Abstract
“Candidatus Neoehrlichia mikurensis” was detected by PCR in 4.0% (34/841) of the rodents tested in this study. The 34 rodents represented nine species from seven regions of China. Phylogenetic analyses based on the partial groEL and nearly entire 16S rRNA gene sequences of the agent revealed genetic diversity, which was correlated with its geographic origins.
TEXT
Anovel Ehrlichia-like pathogen was originally detected in Ixodes ricinus ticks from the Netherlands in 1999 (1). In 2004, this agent was isolated by inoculating laboratory rats with naturally infected wild Rattus norvegicus rats in Japan and named “Candidatus Neoehrlichia mikurensis,” representing a new member of the family Anaplasmataceae (2). Since 2010, “Candidatus Neoehrlichia mikurensis” has been found in several human patients from Sweden, Germany, Switzerland, and the Czech Republic, which suggests that this could be an emerging pathogen in Europe (3–6).
In China, “Candidatus Neoehrlichia mikurensis” was primarily detected in Rattus norvegicus from the southern part of the country (7). Most recently, in a hospital-based surveillance study in northeastern China, we identified “Candidatus Neoehrlichia mikurensis” infection in febrile patients with recent histories of tick bites (8). The ticks and rodents collected around the patients' residences were also found to be positive for the agent; therefore, the local natural foci were established (8). These findings suggest the potential wide existence of infection with “Candidatus Neoehrlichia mikurensis,” and our understanding of the ecologic characteristics of this new agent remains far from complete. In the present study, we identified the infection of “Candidatus N. mikurensis” in wild rodents from China. The geographic distribution was described, and the genetic characteristics were investigated.
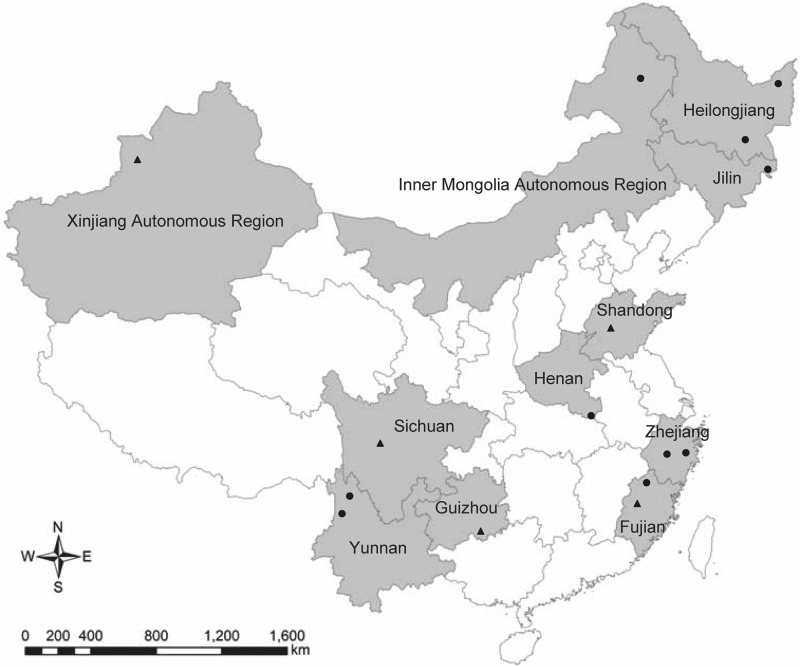
Between 2005 and 2009, rodents were collected at 15 study sites in 11 regions at local peak seasons of tick activities (Fig. 1). All the study sites featured forest or cropland landscapes, where local residents were frequently exposed to rodents and ticks. Rodents were captured using snap traps and then identified by morphological features to the species level. After the species was identified, spleen samples were collected for DNA extraction and PCR.
Fig 1.
Geographic location of tested rodents. The circles represent positive sites, and the triangles represent negative sites. (Map constructed using ArcGIS software with data from the Data Sharing Infrastructure of Earth System Science [http://eng.geodata.cn:8080/geonetwork/srv/en/main.home].)
DNA was extracted by using the DNeasy tissue kit (Qiagen, Germantown, MD). “Candidatus Neoehrlichia mikurensis” was detected by a nested PCR specific for the 60-kDa heat shock protein-encoding gene (groEL) using the primers CNM-out1 (5′-TGGCAAATGTAGTTGTAACAGG-3′) and CNM-out2 (5′-TCTACTTCACTTGAACCGCCA-3′) in the first run and CNM-in1 (5′-GCTATTAGTAAGCCTTATGGTAC-3′) and CNM-in2 (5′-GAAGAATTACTATCTACGCTACC-3′) in the second. Positive samples were confirmed by amplifications of the entire 16S rRNA gene (rrs) as described previously (8).
The sequence was aligned by using ClustalW software. Phylogenetic analysis (neighbor joining) based on the rrs and groEL nucleotide sequences were conducted with MEGA4.0 (9) using the maximum composite likelihood method and a bootstrap analysis of 1,000 replicates. A chi-square test or Fisher's exact test was performed to determine the statistical differences between the prevalence of “Candidatus Neoehrlichia mikurensis” and rodent species, as well as geographic regions. The differences were considered statistically significant at P values of <0.05.
In total, 841 rodents of 22 species from 10 genera were captured and examined (Table 1). The predominant species were Apodemus agrarius (14.6%), and the other species accounted for 0.8% to 10.3%. Each region had 3 to 9 species of rodents sampled. “Candidatus Neoehrlichia mikurensis” was detected in 34 (4.0%) of the 841 rodents by amplifications of both groEL and rrs (Table 1). The 34 rodents with positive results represented 9 species, with the prevalence in each species ranging from 1.1% to 25.0%. When each rodent species was compared with all others (see Table S1 in the supplemental material), the infection rates in Eothenomys custos (25.0%), A. agrarius (12.0%), and Apodemus sylvaticus (12.5%) were significantly higher (P = 0.038, P < 0.001, and P = 0.019, respectively).
Table 1.
Prevalence of “Ca. Neoehrlichia mikurensis” in rodents of different species from 11 regions in Chinaa
| Rodent species | No. positive/no. tested |
Total no. positive/total no. tested (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jilin | Heilongjiang | IMAR | XJAR | Shandong | Henan | Fujian | Zhejiang | Sichuan | Guizhou | Yunnan | ||
| Apodemus agrarius | 11/42 | 2/23 | 0/20 | 0 | 0/1 | 1/31 | 0 | 0 | 0 | 0 | 0 | 14/117 (12.0) |
| Apodemus sylvaticus | 0 | 0 | 0 | 0 | 0 | 0 | 0/4 | 5/32 | 0/4 | 0 | 0 | 5/40 (12.5) |
| Apodemus chevrieri | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/49 | 0/49 |
| Apodemus draco | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1/7 | 1/7 (14.3) |
| Apodemus peninsulae | 3/36 | 2/18 | 0/3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5/57 (8.8) |
| Eothenomys miletus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/22 | 0/22 |
| Eothenomys custos | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2/8 | 2/8 (25.0) |
| Microtus clarkei | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/17 | 0/17 |
| Microtus maximowiczii | 0 | 0 | 0/18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/18 |
| Mus musculus | 0/6 | 0 | 0/6 | 0/30 | 0/13 | 0 | 0 | 0 | 0 | 0/8 | 0 | 0/63 |
| Mus pahari | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/7 | 0/2 | 0/9 |
| Myodes rufocanus | 1/20 | 2/58 | 1/5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4/83 (4.8) |
| Myodes rutilus | 0 | 0/12 | 0/7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/19 |
| Niviventer confucianus | 0 | 0 | 0 | 0 | 0 | 0/6 | 1/29 | 0/15 | 0/2 | 0 | 0 | 1/52 (1.9) |
| Niviventer fulvescens | 0 | 0 | 0 | 0 | 0 | 0 | 0/9 | 0/1 | 0/6 | 0 | 0 | 0/16 |
| Niviventer coxingi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/1 | 0 | 0 | 0/5 | 0/6 |
| Rattus norvegicus | 0/5 | 0/10 | 0/6 | 0 | 0/28 | 1/12 | 0 | 0/5 | 0/12 | 0/9 | 0 | 1/87 (1.1) |
| Rattus flavipectus | 0 | 0 | 0 | 0 | 0 | 0 | 0/4 | 0 | 0/11 | 0/10 | 0/2 | 0/27 |
| Rattus losea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/52 | 0/6 | 0/8 | 0/2 | 0/68 |
| Meriones unguiculatus | 0 | 0 | 0 | 0/20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/20 |
| Tamias sibiricus | 1/4 | 0 | 0/3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1/7 (14.3) |
| Tscherskia triton | 0/3 | 0/2 | 0 | 0/8 | 0 | 0/2 | 0/5 | 0/29 | 0 | 0 | 0 | 0/49 |
| Total | 16/116 | 6/123 | 1/68 | 0/58 | 0/42 | 2/51 | 1/51 | 5/135 | 0/41 | 0/42 | 3/114 | 34/841 (4.0) |
IMAR, Inner Mongolia Autonomous Region; XJAR, Xinjiang Autonomous Region.
“Candidatus Neoehrlichia mikurensis” was detected in 10 of the 15 study sites, representing Heilongjiang, Jilin, Henan, Zhejiang, Fujian, and Yunnan Provinces and the Inner Mongolia Autonomous Region (IMAR) (Table 1). The prevalence of “Candidatus Neoehrlichia mikurensis” in the 7 regions ranged from 1.5% to 13.8% and showed significant differences (P < 0.001). Rodents collected from Jilin Province were at a significantly higher risk for infection in contrast to other regions (P < 0.001).
All PCR amplicons were successfully sequenced. Consequently, fragments of groEL (914 bp) and rrs (1,501 bp) were obtained from each positive sample. Sequence comparisons revealed three different “Candidatus Neoehrlichia mikurensis” variants that were consistent with the three geographic regions, Northeast China (Heilongjiang and Jilin Provinces and IMAR), Southwest China (Yunnan Province), and Southeast China (Henan, Zhejiang, and Fujian Provinces). Nucleotide sequence analysis of the three variants showed 93.8% to 96.5% similarities of groEL and 99.0% to 99.3% similarities of rrs.
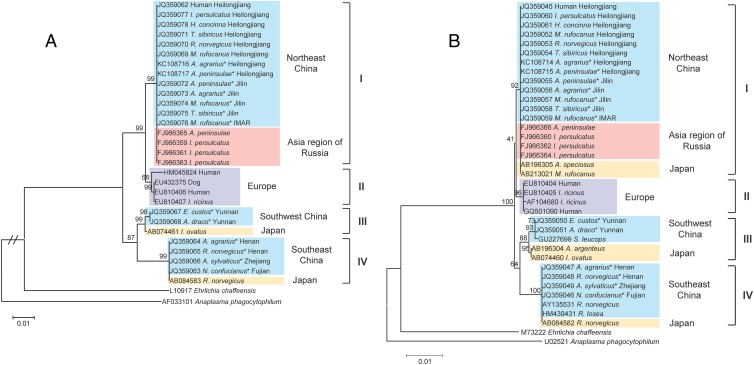
Phylogenetic analysis based on all available and comparably long groEL nucleotide sequences (874 bp) revealed four distinct clusters of “Candidatus Neoehrlichia mikurensis” strains that related to geographic origins (Fig. 2A). Sequences obtained from Northeast China were identical to those from the Asia region of Russia and were included in cluster I. Sequences obtained from Southwest China and Southeast China were separately clustered in cluster III and cluster IV, both together with sequences from Japan. No sequence in the present study was identified in cluster II, which contained only sequences from European countries such as Germany, the Netherlands, and Switzerland.
Fig 2.
Neighbor-joining tree of “Candidatus Neoehrlichia mikurensis” inferred from 874 bp of the groEL gene sequences (A) and 1,303 bp of the rrs gene sequences (B) using the maximum composite likelihood method. Numbers represent the bootstrap support for each node, as a percentage of 1,000 replicates. Sequences identified in the present study are indicated by asterisks. The host species and geographic regions are specified. The genus abbreviations of host species are as follows: A., Apodemus; E., Eothenomys; H., Haemaphysalis; I., Ixodes; M., Myodes; N., Niviventer; R., Rattus; and T., Tamias.
Similar to the phylogenetic analysis of groEL sequences, a neighbor-joining tree based on all available gene sequences and the nearly entire rrs gene sequence (1,303 bp) also separated these sequences into four different clusters (Fig. 2B). The sequences of “Candidatus Neoehrlichia mikurensis” from China were classified into three clusters and markedly related to geographic origins, which was consistent with the results from the phylogenetic analysis of groEL sequences.
The present study detected “Candidatus Neoehrlichia mikurensis” in diverse species of rodents from various regions, indicating the agent is widely distributed in China. These results were consistent with previous studies, which also found the agent in several different rodent species from Sweden, Japan, and the Asia region of Russia (10–13). This finding suggests that a variety of rodent species may be involved in the enzootic maintenance and transmission of “Candidatus Neoehrlichia mikurensis.”
Three genetically different “Candidatus Neoehrlichia mikurensis” variants were detected in rodents from China and one was identical to the strains causing human infection (8). In other regions where no human cases were reported, the presence of “Candidatus Neoehrlichia mikurensis” in rodents may imply a potential threat to humans. We also found that the “Candidatus Neoehrlichia mikurensis” gene clusters correlated with distinct geographic origins. Different gene clusters were found in the same rodent species from different regions (Fig. 2), suggesting that the genetic diversity might not be associated with host species. Further epidemiological and evolutionary studies, representing many more geographic locations, rodent species, and genetic information, would help to improve our understanding of the evolutionary and geographical relationships of “Candidatus Neoehrlichia mikurensis.”
In conclusion, this study contributes to a better understanding of the geographic distribution and genetic diversity of the novel human pathogen “Candidatus Neoehrlichia mikurensis” in rodents. Insights from naturally occurring infection in rodents might enhance understanding of human diseases with “Candidatus Neoehrlichia mikurensis” infection due to the underidentification of human cases.
Nucleotide sequence accession numbers.
The nucleotide sequences of “Candidatus Neoehrlichia mikurensis” determined in the present study were submitted to GenBank under the accession numbers KC108716, KC108717, JQ359063, JQ359064, JQ359065, JQ359066, JQ359067, JQ359068, JQ359072, JQ359073, JQ359074, JQ359075, and JQ359076 for groEL and KC108714, KC108715, JQ359046, JQ359047, JQ359048, JQ359049, JQ359050, JQ359051, JQ359055, JQ359056, JQ359057, JQ359058, and JQ359059 for rrs (see Table S2 in the supplemental material).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of China (grants 81130086, 81222037, 81172729, and 3086250) and by the Special Fund for Health Research in the Public Interest (grant 201202019).
Footnotes
Published ahead of print 26 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02917-12.
REFERENCES
- 1. Schouls LM, Van De Pol I, Rijpkema SG, Schot CS. 1999. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J. Clin. Microbiol. 37:2215–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kawahara M, Rikihisa Y, Isogai E, Takahashi M, Misumi H, Suto C, Shibata S, Zhang C, Tsuji M. 2004. Ultrastructure and phylogenetic analysis of “Candidatus Neoehrlichia mikurensis” in the family Anaplasmataceae, isolated from wild rats and found in Ixodes ovatus ticks. Int. J. Syst. Evol. Microbiol. 54:1837–1843 [DOI] [PubMed] [Google Scholar]
- 3. Fehr JS, Bloemberg GV, Ritter C, Hombach M, Luscher TF, Weber R, Keller PM. 2010. Septicemia caused by tick-borne bacterial pathogen Candidatus Neoehrlichia mikurensis. Emerg. Infect. Dis. 16:1127–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pekova S, Vydra J, Kabickova H, Frankova S, Haugvicova R, Mazal O, Cmejla R, Hardekopf DW, Jancuskova T, Kozak T. 2011. Candidatus Neoehrlichia mikurensis infection identified in 2 hematooncologic patients: benefit of molecular techniques for rare pathogen detection. Diagn. Microbiol. Infect. Dis. 69:266–270 [DOI] [PubMed] [Google Scholar]
- 5. von Loewenich FD, Geissdorfer W, Disque C, Matten J, Schett G, Sakka SG, Bogdan C. 2010. Detection of “Candidatus Neoehrlichia mikurensis” in two patients with severe febrile illnesses: evidence for a European sequence variant. J. Clin. Microbiol. 48:2630–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Welinder-Olsson C, Kjellin E, Vaht K, Jacobsson S, Wenneras C. 2010. First case of human “Candidatus Neoehrlichia mikurensis” infection in a febrile patient with chronic lymphocytic leukemia. J. Clin. Microbiol. 48:1956–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan H, Liu S, Ma Y, Tong S, Sun Y. 2003. Ehrlichia-like organism gene found in small mammals in the suburban district of Guangzhou of China. Ann. N. Y. Acad. Sci. 990:107–111 [DOI] [PubMed] [Google Scholar]
- 8. Li H, Jiang JF, Liu W, Zheng YC, Huo QB, Tang K, Zuo SY, Liu K, Jiang BG, Yang H, Cao WC. 2012. Human infection with Candidatus Neoehrlichia mikurensis, China. Emerg. Infect. Dis. 18:1636–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 10. Andersson M, Raberg L. 2011. Wild rodents and novel human pathogen Candidatus Neoehrlichia mikurensis, Southern Sweden. Emerg. Infect. Dis. 17:1716–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naitou H, Kawaguchi D, Nishimura Y, Inayoshi M, Kawamori F, Masuzawa T, Hiroi M, Kurashige H, Kawabata H, Fujita H, Ohashi N. 2006. Molecular identification of Ehrlichia species and “Candidatus Neoehrlichia mikurensis” from ticks and wild rodents in Shizuoka and Nagano Prefectures, Japan. Microbiol. Immunol. 50:45–51 [DOI] [PubMed] [Google Scholar]
- 12. Rar VA, Livanova NN, Panov VV, Doroschenko EK, Pukhovskaya NM, Vysochina NP, Ivanov LI. 2010. Genetic diversity of Anaplasma and Ehrlichia in the Asian part of Russia. Ticks Tick Borne Dis. 1:57–65 [DOI] [PubMed] [Google Scholar]
- 13. Tabara K, Arai S, Kawabuchi T, Itagaki A, Ishihara C, Satoh H, Okabe N, Tsuji M. 2007. Molecular survey of Babesia microti, Ehrlichia species and Candidatus Neoehrlichia mikurensis in wild rodents from Shimane Prefecture, Japan. Microbiol. Immunol. 51:359–367 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.