Abstract
Balancing the flux of a heterologous metabolic pathway by tuning the expression and properties of the pathway enzymes is difficult, but it is critical to realizing the full potential of microbial biotechnology. One prominent example is the metabolic engineering of a Saccharomyces cerevisiae strain harboring a heterologous xylose-utilizing pathway for cellulosic-biofuel production, which remains a challenge even after decades of research. Here, we developed a combinatorial pathway-engineering approach to rapidly create a highly efficient xylose-utilizing pathway for ethanol production by exploring various combinations of enzyme homologues with different properties. A library of more than 8,000 xylose utilization pathways was generated using DNA assembler, followed by multitiered screening, which led to the identification of a number of strain-specific combinations of the enzymes for efficient conversion of xylose to ethanol. The balancing of metabolic flux through the xylose utilization pathway was demonstrated by a complete reversal of the major product from xylitol to ethanol with a similar yield and total by-product formation as low as 0.06 g/g xylose without compromising cell growth. The results also suggested that an optimal enzyme combination depends on not only the genotype/phenotype of the host strain, but also the sugar composition of the fermentation medium. This combinatorial approach should be applicable to any heterologous pathway and will be instrumental in the optimization of industrial production of value-added products.
INTRODUCTION
Coordinating the intracellular activities of multiple enzymes in a heterologous metabolic pathway is crucial in order to maximize the flux through the pathway without the accumulation of undesirable intermediates (1–3). The intracellular enzyme activity can be modulated by changing enzyme expression at transcriptional (promoter-engineering) or translational (engineering of the ribosomal binding sites) levels (2, 4, 5). The method of tunable intergenic regions (TIGRs) exploits the prokaryotic operon system to coordinate the expression of multiple genes to regulate several genes simultaneously. The importance of the balanced metabolic pathway was well demonstrated in taxadiene synthesis (1). The metabolic pathway converting glyceraldehyde 3-phosphate and pyruvate into taxadiene was divided into two modules, the methylerythritol-phosphate (MEP) pathway forming isopentenyl pyrophosphate (four genes) and the heterologous downstream terpenoid-forming pathway, and balancing the two modules using various promoters and gene copy numbers substantially increased taxadiene production with minimal accumulation of by-product (1).
Balancing of the pathway flux by modulating transcriptional expression was shown to be a critical element in pathway construction for efficient xylose utilization by Saccharomyces cerevisiae and isoprenoid pathway optimization for paclitaxel precursor overproduction in Escherichia coli (1, 2). In previous studies, the expression levels of the genes constituting the pathways were varied by manually shuffling a small number of promoters of different strengths, which significantly limited the ranges of enzyme expression and their combinations. Here, we report a combinatorial pathway-engineering approach that allows the identification of balanced heterologous metabolic pathways at the translational level by generating a library of pathways followed by high-throughput screening or selection. While the pathway optimization strategies involving gene expression at the transcription level, such as promoter and TIGR engineering, aim to balance metabolic pathways by controlling enzyme concentrations, combinatorial pathway assembly explores various combinations of enzymes with different enzymatic properties, such as catalytic activity, cofactor specificity, stability, and substrate specificity, providing balanced metabolic pathways optimized to the metabolic environment of the host strain.
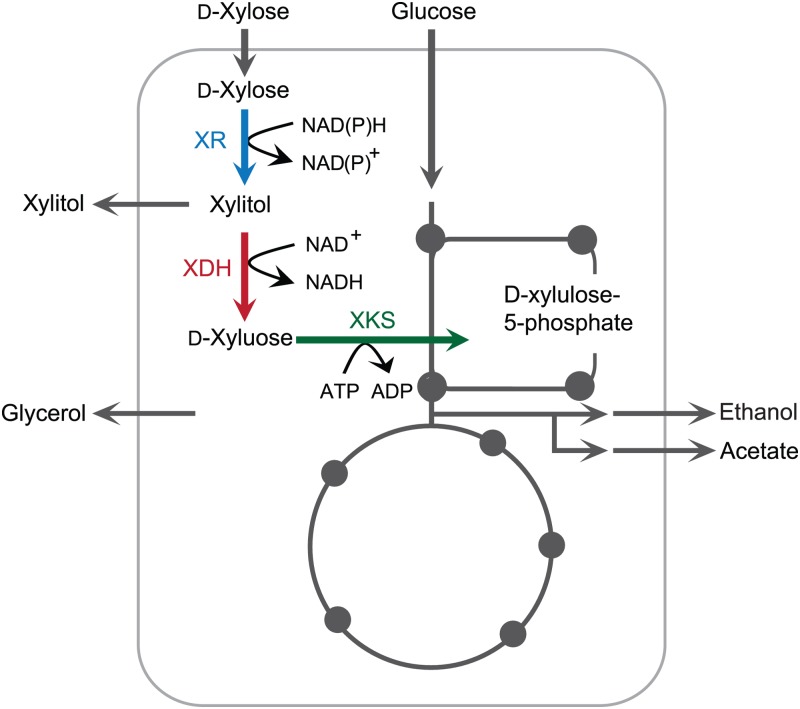
To demonstrate the efficiency and versatility of the method, we applied our combinatorial pathway-engineering approach to a fungal xylose utilization pathway. Xylose is the most abundant pentose sugar (up to 25%) in lignocellulosic biomass (6), and its efficient microbial conversion is a critical step toward the production of value-added products from renewable resources. In S. cerevisiae, xylose catabolism is mediated by a heterologously expressed fungal pathway consisting of xylose reductase (XR), xylitol dehydrogenase (XDH), and xylulose kinase (XKS), producing a pentose phosphate pathway (PPP) intermediate, xylulose-5-phosphate, which can be further metabolized to ethanol (Fig. 1). This pathway is coupled with the endogenous metabolic network by the requirement for NAD(P)H for XR, NAD+ for XDH, and ATP for XKS (Fig. 1). Alternatively, heterologous expression of the bacterial xylose isomerase (XI) can be used to covert xylose into xylulose (7–10). This pathway allows circumventing of the cofactor imbalance problem caused by the different cofactor preferences of XR and XDH. The low expression level of bacterial genes often remains an obstacle, and combination with directed evolution at a protein (isomerase) or organism level was shown to be effective for improved xylose utilization (8, 11–13). In fungal pathways, unbalanced XR and XDH activities and cofactor usage result in xylitol production, reducing the final ethanol yield (14, 15), and XKS activity that is too low or too high hampers cell growth, yielding lower ethanol productivity (16, 17). This suggests that the enzymatic properties of XR, XDH, and XKS need to be carefully tuned so that xylose can be directed to ethanol with unimpaired cell growth and minimized xylitol production.
Fig 1.
Overview of the xylose utilization pathway.
Various fungal and yeast strains, such as Neurospora crassa, Penicillum chrysogenum, and Kluyveromyces lactis, can utilize xylose in nature, suggesting the existence of functional xylose pathways in the organisms, which leads us to the hypothesis that well-balanced xylose pathways could be identified if combinations of the three enzyme homologues from various species or strains could be generated and properly screened. Because of the large number of enzyme homologues, it would be tremendously laborious and time-consuming to construct and test all possible combinations manually. Even worse, there is no guarantee that the best combination found in a strain could be transferrable to other strains where genetic and metabolic differences exist. Our pathway assembly provides an efficient platform for the exploration of the large pools of randomly assembled enzyme combinations using DNA assembler. Superior combinations or pathways for bioethanol production could be isolated by screening based on cell growth, productivity, or yield.
MATERIALS AND METHODS
Pathway library generation by combinatorial assembly.
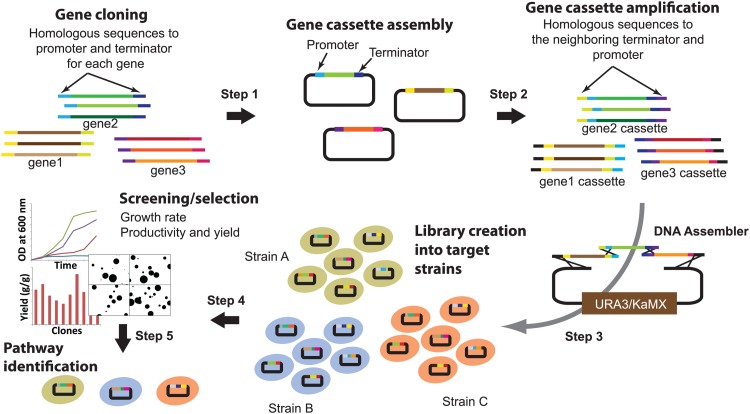
As shown in Fig. 2, bioinformatic tools are used to identify multiple homologues of individual enzymes from a wide variety of organisms. The resulting genes encoding the (putative) enzymes in the pathway are cloned into expression cassettes containing unique promoters and terminators for each set of homologous genes (step 1). Gene cassettes flanked by the sequences homologous to adjacent fragments are amplified by PCR (step 2), and a library of pathways consisting of random combinations of the gene cassettes is generated in a target strain by the DNA assembler method (18) (step 3). The resultant library is subjected to high-throughput screening or selection, depending on culture conditions (step 4), which leads to the identification of improved pathways optimized for different genotypic and phenotypic backgrounds (step 5).
Fig 2.
Overall scheme of the combinatorial pathway engineering approach. In step 1, genes encoding enzyme homologues from various organisms are cloned into gene expression cassette plasmids containing a promoter and terminator pair. During PCR amplification, flanking sequences (30 to 40 nt each) homologous to the end and beginning of the promoter and terminator sequences in the expression cassette plasmids are added to each gene. In steps 2 and 3, the entire expression cassette of each gene is amplified with flanking sequences homologous to the neighboring regions of the recipient plasmids or gene cassettes, assembled, and directed into the host S. cerevisiae strains. In steps 1 and 3, assembly is achieved by endogenous homologous recombination, and the recombination efficiency is controlled by the flanking sequences (80 to 100 nt), identical for each homologue. In steps 4 and 5, the libraries containing all possible combinations of the genes are subjected to screening/selection, and the pathway with balanced enzyme properties is identified for each strain.
Strains, media, and culture conditions.
The Classic Turbo Yeast (CTY) brewing strain was obtained from Still Sprits (Everett, WA), and the diploid laboratory yeast strain INVSc1 (MATα his3-Δ1 leu2 trp1-289 ura3-52) was purchased from Life Technologies (Grand Island, NY). All the other fungal and yeast strains were obtained from the NRRL (ARS Culture Collection, Peoria, IL), ATCC (Manassas, VA), or DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). The genomic DNA of Candida dubliniensis was prepared in Hoyer L. Lois's laboratory (University of Illinois at Urbana-Champaign, Urbana, IL). E. coli DH5α (Cell Media Facility, University of Illinois at Urbana-Champaign, Urbana, IL) was used for recombinant-DNA manipulation. Yeast strains were cultivated in either synthetic dropout medium (1.7 g/liter Difco yeast nitrogen base without amino acids and ammonium sulfate, 5 g/liter ammonium sulfate, 0.8 g/liter amino acid dropout mix) or complex medium supplemented with sugar as a carbon source (1% yeast extract, 2% peptone, 1.0 g/liter adenine hemisulfate). E. coli strains were cultured in Luria broth (LB) (Fisher Scientific, Pittsburgh, PA). S. cerevisiae strains were grown at 30°C and 100 or 250 rpm in nonbaffled and baffled flasks, respectively. Lower rpm and nonbaffled flasks were used to reduce aeration for a higher ethanol yield. E. coli strains were grown at 37°C and 250 rpm. A Transcriptor First Strand cDNA synthesis kit was purchased from Roche (Mannheim, Germany). A Wizard Genomic DNA purification kit was purchased from Promega (Madison, WI), and Zymoprep II was purchased from Zymo Research (Irvine, CA). Phusion DNA polymerase and restriction enzymes were purchased from New England BioLabs (Beverly, MA). The QIAprep spin plasmid miniprep kit, QIAquick PCR purification kit, QIAquick gel purification kit, and RNeasy miniprep kit were purchased from Qiagen (Valencia, CA). Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA). d-Xylose was obtained from Acros Organics (Geel, Belgium), and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA).
Cloning of homologous genes.
Nucleotide BLAST searching (http://www.ncbi.nlm.nih.gov) was used to identify homologues of XR, XDH, and XKS of Scheffersomyces stipitis. Sequence identity and the presence of functional domains in (predicted) protein sequences were used as criteria for the homologue search, and the search was repeated recursively with newly identified genes as templates. Genomic DNA or cDNA synthesized from mRNA was used to clone the genes, depending on the presence of introns. Genomic DNA and mRNA preparation and cDNA synthesis were performed according to the procedures recommended by the manufacturers.
Plasmid and pathway construction.
Most of the cloning and pathway construction was done with DNA assembler (18) unless otherwise noted. Primers were designed so that the fragments to be assembled were flanked with sequences identical to those of adjacent DNA fragments. The overlapping regions were designed to be 80 to 100 nucleotides (nt) long between insert fragments and 100 to 200 nt long between linearized backbone plasmid and insert fragments. Genes encoding homologues of XR, XDH, and XKS were assembled into helper plasmids containing pairs of promoters and terminators for each gene and DNA sequence homologous to the upstream and downstream genes. The ADH1 promoter and ADH1 terminator, PGK1 promoter and CYC1 terminator, and PYK1 promoter and HXT7 terminator were used for the construction of XR, XDH, and XKS cassettes, respectively. The helper plasmids were constructed by assembling promoter and terminator fragments into a pRS414 single-copy plasmid using DNA assembler. During the helper plasmid construction, a unique XhoI site was inserted between the promoter and terminator for linearization. Correct construction of the gene cassettes was confirmed by diagnostic PCR using primers annealing to the end of the promoter and the beginning of the terminator (see Table S1 in the supplemental material for the primer sequences). The final assembly fragments were amplified by PCR, and the sizes of the fragments were confirmed using agarose gel electrophoresis (see Table S2 in the supplemental material for the primer sequences). The confirmed fragments of each gene cassette were purified using a PCR purification kit and cotransferred into the target strains with linearized pRS416 plasmid (100 ng of each fragment) to create xylose pathway libraries. The resulting library size was larger than 1.3 × 104 in all tested strains, and xylose pathway constructs were confirmed by diagnostic PCR and restriction digestion analysis. The combinations of the genes in the pathways of selected recombinants were identified by DNA sequencing (ATCG, Inc., Wheeling, IL). When a larger library was needed, multiple transformations were performed, and the final transformants were combined.
Screening.
After library creation, the transformed cells were plated on xylose plates. The library screening was performed on synthetic complete (SC) medium plates supplemented with 20 g/liter xylose. The cell densities on the plates were adjusted so that the total number of colonies on the plates was larger than all possible combinations of XR, XDH, and XKS (20 × 22 × 19). After 3 days, 50 or 80 clones with the largest colony sizes were selected and inoculated into proper selection medium: SC-dextrose without uracil (SCD-URA) for INVSc1 and SC-dextrose (SCD) plus G418 (5 g/liter) for the ATCC 4124 and CTY strains. Selected clones with large colony size were grown in selection medium for 36 h and inoculated into tubes containing 3 ml of yeast extract-peptone-xylose (YPX) (2% xylose) at an optical density (OD) of 0.1. The OD was measured at three time points (20, 32, and 44 or 48 h), and two OD readings in the exponential growth phase (20 and 32 h) were used to estimate the specific growth rates. The final OD reading (44 or 48 h) was used to ensure proper biomass yield, as well as the high specific growth rates required for high ethanol productivity. The top 10 recombinants with the highest growth rates were selected for the next round of screening. These top 10 fast growers were inoculated into shake flasks (50 ml) containing 10 ml of YPX (2% xylose) at an OD of 1.0. Cell growth was monitored by measuring the OD after 24, 36, and 48 h, and the culture supernatant was analyzed by high-performance liquid chromatography (HPLC) for xylose consumption and product yield. The best recombinant out of 10 was selected based on ethanol yield and productivity.
Fermentation.
For seed cultures, cells were inoculated into selection medium (3 ml) and grown for 24 h at 30°C and 250 rpm. A small volume of the culture (200 μl) was transferred into baffled shake flasks (125 ml) containing 25 ml of selection medium and grown overnight at 30°C and 250 rpm. The cells in the flask cultures were harvested in mid-exponential growth phase, washed with water, and used to inoculate fermentation cultures at an OD of 1.0 (approximately 0.3 g/liter cell dry weight [CDW]). Fermentations were performed in nonbaffled shake flasks (125 ml) with 25 ml of YPX or yeast extract-peptone-dextrose-xylose (YPDX) medium under reduced-aeration conditions (30°C; 100 rpm). Cell growth was monitored by measuring the OD at 600 nm (Cary 300 UV-visible spectrophotometer; Agilent Technologies, Santa Clara, CA). The concentrations of glucose, xylose, xylitol, acetate, glycerol, and ethanol were analyzed by HPLC. An HPLC system equipped with a refractive-index detector (Shimadzu Scientific Instruments, Columbia, MD) and an HPX-87H column (Bio-Rad, Hercules, CA) was operated at a flow rate of 0.6 ml/min at 60°C, using 5 mM sulfuric acid as the mobile phase. The HPLC chromatogram was analyzed using LC Solution Software (Shimadzu Scientific Instruments, Columbia, MD). In the analysis of fermentation data, the time required to consume more than 95% of the total sugar was used to determine the sugar consumption rate, yield, and productivity. For all the selected recombinants, the plasmids were isolated and amplified in E. coli DH5α by transformation. Then, the constructs were isolated from E. coli and used to transform a fresh host strain again to eliminate any metabolic interference from the genomic change(s). The xylose fermentation of the three clones among the 10 fast growers of strain INVSc1 were tested as screened and after retransformation (n = 2), and no significant changes were found (see Fig. S1 in the supplemental material). All the fermentation and enzyme activity data were acquired with the retransformed clones.
Enzymatic activity assay.
The crude lysate was used for enzyme activity analysis in a 96-well plate format using a Biotek Synergy2 microplate reader (Winooski, VT). The activities were measured by monitoring the absorbance at 340 nm for depletion or generation of NAD(P)H as described previously (19–22). A negative control was included, consisting of the yeast with an empty plasmid. Cultures in SC medium were inoculated to an initial OD of 0.2 in culture tubes and were grown for 48 h at 30°C and 250 rpm. Cell mass equivalent to an OD of 20 was collected for lysis with YPER Extraction Reagent (Pierce, Rockford, IL). Bicinchoninic acid (BCA) protein assay reagent (Pierce) was used to determine the protein concentrations, and albumin standard (Pierce) was used for the protein concentration standard curve. Specific activities are expressed as units per milligram of protein. Units are defined as micromoles of NAD(P)H reduced or oxidized per minute. The XR reaction mixture contained 50 mM KH2PO4 buffer (pH 7), 0.2 mM NADPH, and 200 mM xylose. The XDH reaction mixture contained 50 mM KH2PO4 buffer (pH 7), 50 mM MgCl2, 1 mM NAD+, and 200 mM xylitol. The XKS assay was coupled to pyruvate kinase and lactate dehydrogenase as described by Simpson (23), and NADH depletion was monitored. For higher throughput, a modified procedure was used based on the glycerol kit manufactured by R-Biopharm (Darmstadt, Germany). The standard protocol was followed, substituting the crude enzyme XKS for galactose kinase, and 5 mM d-xylulose was used as the substrate, substituting for glycerol.
RESULTS
Library generation and screening.
Two small libraries of xylose-utilizing pathways were created by DNA assembler in order to evaluate the efficiency of library generation and the diversity of the library. The total number of possible combinations in each library was 480 (8 XR × 10 XDH × 6 XKS). The three genes in 16 and 12 pathways isolated from these two independent test libraries, respectively, were all different without significant bias toward specific combinations or genes (Table 1; see Table S3 in the supplemental material). Eight randomly picked clones were tested for their growth on xylose as a sole carbon source. Among the eight clones, two clones showed limited growth up to an OD600 of 1 to 2, and the other six clones grew up to an OD600 of 6 to 14 at different rates (see Fig. S1 in the supplemental material). These results indicated that the functionality (growth on xylose) and efficiency of xylose utilization (growth rate) are directly correlated with the combination of the three enzymes.
Table 1.
Xylose pathways identified in the test libraries
| Clone no.a | Origin of enzyme homologuesb |
||
|---|---|---|---|
| XR | XDH | XKS | |
| 1 | K. lactis | Pachysolen tannophilus | S. stipitis |
| 2 | N. crassa | A. oryzae | S. stipitis |
| 3 | Aspergillus oryzae | C. tropicalis | S. stipitis |
| 4 | A. oryzae | C. tropicalis | P. chrysogenum |
| 5 | Candida tropicalis | P. tannophilus | P. chrysogenum |
| 6 | K. lactis | Talaromyces stipitatus | S. stipitis |
| 7 | Pichia guilliermondii | T. stipitatus | Aspergillus niger |
| 8 | N. crassa | N. crassa | S. stipitis |
| 9 | P. guilliermondii | T. stipitatus | P. chrysogenum |
| 10 | N. crassa | Candida shehatae | C. tropicalis |
Ten combinations of the three xylose pathway genes in 28 randomly picked clones from two independent test libraries are presented (see Table S3 in the supplemental material for the complete list of combinations).
Coverage (number of homologues found in 28 clones/total number of homologues used for library creation) was 7/8 for XR, 8/10 for XDH, and 6/6 for XKS.
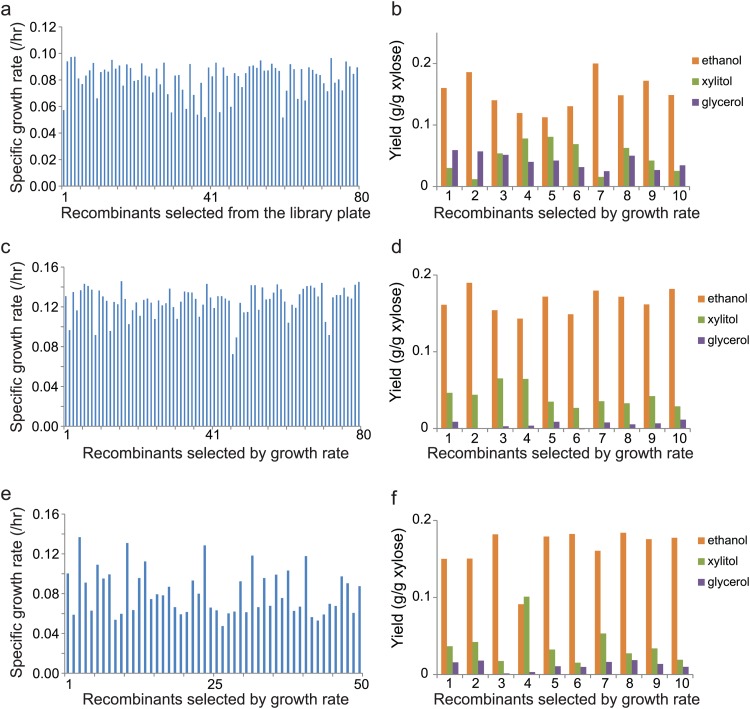
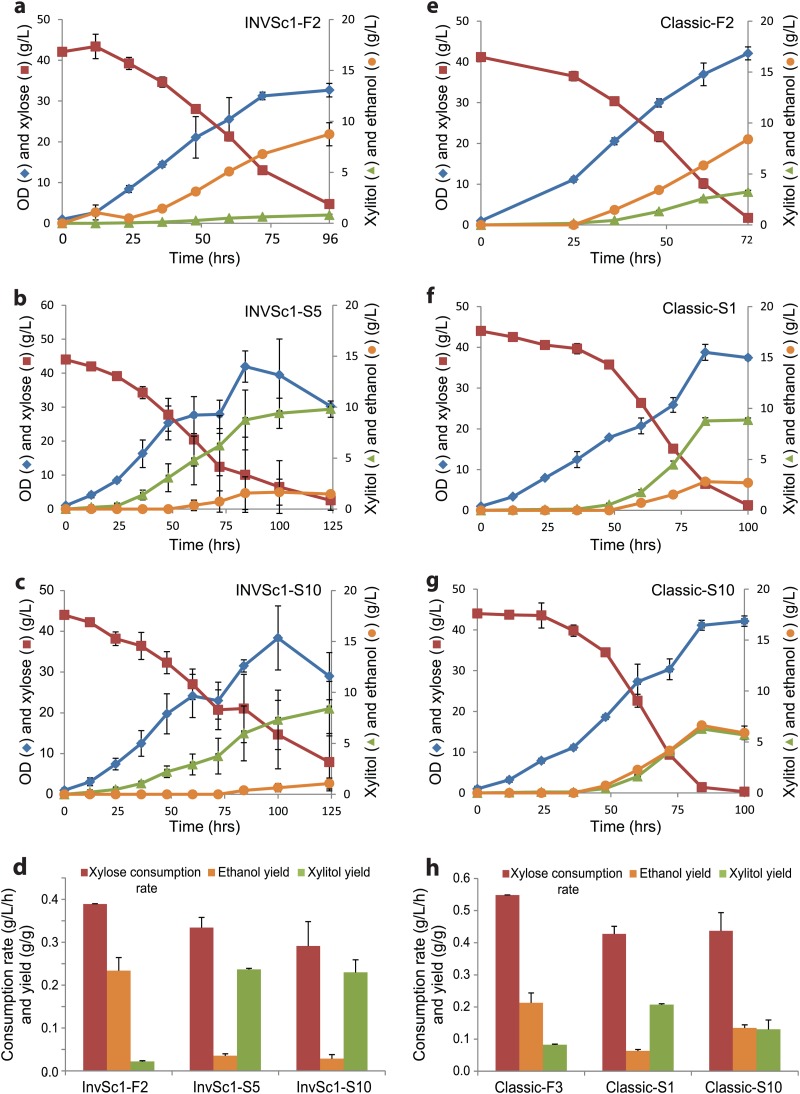
With proven random combination of the three genes and their correlation with xylose utilization, a library of xylose-utilizing pathways with a theoretical size of 8,360 was generated in three S. cerevisiae strains, including a laboratory strain, INVSc1, and two industrial strains, ATCC 4124 and CTY (see Table S1 in the supplemental material). The libraries were prescreened based on the colony size on the selection plates containing xylose as a sole carbon source, and this step ensured the functionality of the selected xylose pathways. In the first-round screening, the prescreened recombinants (80 for INVSc1 and ATCC 4124 and 50 for CTY strains) were ranked by specific growth rate, and the fastest and slowest 10 recombinants were isolated for further characterization. Here, the strains are described with the name of the host strain followed by F or S for the fast or slow grower, respectively, and a number representing the rank of the specific growth rate among the 10 fast or slow growers selected in the first-round screening. For example, INVSc1-F1 and INVSc1-S10 represent the fastest and slowest recombinants among the 10 fast and slow growers selected in the first-round screening of the INVSc1 library. As in the small test library, no combination was duplicated in all tested recombinants (30 fast growers and 10 slow growers) (see Table S4 in the supplemental material). It was noticeable that the XR of the 10 fast growers of the CTY strain was enriched with Aspergillus flavus XR (7/10) (see Table S4 in the supplemental material). No such strong bias toward a specific homologue was found elsewhere. The specific growth rates of the 10 fast growers of INVSc1, ATCC 4124, and CTY were in the ranges of 0.09 to 0.1, 0.14 to 0.15, and 0.09 to 0.14 h−1, respectively (Fig. 3). The metabolic effects of the different combinations were manifested in xylose fermentation under reduced-aeration conditions. Among the 10 fast growers of the laboratory strain (INVSc1), the ethanol yields differed by almost 2-fold (0.11 and 0.20 g/g xylose by recombinant INVSc1-F5 and -F7, respectively), and major by-products also varied with different enzyme combinations. INVSc1-F1 and -F2 produced glycerol, and INVSc1-F4, -F5, and -F6 produced xylitol as a major by-product (Fig. 3). For example, INVSc1-F2 containing Aspergillus nidulans XR, Candida albicans XDH, and S. cerevisiae XKS produced ethanol, xylitol, and glycerol at yields of 0.13, 0.07, and 0.03 g/g xylose, respectively, while INVSc1-F6, consisting of A. nidulans XR, S. stipitis XDH, and Podospora anserina XKS produced ethanol, xylitol, and glycerol at yields of 0.19, 0.01, and 0.06 g/g xylose, respectively (Fig. 3b). The selected ATCC 4124 recombinants had higher specific growth rates than INVSc1 (Fig. 3a and c). Unlike strain INVSc1, xylitol was a major by-product, and the production of glycerol was minimal in the xylose fermentation of all 10 fast growers of strains ATCC 4124 and CTY (Fig. 3d and f). Noticeably, Classic-F4 produced more xylitol than ethanol (Fig. 3f). The combination of enzymes determined the xylose consumption rate and product distribution (Fig. 4). In a comparison between fast and slow growers, INVSc1-F2 produced 8.7 ± 1.3 g/liter ethanol and 1.60 ± 0.04 g/liter glycerol from 38 g/liter of xylose in 96 h, and two slow growers, INVSc1-S5 and -S10, showed an opposite product distribution in xylose fermentations under the reduced-aeration conditions (4%) (Fig. 4a to c). INVSc1-S10 produced 8.4 ± 2.7 g/liter xylitol and 1.0 ± 0.5 g/liter ethanol from 38 g/liter xylose in 125 h (Fig. 4d). Similarly, two pathways, Classic-F3 and -S1, showed opposite product and by-product distributions, and Classic-S10 produced equal amounts of ethanol and xylitol (5.7 ± 0.1 and 5.9 ± 0.7 g/liter, respectively) (Fig. 4e to h). These results clearly demonstrate that the combination of enzymes with different properties has significant effects on xylose utilization, and balanced combinations could be identified using a screening strategy optimized for the target compound.
Fig 3.
Library screening for fast growth and high product yield. (a, c, and e) Specific growth rates of 80 (INVSc1; ATCC 4124) and 50 (Classic Turbo Yeast) recombinants selected from xylose plates. (b, d, and f) Fermentation product profiles of the 10 recombinants (second-round screening) of INVSc1, Classic, and ATCC 4124 strains. The 10 recombinants were selected based on the growth rates determined in the 1st-round screening.
Fig 4.
Xylose fermentation and product distribution of the selected fast and slow growers of INVSc1 and Classic strains. (a to c) Xylose fermentation profiles of one fast grower (INVSc1-F2 [a]) and two slower growers (INVSc1-S5 [b] and INVSc1-S10 [c]). (d) Volumetric xylose consumption rates (g/liter/h) and product yields (g/g consumed xylose) in the fermentation of INVSc1-F2, -S5, and -S10. (e to g) Xylose fermentation profiles of one fast grower (Classic-F3 [e]) and two slower growers (Classic-S1 [f] and Classic-S10 [g]). (h) Volumetric xylose consumption rates (g/liter/h) and product yields (g/g consumed xylose) in the fermentations of Classic-F3, -S1, and -S10. The error bars represent standard deviations (n = 3).
Sugar utilization of the screened recombinants.
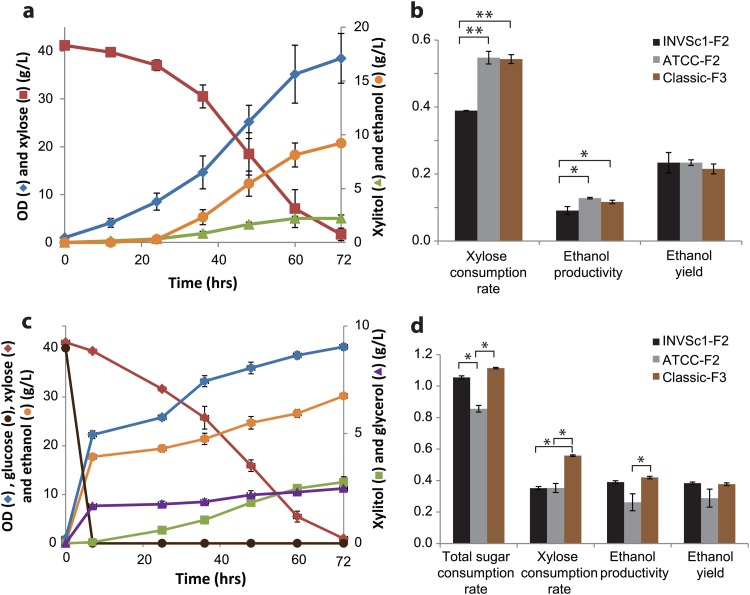
Among the 10 fast growers of each strain, INVSc1-F2, ATCC 4124-F2, and Classic-F3 were selected based on their high ethanol yields in the second-round screening (Fig. 3), and their xylose fermentation properties were determined in detail. In single-sugar (xylose) fermentations, the two industrial strains (ATCC 4124-F2 and Classic-F3) showed equivalent xylose utilization and were more efficient than the laboratory strain (INVSc1-F2) (Fig. 5a and b). INVSc1-F2 consumed 40 g/liter of xylose in 96 h, while ATCC 4127-F2 and Classic-F3 required 72 h, with equivalent ethanol yields. ATCC 4124-F2 and Classic-F3 showed significantly higher xylose consumption rates (0.55 ± 0.02 and 0.54 ± 0.01 g/liter/h, respectively) than INVSc1-F2 (0.39 ± 0.00 g/liter/h; P < 0.01) and higher ethanol productivities (0.13 ± 0.00 and 0.12 ± 0.00 g/liter/h) than INVSc1-F2 (0.09 ± 0.01 g/liter/h). All three recombinant strains showed comparable final ethanol yields in the range of 0.20 and 0.23 g/g xylose (Fig. 5b). Despite the comparable xylose fermentation performances of the two industrial strains, Classic-F3 showed markedly higher total sugar consumption rate, ethanol productivity, and ethanol yield than ATCC 4124-F2 (Fig. 5c and d). Classic-F3 consumed 40 g/liter xylose within 72 h in single-sugar fermentation and cofermentation with 40 g/liter glucose. On the other hand, the xylose consumption of ATCC 4124-F2 was significantly reduced, from 0.55 ± 0.02 to 0.35 ± 0.01 g/liter/h (P < 0.005; n = 3), and its ethanol productivity and yield were even lower than those of the laboratory strain (INVSc1-F2) in mixed-sugar fermentations (Fig. 5c and d).
Fig 5.
Xylose fermentation and cofermentation with glucose of the screened recombinants. (a and b) Xylose (4%) fermentation profile of ATCC 4124-F2 and comparison between selected recombinants of the three strains. (c and d) Glucose (4%) and xylose (4%) cofermentation profiles of Classic-F3 and comparison between selected recombinants of the three strains. In panels b and d, the unit for the consumption and production rate was g/liter/h and the unit for the ethanol yield was g/g sugar consumed. The error bars represent standard deviations (n = 3). Statistical significance: *, n = 3, P < 0.05; **, n = 3, P < 0.005.
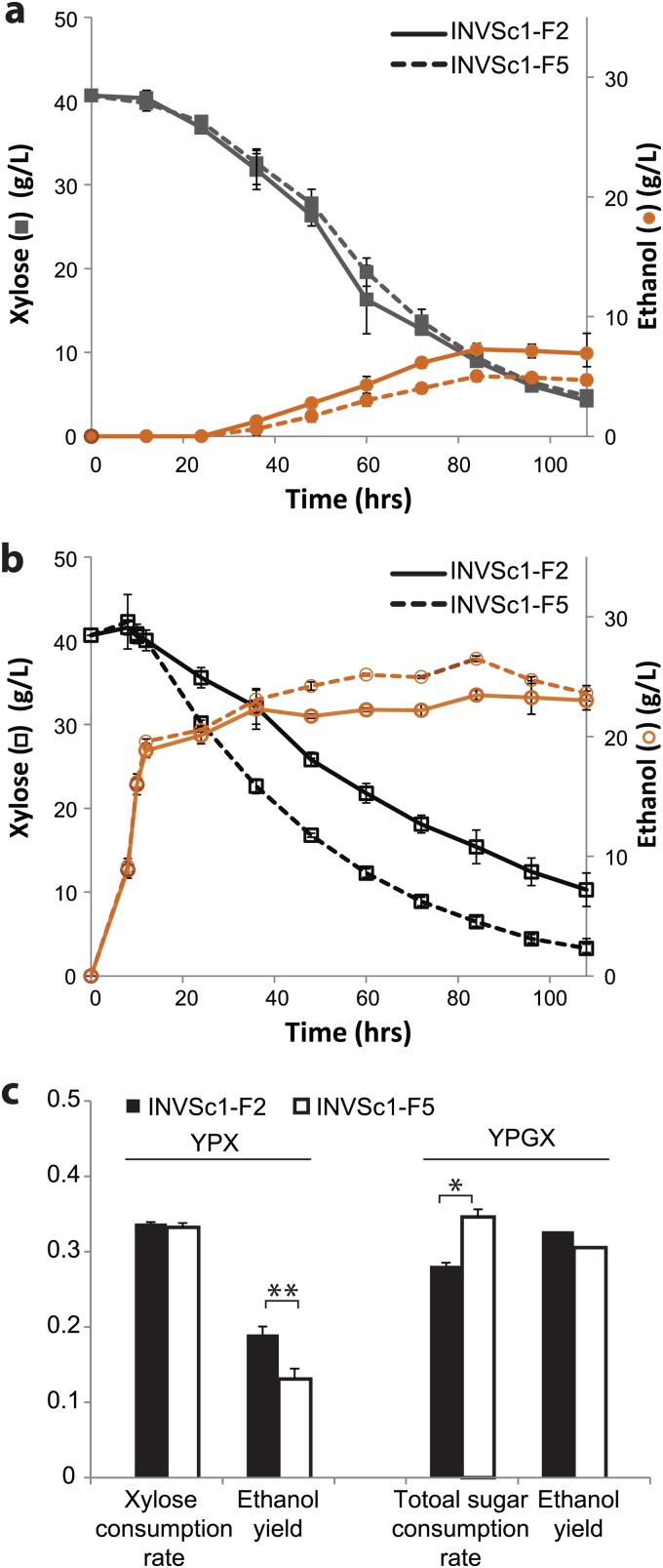
In addition to strain-dependent sugar coutilization, the xylose utilization rates of the recombinants derived from the same host strain were also dependent on the combination of the enzyme homologues in single- and mixed-sugar fermentations. Among the 10 fast growers of INVSc1 recombinants, INVSc1-F2 and -F7 showed higher xylose consumption rates and ethanol yields in the screening in YPX medium (20 g/liter xylose) (see Fig. S2a in the supplemental material). When the same fast growers were subjected to screening in YPGX medium (40 g of glucose and 40 g of xylose), INVSc1-F5 showed the fastest xylose consumption and highest ethanol yield (see Fig. S2b in the supplemental material). Consistent with the screening results, INVSc1-F2 and INVSc1-F5 showed comparable xylose consumption rates, with INVSc1-F2 having a higher ethanol yield in xylose fermentation (4% YPX) (Fig. 6a) (P < 0.005; n = 3). In cofermentation with glucose, the xylose consumption rate of INVSc1-F5 was significantly higher than that of INVSc1-F2, with the same glucose consumption rate and ethanol yield (P < 0.05; n = 3) (Fig. 6b and c).
Fig 6.
Effects of glucose cofermentation on the xylose utilization rate. (a and b) Xylose fermentation (a) and cofermentation (b) profiles of INVSc1-F2 and INVSc1-F5. (c) Consumption rate and ethanol yield comparison. The unit for the consumption and production rates was g/liter/h, and the unit for the ethanol yield was g/g sugar consumed. The error bars represent standard deviations (n = 3). Statistical significance: *, n = 3, P < 0.05; **, n = 3, P < 0.005.
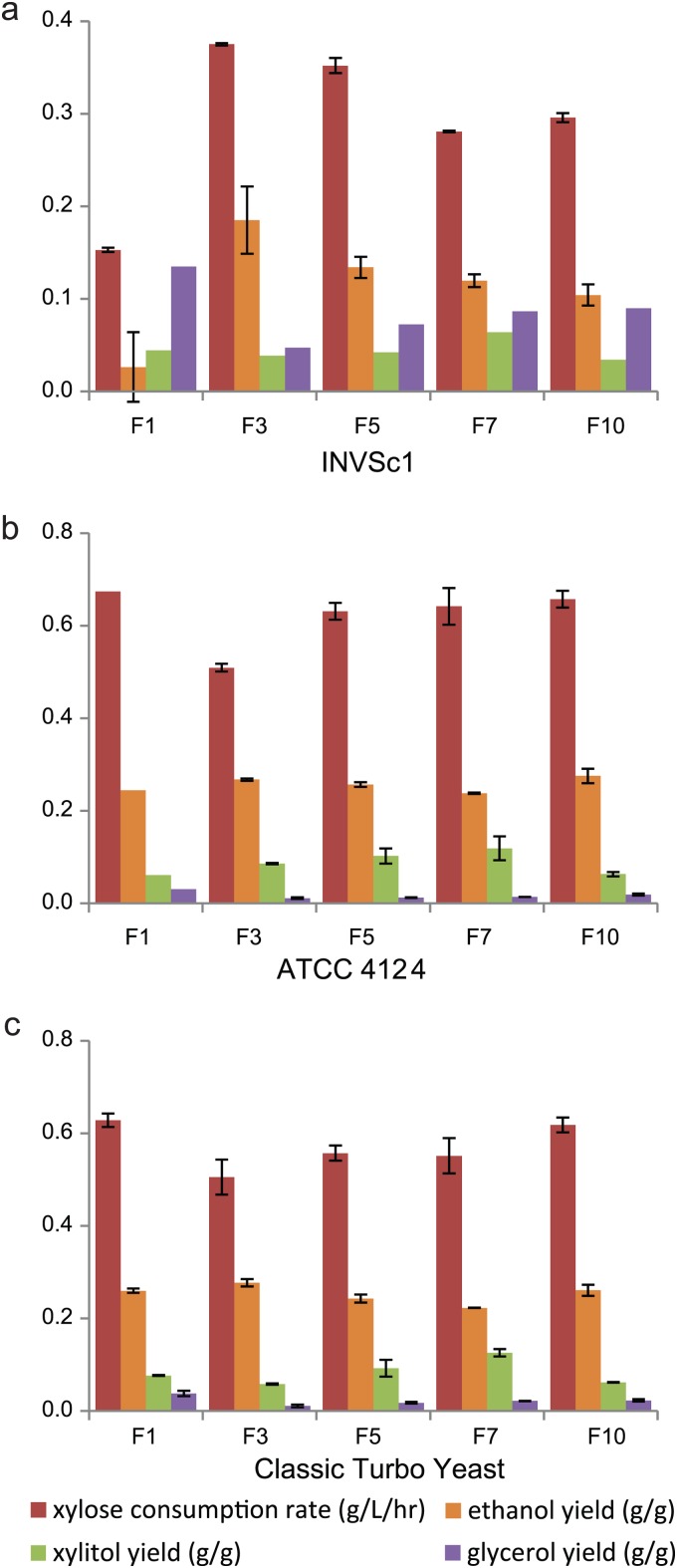
The enzyme combinations identified through screening were strain specific. The five pathways screened from the library of the CTY strain (F1, F3, F5, F7, and F10) were transferred to strains INVSc1and ATCC 4124 to determine the effects of the strain background (Fig. 7). The xylose consumption rates and ethanol yields of all five CTY pathways were substantially reduced, with concurrent increase of by-product formation in a laboratory strain, INVSc1, while their xylose utilization rates were almost identical in two industrial strains (Fig. 7). For example, the ethanol yield of the Classic-F1 pathway dropped by approximately 10-fold, from 0.25 to 0.026 g/g xylose, in the INVSc1 strain. Also, the least efficient Classic-F3 pathway in the two industrial strains was the most efficient in the laboratory strain among the five pathways tested (Fig. 7).
Fig 7.
Xylose utilization efficiencies of the pathways in different strain backgrounds. Shown are the xylose consumption rates and ethanol yields of the INVSc1 (a), ATCC 4124 (b), and CTY (c) recombinants transformed with 5 pathways identified by screening of the CTY library. The error bars indicate standard deviations (n = 3).
Enzyme activities.
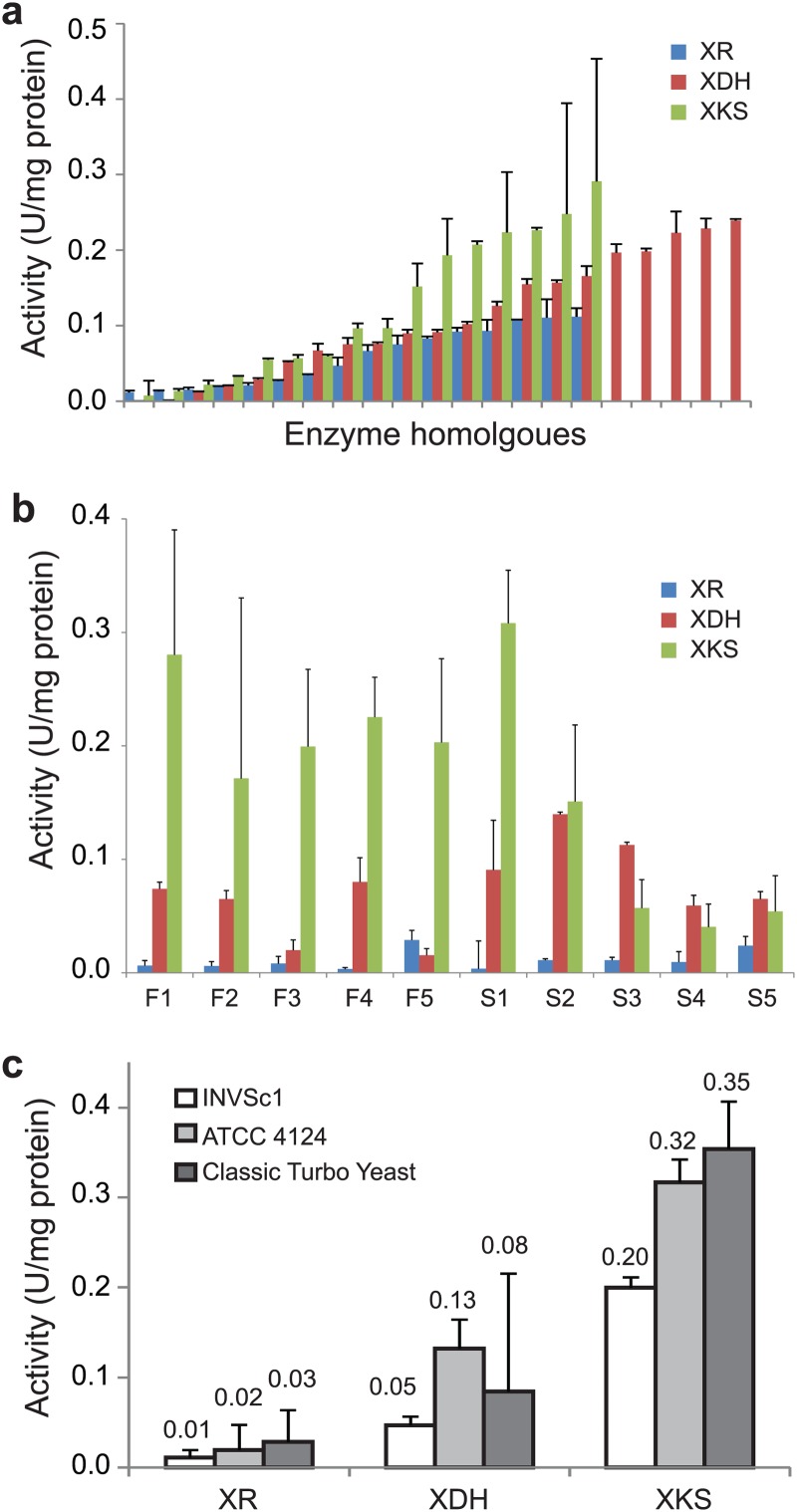
Before the combinatorial assembly of the three enzyme homologues, their individual activities were measured in strain INVSc1. The activities of the XR, XDH, and XKS homologues were in the range of near zero to 0.11, 0.24, and 0.29 U/mg protein and varied by 17-, 19-, and 12-fold for each homologue, respectively (Fig. 8a; see Table S5 in the supplemental material). As was found in the combination of the enzymes and consequent xylose utilization efficiency, various enzyme activities and their ratios were found in the 10 fast and slow growers of the three strains (Fig. 8b; see Fig. S3 in the supplemental material). However, the activities and their ratios were not as diverse as the combinations of the homologues. When the enzyme activities of a total of 60 recombinant pathways (10 fast and 10 slow growers of each strain) were compared, higher XDH than XR activity and higher XKS than XDH activity by 2- to 5-fold were most frequently found regardless of the strain background (52 out of 60; 87%) (see Fig. S3 and S4 in the supplemental material). The average activities of all three enzymes were higher in the industrial strains than in the laboratory strain (Fig. 8c).
Fig 8.
Enzyme activities of XR, XDH, and XKS used for library generation and in the screened xylose pathways. (a) Enzyme activity distributions of XR, XDH, and XKS homologues used for library generation. (b) Measured enzyme activities of five fast growers (INVSc1-F1 to -F5) and five slow growers (INVSc1-S1 to -S5). (c) Average enzyme activities of the 10 fast growers of each strain. The error bars represent standard deviations (n = 2).
DISCUSSION
Combinatorial assembly using DNA assembler was demonstrated to be efficient for generating libraries of metabolic pathways, using xylose utilization pathways as a proof of concept. DNA assembler enabled random combination of three enzyme homologues, and a library size of 104 to 105, larger than the maximum theoretical library size (8,360), could be achieved in a single yeast transformation. If necessary, the library size could be easily increased by increasing the number of cells and amounts of DNA fragments used in the yeast transformation or by performing multiple transformations. The DNA assembler method utilizes yeast homologous recombination for the random assembly of multiple genes. It has an advantage in that the library generation can be performed directly in target strains in a single step, whereas other assembly methods, such as the SLIC (sequence- and ligation-independent cloning) and Gibson methods, require two steps of in vitro assembly and transformation into target host strains (Fig. 1) (24, 25). The DNA assembler-based combinatorial approach requires an expression cassette of each individual gene, consisting of a unique promoter and terminator pair to avoid unwanted homologous recombination. ADH1, PGK1, and PYK1 are all medium-strength promoters and were selected in order not to limit the activity range of any one of the enzymes by using a set of promoters significantly different in transcription strength (26). More than 10 promoters are available for the expression of heterologous enzymes in S. cerevisiae (2, 26, 27). This requirement could be met by carefully designing the expression cassettes, taking into account the promoter strengths under the culture conditions.
The two-step sequential library screening efficiently identified the efficient and balanced xylose utilization pathways for the three strains. The pathway balancing was indicated by the transition of the major products from xylitol and glycerol to ethanol and the activity ratios of XR, XDH, and XKS. The two-step sequential screening was devised due to the nonavailability of a high-throughput screening method for fast growth, high ethanol yield, and productivity. The initial screening screened the library based on growth on xylose as a sole carbon source. Although faster growth is not always associated with high ethanol yield, this strategy has the advantage of screening out all nonfunctional pathways (no growth) from the entire library, efficiently reducing the number of recombinants to be tested for xylose fermentation in the next round of screening. In the 2nd-round screening, all the selected recombinants that had similar growth rates in the 1st-round screening contained unique enzyme combinations and showed markedly different by-product distributions and ethanol yields (Table 1 and Fig. 3 and 4). The complete conversion of the major product between ethanol and xylitol in the xylose fermentations of INVSc1 and CTY recombinants showed the functional, as well as combinatorial, diversity of the xylose metabolic pathways in the libraries and the effects of pathway balancing (Fig. 4). Xylitol production results from the different cofactor preferences and activities of XR and XDH (22, 28, 29), and glycerol is a common by-product in anaerobic glucose fermentation and is related to the regeneration of NAD+ (30). Although the limited oxygen supply and growth in complex media can alleviate the cofactor imbalance reducing xylitol production, the significant reduction of the by-products and higher ethanol yield among the selected recombinants demonstrates the effects of pathway balancing. In particular, a wide range of xylitol yields of INVSc1 recombinants, varying from 0.22 to 0.01, showed the metabolic imbalance and transition from unbalanced to balanced xylose conversion. This pathway balancing was also indicated in the activity ratios. The activities of the individual homologues of XR, XDH, and XKS and their random assembly predicted that about 42% of the combinations would have the observed activity ratio (XKS > XDH > XR) (see Fig. S3 and S4 in the supplemental material). However, the frequency of that ratio among the selected recombinants of three different strain backgrounds was 87% (52 out of 60), which indicated that the combination of the enzymes with this activity ratio was enriched throughout the screening steps. Although these in vitro enzyme activities might not reflect in vivo activities, the more frequent occurrence of the ratio than predicted, by more than 2-fold, suggested that this would be a preferred activity ratio for balanced xylose utilization. XDH activity higher than that of XR was also found in a study in which the activity ratio was modulated by changing the copy numbers of XDH (14).
The by-product yields of the recombinants found in the second-round screening were equivalent to or lower than those of the reference strains under the same aeration and complex-medium conditions (Table 2; see Table S6 in the supplemental material). The xylitol yield of INVSc1-F2 (0.01 to 0.02 g/g xylose) was 5 to 10 times lower than that of the strain engineered from the same strain (INVSc1) by Matsushika et al. (0.1 g/g xylose) (see Table S6 in the supplemental material) (31). Among all the recombinants tested for xylose fermentation, ATCC 4124-S10, which was screened from the 10 slow growers, produced the lowest total by-product yields (0.04 and 0.05 g/g xylose) and the highest ethanol yields (0.27 and 0.31 g/g xylose) in 40- and 80-g/liter xylose fermentations (Table 2; see Table S6 in the supplemental material). Even with the low by-product formation, the ethanol yields were compromised by the high biomass yield, and the ethanol yields obtained were not higher than the reported values. INVSc1-F2 produced 11.0 g/liter of biomass from 40 g/liter of xylose, while an INVSc1 recombinant engineered by Matsushika (S. stipitis XR and XDH and S. cerevisiae XKS) produced only 4.5 g/liter of biomass from 45 g/liter xylose (see Table S5 in the supplemental material) (31). This high biomass yield could explain the lower ethanol yield (0.24 versus 0.36 g/g xylose) (see Table S6 in the supplemental material). The higher biomass yield found across the strains and sugar compositions might result from the growth-based screening criteria used in the first-round screening.
Table 2.
Xylose and mixed-sugar fermentation properties of selected recombinants
| Host strain | Strain | Genotype | OD0a | Duration (h) | Sugar consumption rate (g/liter/h) | Ybmb (g/g) | Yetohc (g/g) | Yxyld (g/g) | Yglyce (g/g) | Yacetf (g/g) |
|---|---|---|---|---|---|---|---|---|---|---|
| Xylose (40 g/liter) | ||||||||||
| INVSc1 | INVSc1-F2 | anidXR caXDH scXKS1 | 1 | 96 | 0.39 | 0.25 | 0.23 | 0.02 | 0.04 | 0.02 |
| Classic | Classic-F3 | anidXR anXDH pcXKS | 1 | 72 | 0.54 | 0.31 | 0.21 | 0.08 | 0.03 | 0.00 |
| ATCC 4124 | ATCC-F2 | pgXR pcXDH aoXKS | 1 | 72 | 0.55 | 0.28 | 0.23 | 0.06 | 0.01 | 0.00 |
| ATCC 4124 | ATCC-S10 | ssXR ssXDH nhXKS | 1 | 72 | 0.62 | 0.28 | 0.27 | 0.03 | 0.01 | 0.00 |
| Glucose (40 g/liter) and xylose (40 g/liter) | ||||||||||
| INVSc1 | INVSc1-F2 | anidXR caXDH scXKS1 | 1 | 72 | 1.05 | 0.13 | 0.37 | 0.06 | 0.03 | 0.03 |
| Classic | Classic-F3 | anidXR anXDH pcXKS | 1 | 72 | 1.11 | 0.15 | 0.38 | 0.07 | 0.03 | 0.00 |
| ATCC 4124 | ATCC-F2 | pgXR pcXDH aoXKS | 1 | 72 | 0.86 | 0.19 | 0.29 | 0.26 | 0.08 | 0.03 |
| Glucose (70 g/liter) and xylose (40 g/liter) | ||||||||||
| Classic | Classic-F3 | anidXR anXDH pcXKS | 5 | 60 | 1.64 | 0.13 | 0.39 | 0.05 | 0.04 | 0.02 |
OD0, initial OD (600 nm).
Ybm, biomass yield, calculated assuming 0.3 g CDW/OD600.
Yetoh, ethanol yield, calculated based on the amount of total sugar consumption.
Yxyl, xylitol, yield calculated based on the amount of consumed xylose.
Yglyc, glycerol yield, calculated based on the amount of total sugar consumption.
Yacet, acetate yield, calculated based on the total amount of sugar consumption.
Correlated with the combination of the enzyme homologues with a given set of promoters and terminators, the differences in the strain backgrounds and sugar compositions significantly affected the combination of the enzymes and their sugar utilization rates and product distributions (Fig. 5 to 7). The more significant reduction in the xylose consumption rate of ATCC 4124-F2 than in those of the INVSc1 and CTY recombinants in mixed-sugar fermentation suggested that glucose repression was affected to different extents depending on the strain background. The two pathways INVSc1-F2 and INVSc1-F5, equivalent in xylose fermentation, differed in xylose consumption in the presence of glucose (Fig. 6), and the same five pathways showed different xylose consumption rates and product distributions in three different strains (Fig. 7). These results, taken together, demonstrated the importance of the strain background and the advantage of our combinatorial approach, which could efficiently identify balanced metabolic pathways optimized for the metabolic environments of host strains. Among numerous factors involved in strain-specific properties, the transcription of XR, XDH, and XKS under various fermentation conditions and in different endogenous metabolic environments, such as the intracellular cofactor and ATP concentrations, would directly affect the enzyme activities and eventually xylose utilization. The higher enzyme activities and resulting faster xylose consumption with higher ethanol yields of the industrial strains and more efficient mixed-sugar fermentation of CTY recombinants over ATCC strains also demonstrate the effects of the strain background. The xylose consumption rates and ethanol yields of ATCC 4124-S2 (0.62 g/liter/h) in single-sugar fermentation and the total sugar consumption rate of CTY-F3 (1.63 g/liter/h) in mixed-sugar fermentation were close to the values produced under the same fermentation conditions by highly efficient xylose-utilizing strains (Table 1; see Fig. S5 and Table S6 in the supplemental material). In this study, the xylose pathways were expressed on single-copy plasmids to eliminate the effects of a possible variation of the copy number. Once the balanced pathways are identified, the amplification of the balanced metabolic pathways using a multicopy expression system, genomic integration followed by evolutionary engineering, or stabilized pathway duplication by chemically induced chromosomal evolution (CIChE) would enable further improvement of xylose utilization (32).
Conclusion.
The combinatorial pathway-engineering approach enables the rapid exploration of a wide range of combinations of the enzymes required for xylose metabolism. Balancing a specific xylose pathway for minimal xylitol formation would require a significant amount of time and effort, even for apparently simple metabolic pathways. We have demonstrated that the combinatorial approach would be a useful approach to quickly identify balanced metabolic pathways without prior knowledge of the details of the enzymatic properties. The combinatorial approach could be used to screen host strains and strain-dependent pathways compatible with the endogenous metabolism of host strains, as well. Various heterologous metabolic pathways, including terpenoid, isoprenoid, and benzylisoquinoline biosynthetic pathways, have been investigated for the industrial production of cosmetic products, food additives, and pharmaceutically active compounds, and balancing these pathways by optimizing protein expression levels and enzymatic properties was found to be critical in improving product yield, selectivity, and productivity (1, 33, 34). Because the combinatorial pathway-engineering method takes advantage of the diversities of isofunctional enzymes and their random combinations, it can be readily applied to the rapid identification and optimization of efficient heterologous pathways in various host organisms to produce a wide variety of value-added chemical products in a highly customized manner.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the BP Energy Biosciences Institute.
We declare that we have no competing financial interests.
Footnotes
Published ahead of print 26 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02736-12.
REFERENCES
- 1. Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. 2010. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330:70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu CF, Jeffries T. 2007. Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 73:6072–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramon A, Smith HO. 2011. Single-step linker-based combinatorial assembly of promoter and gene cassettes for pathway engineering. Biotechnol. Lett. 33:549–555 [DOI] [PubMed] [Google Scholar]
- 4. Alper H, Fischer C, Nevoigt E, Stephanopoulos G. 2006. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U. S. A. 103:3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salis HM, Mirsky EA, Voigt CA. 2009. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27:946–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carroll A, Somerville C. 2009. Cellulosic biofuels. Annu. Rev. Plant Biol. 60:165–182 [DOI] [PubMed] [Google Scholar]
- 7. Bettiga M, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2008. Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnol. Biofuels 1:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MSM, de Laat WTAM, den Ridder JJJ, Op den Camp HJM, van Dijken JP, Pronk JT. 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 4:69–78 [DOI] [PubMed] [Google Scholar]
- 9. Kuyper M, Hartog MMP, Toirkens MJ, Almering MJH, Winkler AA, van Dijken JP, Pronk JT. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5:399–409 [DOI] [PubMed] [Google Scholar]
- 10. Miller KP, Gowtham YK, Henson JM, Harcum SW. 2012. Xylose isomerase improves growth and ethanol production rates from biomass sugars for both Saccharomyces pastorianus and Saccharomyces cerevisiae. Biotechnol. Prog. 28:669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gardonyi M, Hahn-Hagerdal B. 2003. The Streptomyces rubiginosus xylose isomerase is misfolded when expressed in Saccharomyces cerevisiae. Enzyme Microbiol. Technol. 32:252–259 [Google Scholar]
- 12. Lee S-M, Jellison T, Alper HS. 2012. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 78:5708–5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou H, Cheng J-S, Wang BL, Fink GR, Stephanopoulos G. 2012. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng. 14:611–622 [DOI] [PubMed] [Google Scholar]
- 14. Jin YS, Jeffries TW. 2003. Changing flux of xylose metabolites by altering expression of xylose reductase and xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 105:277–286 [DOI] [PubMed] [Google Scholar]
- 15. Karhumaa K, Fromanger R, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2007. High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 73:1039–1046 [DOI] [PubMed] [Google Scholar]
- 16. Jin YS, Ni HY, Laplaza JM, Jeffries TW. 2003. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl. Environ. Microbiol. 69:495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsushika A, Watanabe S, Kodaki T, Makino K, Sawayama S. 2008. Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP+-dependent xylitol dehydrogenase, and xylulokinase. J. Biosci. Bioeng. 105:296–299 [DOI] [PubMed] [Google Scholar]
- 18. Shao ZY, Zhao H, Zhao HM. 2009. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 37:e16 doi:10.1093/nar/gkn991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bera AK, Sedlak M, Khan A, Ho NWY. 2010. Establishment of l-arabinose fermentation in glucose/xylose co-fermenting recombinant Saccharomyces cerevisiae 424A(LNH-ST) by genetic engineering. Appl. Microbiol. Biotechnol. 87:1803–1811 [DOI] [PubMed] [Google Scholar]
- 20. Bergmyer HU. (ed). 1974. Methods of enzymatic analysis, vol 1 Academic Press, New York, NY [Google Scholar]
- 21. Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsushika A, Watanabe S, Kodaki T, Makino K, Inoue H, Murakami K, Takimura O, Sawayama S. 2008. Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81:243–255 [DOI] [PubMed] [Google Scholar]
- 23. Simpson FJ. 1966. d-Xylulokinase. Methods Enzymol. 9:454–458 [Google Scholar]
- 24. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345 [DOI] [PubMed] [Google Scholar]
- 25. Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4:251–256 [DOI] [PubMed] [Google Scholar]
- 26. Sun J, Shao ZY, Zhao H, Nair N, Wen F, Xu JH, Zhao HM. 2012. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109:2082–2092 [DOI] [PubMed] [Google Scholar]
- 27. Nevoigt E, Kohnke J, Fischer CR, Alper H, Stahl U, Stephanopoulos G. 2006. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 72:5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bengtsson O, Hahn-Hagerdal B, Gorwa-Grauslund MF. 2009. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hagerdal R, Gorwa-Grauslund MF. 2006. The expression of a Pichia stipitis xylose reductase mutant with higher Km for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 93:665–673 [DOI] [PubMed] [Google Scholar]
- 30. Guo ZP, Zhang LA, Ding ZY, Shi GY. 2011. Minimization of glycerol synthesis in industrial ethanol yeast without influencing its fermentation performance. Metab. Eng. 13:49–59 [DOI] [PubMed] [Google Scholar]
- 31. Matsushika A, Inoue H, Murakami K, Takimura O, Sawayama S. 2009. Bioethanol production performance of five recombinant strains of laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Bioresour. Technol. 100:2392–2398 [DOI] [PubMed] [Google Scholar]
- 32. Tyo KEJ, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 27:760–765 [DOI] [PubMed] [Google Scholar]
- 33. Hawkins KM, Smolke CD. 2008. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 4:564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leonard E, Ajikumar PK, Thayer K, Xiao WH, Mo JD, Tidor B, Stephanopoulos G, Prather KLJ. 2010. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc. Natl. Acad. Sci. U. S. A. 107:13654–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.