Abstract
The rhcJ and ttsI mutants of Bradyrhizobium japonicum USDA122 for the type III protein secretion system (T3SS) failed to secrete typical effector proteins and gained the ability to nodulate Rj2 soybean plants (Hardee), which are symbiotically incompatible with wild-type USDA122. This suggests that effectors secreted via the T3SS trigger incompatibility between these two partners.
TEXT
Leguminous plants form root nodule symbioses with nitrogen-fixing soil bacteria from the order Rhizobiales. Association of Bradyrhizobium japonicum with agronomically important soybean cultivars is widespread throughout the world, including the United States (1), Brazil (2), and Japan (3). Several soybean genotypes restrict nodulation with specific rhizobial strains; these include Rj2, Rj3, Rj4, and Rfg1 (allelic to Rj2), which behave as single dominant genes (4, 5). In particular, the soybean cultivar Hardee (Rj2) is incompatible with B. japonicum USDA122 (6, 7). The Rj2/Rfg1 gene encodes a member of the Toll-interleukin receptor/nucleotide-binding site/leucine-rich repeat (TIR-NBS-LRR) class of proteins, involved in resistance to microbial pathogens (5). Different amino acid substitutions in Rj2 and Rfg1 restrict nodulation with B. japonicum USDA122 and Sinorhizobium fredii USDA257, respectively (5). The type III protein secretion system (T3SS) mutant of S. fredii USDA257, which is unable to secrete effector proteins (8), is capable of nodulating Rfg1 soybean plants. Thus, T3SS effectors may negatively affect symbiosis with Rfg1 soybean plants (5). Since the cell surface components of B. japonicum Is-1 and S. fredii USDA257 were suggested to be involved in the incompatibility with Rj2 soybean plants (8, 9), the question still remains how Rj2 prevents effective nodulation. Thus, the aim of the present work was to test whether functional T3SS causes the symbiotic incompatibility between B. japonicum USDA122 and Rj2 soybean plants.
The bacterial strains and plasmids used in this work are listed in Table S1 in the supplemental material. B. japonicum cells were grown at 30°C in HM salt medium (supplemented with 0.1% arabinose and 0.025% [wt/vol] yeast extract [Difco, Detroit, MI]) for DNA isolation and plant inoculation (see Table S2 in the supplementary material) or at 28°C in arabinose gluconate (AG) medium (1) for analysis of extracellular proteins (10). Escherichia coli cells were grown at 37°C in Luria-Bertani medium (11). Isolation of plasmids, DNA ligation, and transformation of E. coli were performed as described by Sambrook et al. (11). Total DNA of B. japonicum USDA122 was prepared as described previously (12). The draft genomic sequences were obtained by using a 454 GS-FLX Titanium pyrosequencer (454 Life Sciences, Branford, CT) (13).
A 3.7-kb DNA fragment carrying rhcJ was excised from prb01211 (14) with EcoRI and inserted into the EcoRI site of pK18mob, resulting in pSE11 (see Table S1 in the supplemental material). A 2.0-kb Ω cassette derived from pHP45 (15) was inserted into a MluI site within rhcJ in pSE11 (Fig. 1B). A 3.5-kb DNA fragment carrying ttsI was excised from brp13612 (14) with HindIII and inserted into the HindIII site of pK18mob, resulting in pSE12 (see Table S1). The Ω cassette was then inserted into a BstXI site within ttsI in pSE12 (Fig. 1C). Triparental mating with USDA122 and pRK2013 was conducted on HM agar plates using pSE11 and pSE12, resulting in 122ΔrhcJ and 122ΔttsI, respectively (see Table S1). Double-crossover events were verified by PCR.
Fig 1.
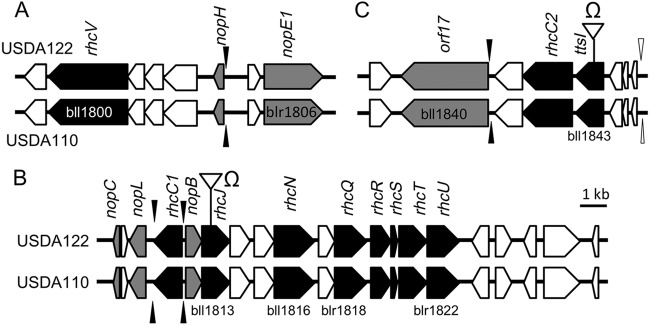
Comparison of gene clusters for the type III secretion system (T3SS) in the USDA122 and USDA110 strains of Bradyrhizobium japonicum. Three major loci within the tts clusters are shown (panels A, B, and C). The open reading frames encoding the T3SS structural components and putative effector proteins are shown in black and gray, respectively. The positions of nod and tts boxes are indicated by open and black arrowheads, respectively. The insertion positions of Ω cassettes (see Table S1 in the supplemental material) in USDA122 are also shown.
B. japonicum USDA122 and the mutant 122ΔrhcJ were grown at 28°C for 48 h in 500 ml AG medium in the absence or in the presence of genistein (10 μM). Extracellular proteins were prepared as described previously (10). The protein samples were subjected to SDS-PAGE (5 to 20% gradient gel in Tris-glycine buffer). Small pieces of the Coomassie-stained gel corresponding to bands of interest were digested with trypsin. Peptides were analyzed by using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS/MS) (TOF/TOF 5800 system; AB Sciex, Foster City, CA). Proteins were identified by the MS-Fit software program (http://prospector.ucsf.edu/prospector/mshome.htm) with the USDA110 database as a reference (14). The mass fragments were further identified by MS/MS analysis.
Surface-sterilized soybean seeds (Glycine max cultivars Hardee [Rj2] and Lee [rj2]) were germinated in sterile vermiculite for 4 days at 25°C. The seedlings were then transplanted into Leonard jar pots (one plant per pot) with sterile vermiculite and nitrogen-free nutrient solution (16) and inoculated with B. japonicum (1 × 107 cells per seedling). Plants were grown in a phytotron (Koito Industries, Tokyo, Japan) for 30 to 40 days at 25/20°C under a 16-h light/8-h dark cycle (16).
Genes implicated in T3SS were organized similarly in the genomes of B. japonicum USDA122 and USDA110 (Fig. 1) (17). Their DNA sequences were also highly conserved. In particular, the amino acid sequences of the regulatory (TtsI) and structural (RhcV, RhcC1, RhcC2, RhcJ, RhcN, RhcQ, RhcR, RhcT, RhcU, and RhcS) components of T3SS were identical between USDA122 and USDA110, although synonymous nucleotide substitutions occurred in genes encoding four T3SS components (RhcV, RhcC2, RhcR, and RhcT) (see Table S3 in the supplemental material). In addition, a nod box upstream of ttsI and 5 tts boxes were highly conserved between USDA122 and USDA110 (Fig. 1) (17, 18). In USDA110, the T3SS gene cluster can be induced by flavonoids and is controlled via the following regulatory cascade: flavonoid signal → NodD1 → nod box → TtsI → tts box (17–19).
To examine whether T3SS is active in USDA122, we compared extracellular proteins in wild-type USDA122 and in 122ΔrhcJ cultures (since rhcJ is required for a functional T3SS in USDA110 [17, 20]). Since flavonoids induce T3SS, we also analyzed extracellular proteins in cell cultures treated or not treated with the isoflavone genistein. Several bands were unique to genistein-induced wild-type USDA122 (Fig. 2).
Fig 2.
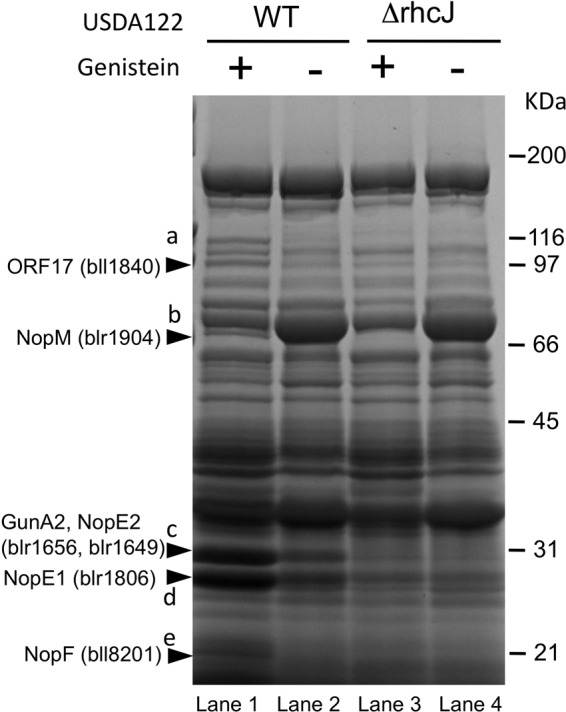
Secreted proteins of the wild-type strain (WT) and the rhcJ mutant (ΔrhcJ) of B. japonicum USDA122 in the presence (+) or absence (−) of 10 μM genistein. Proteins induced by genistein in wild-type USDA122 but not in the rhcJ mutant (bands a to e) were identified by MALDI-TOF-MS/MS analysis (see Fig. S1 to S5 in the supplemental material).
The excised pieces of gel corresponding to individual bands (a to e in lanes 1 to 3 in Fig. 2) were subjected to MALDI-TOF-MS/MS analysis (see Fig. S1 to S5 in the supplemental material). The peptides derived from ORF17 (bll1840) were detected in genistein-induced wild-type USDA122 (Fig. 2, WT+) and to a lesser extent in untreated wild-type USDA122 (WT−) but not in the genistein-induced 122ΔrhcJ mutant (ΔrhcJ+) (band a in Fig. 2). Band “a” also contained the bll4853 protein, but the intensities of its peptide signals were similar in all 3 samples (see Fig. S1). Interestingly, a typical tts box was present upstream of orf17 in USDA122 and bll1840 in USDA110 (Fig. 1C). These results suggested that ORF17 is a T3SS-dependent secreted protein in USDA122.
Based on similar analyses, GunA2 (blr1656), NopE1 (blr1806), NopE2 (blr1649), NopF (bll8201), and NopM (blr1904) were identified as T3SS-dependent secreted proteins in USDA122 (Fig. 2; see also Fig. S2 to S5 in the supplemental material). Most of these proteins (GunA2, NopE1, NopE2, and NopF) were previously identified in USDA110 (20). NopE1 is a true USDA110 effector, because it translocates into the host cells and affects nodulation efficiency on Vigna radiata (21). The peptides of band “d” (approximately 28 kDa) were derived from the C-terminal half of the NopE1 protein (51 kDa) (see Fig. S4), consistent with a self-cleavage activity of NopE1 (21). Overall, these results showed that 122ΔrhcJ failed to secrete typical effector proteins, indicating that the T3SS is active in B. japonicum USDA122 and is inactivated by the loss of RhcJ.
Next, we examined the symbiotic phenotypes of Hardee with 122ΔrhcJ and 122ΔttsI. Both T3SS knockout mutants induced normal nodules with red leghemoglobin in nodule sections, and the host plants had normal green coloring (Fig. 3). In contrast, wild-type USDA122 failed to induce normal nodules, and plants had yellow shoots (Fig. 3). Wild-type USDA122 occasionally induced host cell division and formation of bump structures without bradyrhizobial cells at the lateral root junctions (WT in Fig. 3C) (5). Wild-type USDA122 and both T3SS knockout mutants induced normal nodules on Lee (rj2) soybean plants (see Fig. S6 in the supplemental material).
Fig 3.
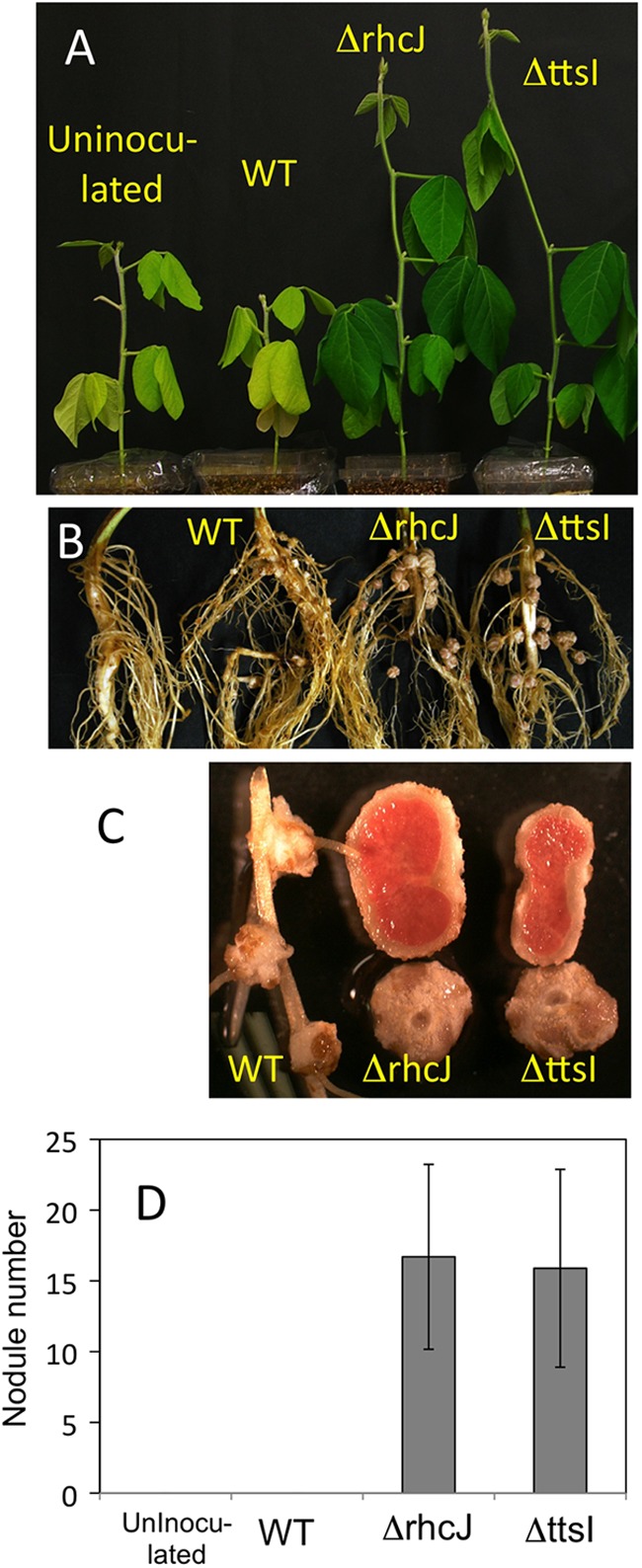
Symbiotic phenotypes of Rj2 soybean cultivar Hardee inoculated with the wild-type strain (WT) and the rhcJ (ΔrhcJ) and ttsI (ΔttsI) mutants of B. japonicum USDA122. Panels A, B, C, and D show plant growth, root systems, nodules, and nodule numbers, respectively. Error bars indicate standard deviations (n = 7 to 10).
Symbiotic nitrogen fixation in Hardee was evaluated by the acetylene reduction assay. Acetylene-reducing activity (ARA) was detected only in the root systems inoculated with the T3SS knockout mutants. ARA levels were similar in root systems inoculated with USDA110 and both USDA122 mutants (see Fig. S7 in the supplemental material). No ARA was detected upon inoculation with wild-type USDA122. Therefore, the loss of T3SS in USDA122 restored nitrogen fixation and nodulation during its interaction with Hardee.
The present work demonstrated that T3SS mediates symbiotic incompatibility between Rj2 soybean Hardee plants and B. japonicum USDA122. Since T3SS functions to secrete effectors during early stages of rhizobial infection (19), it is critical to identify proteins that induce symbiotic incompatibility between Rj2 soybean plants and USDA122. Such effectors can be anticipated to directly or indirectly interact with the Rj2 variants of the Rj2/Rfg1 gene product in soybean plants to restrict nodulation with B. japonicum USDA122 (5). A preliminary survey of putative effectors around the T3SS gene cluster showed that the amino acid substitutions of three gene (nopE1, nopP1, and orf17) products occurred between B. japonicum USDA122 and USDA110 (see Table S4 in the supplemental material).
Little is known about the ecological and evolutionary implications of exploitation of pathogenic secretion systems and effectors by rhizobia for symbiotic purposes (5, 19). Modern rj2 soybean cultivars often show lower nitrogen-fixing activity when inoculated with USDA122 than with USDA110 (3). Thus, it would be fascinating to speculate that Rj2 soybean plants may have evolved incompatibility with B. japonicum USDA122 to prevent its inferior symbionts from using the pathogenic systems. In this regard, the “zigzag” model of pathogen-plant interactions (22) might be adopted to describe rhizobium-plant interactions, because only two amino acid substitutions in and around the NBS domain of the Rj2/Rfg1 gene product define the genetic basis of Rj2 soybean plant incompatibility with B. japonicum USDA122 (5).
Nucleotide sequence accession numbers.
The contig sequences relevant to T3SS gene clusters were identified by BLASTN searches and deposited in the DDBJ/EMBL/GenBank database under accession numbers AB758605 to AB758607.
Supplementary Material
ACKNOWLEDGMENTS
We thank Naoko Higashitani and Atsushi Higashitani (Tohoku University) for MALDI-TOF-MS analysis. We are also grateful to Zyunji Ishizuka (Kyusyu University) for kindly providing seeds of the soybean cultivars Hardee and Lee.
This work was supported in part by grant PMI-0002 from the Ministry of Agriculture, Forestry and Fisheries of Japan; by BRAIN; by two Grants-in-Aid for Scientific Research, (A) 23248052 and (C) 22580074, from the Ministry of Education, Science, Sports and Culture of Japan; by a grant from Genesis Research Institute; and by the Japanese Government Scheme for the Development of Mitigation and Adaptation Techniques to Global Warming in the Sectors of Agriculture, Forestry and Fisheries.
Footnotes
Published ahead of print 30 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03297-12.
REFERENCES
- 1. Sadowsky MJ, Tully RE, Cregan PB, Keyser HH. 1987. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybean. Appl. Environ. Microbiol. 53:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Batista JS, Hungria M, Barcellos FG, Ferreira MC, Mendes IC. 2007. Variability in Bradyrhizobium japonicum and B. elkanii seven years after introduction of both the exotic microsymbiont and the soybean host in a cerrados soil. Microb. Ecol. 53:270–284 [DOI] [PubMed] [Google Scholar]
- 3. Itakura M, Saeki K, Omori H, Yokoyama T, Kaneko T, Tabata S, Ohwada T, Tajima S, Uchiumi T, Honnma K, Fujita K, Iwata H, Saeki Y, Hara Y, Ikeda S, Eda S, Mitsui H, Minamisawa K. 2009. Genomic comparison of Bradyrhizobium japonicum strains with different symbiotic nitrogen-fixing capabilities and other Bradyrhizobiaceae members. ISME J. 3:326–339 [DOI] [PubMed] [Google Scholar]
- 4. Hayashi M, Saeki Y, Haga M, Harada K, Kouchi H, Umehara Y. 2012. Rj(rj) gene involved in nitrogen-fixing root nodule formation in soybean. Breed. Sci. 61:544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang S, Tang F, Gao M, Krishnan HB, Zhu H. 2010. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U. S. A. 107:18735–18740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caldwell BE. 1966. Inheritance of strain-specific ineffective nodulation in soybeans. Crop Sci. 6:427–428 [Google Scholar]
- 7. Caldwell BE, Hinson K, Johnson HW. 1966. A strain-specific ineffective nodulation reaction in the soybean Glycine max L. Merrill. Crop Sci. 6:495–496 [Google Scholar]
- 8. Krishnan HB, Lorio J, Kim WS, Jiang G, Kim KY, DeBoer M, Pueppke SG. 2003. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and export by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant Microbe Interact. 16:617–625 [DOI] [PubMed] [Google Scholar]
- 9. Tsurumaru H, Yamakawa T, Tanaka M, Sakai M. 2008. Tn5 mutants of Bradyrhizobium japonicum Is-1 with altered compatibility with Rj2-soybean cultivars. Soil Sci. Plant Nutr. 54:197–203 [Google Scholar]
- 10. Okazaki S, Zehner S, Hempei J, Lang K, Göttfert M. 2009. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 295:88–95 [DOI] [PubMed] [Google Scholar]
- 11. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 12. Okubo T, Tsukui T, Maita H, Okamoto S, Oshima K, Fujisawa T, Saito A, Futamata H, Hattori R, Shimomura S, Haruta S, Morimoto S, Wang Y, Sakai Y, Hattori M, Aizawa S, Nagashima K, Masuda S, Hattori T, Yamashita A, Bao Z, Hayatsu M, Kajiya-Kanegae H, Yoshinaga I, Sakamoto K, Toyota K, Nakao M, Kohara M, Anda M, Niwa R, Jung-Hwan P, Sameshima-Saito R, Tokuda S, Yamamoto S, Yamamoto S, Yokoyama T, Akutsu T, Nakamura Y, Nakahira-Yanaka Y, Takada-Hoshino Y, Hirakawa H, Mitsui H, Terasawa K, Itakura M, Sato S, Ikeda-Ohtsubo W, Sakakura N, Kaminuma E, Minamisawa K. 2012. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ. 27:306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okubo T, Ikeda S, Yamashita A, Terasawa K, Minamisawa K. 2012. Pyrosequence read length of 16S rRNA gene affects phylogenetic assignment of plant-associated bacteria. Microbes Environ. 27:204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189–197 [DOI] [PubMed] [Google Scholar]
- 15. Prentki P, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313 [DOI] [PubMed] [Google Scholar]
- 16. Inaba S, Ikenishi F, Itakura M, Kikuchi M, Eda S, Chiba N, Katsumata C, Suwa Y, Mitsui H, Minamisawa K. 2012. N2O emission from degraded soybean nodules depends on denitrification by Bradyrhizobium japonicum and other microbes in the rhizosphere. Microbes Environ. 27:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krause A, Doerfel A, Göttfert M. 2002. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 15:1228–1235 [DOI] [PubMed] [Google Scholar]
- 18. Zehner S, Schober G, Wenzel M, Lang K, Göttfert M. 2008. Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol. Plant Microbe Interact. 21:1087–1093 [DOI] [PubMed] [Google Scholar]
- 19. Deakin WJ, Broughton WJ. 2009. Symbiotic use of pathogenetic strategies: rhizobial protein secretion systems. Nat. Rev. Microbiol. 7:312–320 [DOI] [PubMed] [Google Scholar]
- 20. Süss C, Hempel J, Zehner S, Krause A, Patschkowski T, Göttfert M. 2006. Identification of genistein-inducible and type III-secreted proteins of Bradyrhizobium japonicum. J. Biotechnol. 126:69–77 [DOI] [PubMed] [Google Scholar]
- 21. Wenzel M, Friedrich L, Göttfert M, Zehner S. 2010. The type III-secreted protein NopE1 affects symbiosis and exhibits a calcium-dependent autocleavage activity. Mol. Plant Microbe Interact. 23:124–129 [DOI] [PubMed] [Google Scholar]
- 22. Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


