylogenomic study of the OCTase genes in Pseudomonas syringae pathovars: the horizontal rransfer of the argK-tox cluster and the evolutionary history of OCTase genes on their genomes. J. Mol. Evol. 54:437–457 [DOI] [PubMed] [Google Scholar]
- 21. Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7:e1002132 doi:10.1371/journal.ppat.1002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Green S, Studholme DJ, Laue BE, Dorati F, Lovell H, Arnold D, Cottrell JE, Bridgett S, Blaxter M, Huitema E, Thwaites R, Sharp PM, Jackson RW, Kamoun S. 2010. Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum. PLoS One 5:e10224 doi:10.1371/journal.pone.0010224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S. 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385–2394 [DOI] [PubMed] [Google Scholar]
- 24. Reinhardt JA, Baltrus DA, Nishimura MT, Jeck WR, Jones CD, Dangl JL. 2009. De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Res. 19:294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teverson DM. 1991. Genetics of pathogenicity and resistance in the haloblight disease of beans in Africa. Ph.D. thesis University of Birmingham, Birmingham, United Kingdom [Google Scholar]
- 26. Rico A, López R, Asensio C, Aizpún MT, Asensio-S-Manzanera MC, Murillo J. 2003. Nontoxigenic strains of Pseudomonas syringae pv. phaseolicola are a main cause of halo blight of beans in Spain and escape current detection methods. Phytopathology 93:1553–1559 [DOI] [PubMed] [Google Scholar]
- 27. Pérez-Martínez I, Rodriguez-Moreno L, Matas IM, Ramos C. 2007. Strain selection and improvement of gene transfer for genetic manipulation of Pseudomonas savastanoi isolated from olive knots. Res. Microbiol. 158:60–69 [DOI] [PubMed] [Google Scholar]
- 28. Sundin GW, Bender CL. 1996. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 5:133–143 [DOI] [PubMed] [Google Scholar]
- 29. Sundin GW, Demezas DH, Bender CL. 1994. Genetic and plasmid diversity within natural populations of Pseudomonas syringae with various exposures to copper and streptomycin bactericides. Appl. Environ. Microbiol. 60:4421–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, Lykidis A, Trong S, Nolan M, Goltsman E, Thiel J, Malfatti S, Loper JE, Lapidus A, Detter JC, Land M, Richardson PM, Kyrpides NC, Ivanova N, Lindow SE. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tourte C, Manceau C. 1995. A strain of Pseudomonas syringae which does not belong to pathovar phaseolicola produces phaseolotoxin. Eur. J. Plant Pathol. 101:483–490 [Google Scholar]
- 32. Martín-Sanz A, Palomo J, Pérez de la Vega M, Caminero C. 2012. Characterization of Pseudomonas syringae pv. syringae isolates associated with bacterial blight in Lathyrus spp. and sources of resistance. Eur. J. Plant Pathol. 134:205–216 [Google Scholar]
- 33. Young A. 2008. Notes on Pseudomonas syringae pv. syringae bacterial necrosis of mango (Mangifera indica) in Australia. Australas. Plant Dis. Notes 3:138–140 [Google Scholar]
- 34. Sohn KH, Jones JDG, Studholme DJ. 2012. Draft genome sequence of Pseudomonas syringae pathovar syringae strain FF5, causal agent of stem tip dieback disease on ornamental pear. J. Bacteriol. 194:3733–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martín-Sanz A, Palomo JL, Caminero C. 2009. First report of bacterial blight caused by Pseudomonas syringae pv. syringae on common vetch in Spain. Plant Dis. 93:1348. [DOI] [PubMed] [Google Scholar]
- 36. Cazorla FM, Arrebola E, Sesma A, Pérez-García A, Codina JC, Murillo J, de Vicente A. 2002. Copper resistance in Pseudomonas syringae strains isolated from mango is encoded mainly by plasmids. Phytopathology 92:909–916 [DOI] [PubMed] [Google Scholar]
- 37. Gutiérrez-Barranquero JA, Arrebola E, Pérez-García A, Codina JC, Murillo J, de Vicente A, Cazorla FM. 2008. Evaluation of phenotypic and genetic techniques to analyze diversity of Pseudomonas syringae pv. syringae strains isolates from mango trees, p 271–281 In Fatmi MB, Collmer A, Iacobellis NS, Mansfield JW, Murillo J, Schaad NW, Ullrich M. (ed), Pseudomonas syringae pathovars and related pathogens—identification, epidemiology and genomics. Springer Netherlands, Dordrecht, The Netherlands [Google Scholar]
- 38. Moore RA, Starratt AN, Ma SW, Morris VL, Cuppels DA. 1989. Identification of a chromosomal region required for biosynthesis of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Can. J. Microbiol. 35:910–917 [Google Scholar]
- 39. Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, Zaccardelli M, Setubal JC, Morales-Lizcano NP, Bernal A, Coaker G, Baker C, Bender CL, Leman S, Vinatzer BA. 2011. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 7:e1002130 doi:10.1371/journal.ppat.1002130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Almeida NF, Yan S, Lindeberg M, Studholme DJ, Schneider DJ, Condon B, Liu H, Viana CJ, Warren A, Evans C, Kemen E, MacLean D, Angot A, Martin GB, Jones JD, Collmer A, Setubal JC, Vinatzer BA. 2008. A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 22:52–62 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 43. Cazorla FM, Olalla L, Torés JA, Codina JC, Pérez-GarcíA A, de Vicente A. 1997. Pseudomonas syringae pv. syringae as microorganism involved in apical necrosis of mango: characterization of some virulence factors, p 82–87 In Rudolph K, Burr TJ, Mansfield JW, Stead D, Vivian A, von Kietzell J. (ed), Pseudomonas syringae pathovars and related pathogens. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 44. Gasson MJ. 1980. Indicator technique for antimetabolic toxin production by phytopathogenic species of Pseudomonas. Appl. Environ. Microbiol. 39:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 46. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 47. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 48. Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 49. Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. 2005. WebACT—an online companion for the Artemis comparison tool. Bioinformatics 21:3665–3666 [DOI] [PubMed] [Google Scholar]
- 50. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Völksch B, Weingart H. 1998. Toxin production by pathovars of Pseudomonas syringae and their antagonistic activities against epiphytic microorganisms. J. Basic Microbiol. 38:135–145 [PubMed] [Google Scholar]
- 52. Gardan L, Shafik H, Belouin S, Broch R, Grimont F, Grimont PAD. 1999. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49:469–478 [DOI] [PubMed] [Google Scholar]
- 53. Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou L, Liu J, Yuan Q, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Deng W-L, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Tang X, Bender CL, White O, Fraser CM, Collmer A. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 100:10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martín-Sanz A, Pérez de la Vega M, Murillo J, Caminero C. 2012. Genetic, biochemical and pathogenic diversity of Pseudomonas syringae pv. pisi strains. Plant Pathol. 61:1063–1072 [Google Scholar]
- 55. Cazorla FM, Torés JA, Olalla L, Pérez-García A, Farré JM, de Vicente A. 1998. Bacterial apical necrosis of mango in southern Spain: a disease caused by Pseudomonas syringae pv. syringae. Phytopathology 88:614–620 [DOI] [PubMed] [Google Scholar]
- 56. Schaad NW, Cheong SS, Tamaki S, Hatziloukas E, Panopoulos NJ. 1995. A combined biological and enzymatic amplification (BIO-PCR) technique to detect Pseudomonas syringae pv. phaseolicola in bean seed extracts. Phytopathology 85:243–248 [Google Scholar]
- 57. Gironde S, Manceau C. 2012. Housekeeping gene sequencing and multilocus variable-number tandem-repeat analysis to identify subpopulations within Pseudomonas syringae pv. maculicola and Pseudomonas syringae pv. tomato that correlate with host specificity. Appl. Environ. Microbiol. 78:3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hwang MS, Morgan RL, Sarkar SF, Wang PW, Guttman DS. 2005. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 71:5182–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sarkar SF, Guttman DS. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Little EL, Bostock RM, Kirkpatrick BC. 1998. Genetic characterization of Pseudomonas syringae pv. syringae strains from stone fruits in California. Appl. Environ. Microbiol. 64:3818–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guttman DS, Gropp SJ, Morgan RL, Wang PW. 2006. Diversifying selection drives the evolution of the type III secretion system pilus of Pseudomonas syringae. Mol. Biol. Evol. 23:2342–2354 [DOI] [PubMed] [Google Scholar]
- 62. Wang PW, Morgan RL, Scortichini M, Guttman DS. 2007. Convergent evolution of phytopathogenic pseudomonads onto hazelnut. Microbiology 153:2067–2073 [DOI] [PubMed] [Google Scholar]
- 63. Yan S, Liu H, Mohr TJ, Jenrette J, Chiodini R, Zaccardelli M, Setubal JC, Vinatzer BA. 2008. Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Appl. Environ. Microbiol. 74:3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Genka H, Baba T, Tsuda M, Kanaya S, Mori H, Yoshida T, Noguchi M, Tsuchiya K, Sawada H. 2006. Comparative analysis of argK-tox clusters and their flanking regions in phaseolotoxin-producing Pseudomonas syringae pathovars. J. Mol. Evol. 63:401–414 [DOI] [PubMed] [Google Scholar]
- 65. Stavrinides J, Ma W, Guttman DS. 2006. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog. 2:e104 doi:10.1371/journal.ppat.0020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.