Abstract
The discovery of human metapneumovirus and its implications for respiratory tract disease have emphasized the need for a sensitive, specific, and rapid assay to detect this virus in a clinical setting. It recently became clear that human metapneumovirus can be grouped into at least four genetic lineages. Previously described assays for the detection of human metapneumovirus were developed by using limited sequence information and failed to detect viruses from all four genetic lineages with comparable sensitivities. Here we describe the development and evaluation of a real-time reverse transcriptase PCR assay that detects human metapneumovirus from the four known genetic lineages with equal specificities and sensitivities.
The isolation and characterization of a novel paramyxovirus, human metapneumovirus (hMPV), were recently described (13, 14). hMPV was first isolated from children suffering from acute respiratory tract disease (13); however, infections in all other age groups were later identified as well (2, 6, 12). The virus has been circulating in humans for at least 50 years. The clinical signs associated with hMPV infection appear to be similar to those caused by human respiratory syncytial virus (RSV), which is the most common cause of respiratory tract disease in children (3, 4, 15, 17). hMPV appears to be responsible for about 7 to 10% (3, 5, 8, 9, 11, 12, 15) of cases of acute respiratory tract infections in infants. This relatively high incidence of hMPV infections and the fact that hMPV-associated disease may be severe have emphasized the need for a reliable, sensitive, and rapid diagnostic test for the detection of this virus.
In a diagnostic setting, PCR is generally accepted for the detection of viral infections, particularly for viruses that are difficult to isolate in cell cultures, such as hMPV. To date, four distinct genetic lineages of hMPV have been described (16). For the detection of hMPV, a single assay that is equally sensitive for all four hMPV genetic lineages is preferred. Here we describe the development and evaluation of a new and sensitive real-time reverse transcriptase (RT) PCR (RT-PCR) assay which meets these requirements and a comparison of this assay with previously described assays.
MATERIALS AND METHODS
Clinical samples and virus isolates.
hMPV-positive specimens were obtained from nasopharyngeal samples collected from patients with symptoms of respiratory tract disease (13, 15). Viruses were grown on tertiary monkey kidney (tMK) cells, and the isolates were stored at −70°C. For each of the four genetic lineages of hMPV, a single virus isolate was selected as the prototype isolate (16). RNA from these prototype virus isolates was used to test the designed Taqman primers and probes. The nucleoprotein sequence of lineage A1, strain NL/1/00 can be obtained from the GenBank database (accession number AF371337) (see below for the accession numbers of the sequences submitted for this study).
For validation of the Taqman RT-PCR assay, we used nasopharyngeal samples that were collected in the 2000-2001 and 2001-2002 winter seasons in The Netherlands and that had previously tested positive (n = 38) or negative (n = 54) for hMPV by virus isolation or a standard RT-PCR assay based on the L gene (15). Specificity tests were performed on RNA isolated from stocks of other RNA viruses, including measles virus, mumps virus, simian virus 5, Newcastle disease virus, respiratory syncytial viruses A and B, avian pneumoviruses A, B, and C, human parainfluenza viruses 1, 2, 3, and 4, and influenza viruses A and B. Generation of these virus stocks and RNA isolation were performed as described before (7).
Conventional end-point RT-PCR and DNA blotting.
The conventional end-point RT-PCR assay targeting a 170-bp fragment of the L gene of hMPV was performed with the following primer set: L-6 (5′-CATGCCCACTATAAAAGGTCAG-3′) and L-7 (5′-CACCCCAGTCTTTCTTGAAA-3′). The reaction mixture (total volume, 50 μl) contained 5 μl of RNA, 50 mM Tris-Cl (pH 8.5), 50 mM NaCl, 4 mM MgCl2, 2 mM dithiothreitol, 600 μM deoxynucleoside triphosphates, 200 nM each primer, 20 U of RNasin, 5 U of Taq polymerase (Perkin-Elmer), and 10 U of avian myeloblastosis virus RT (Promega, Leiden, The Netherlands). The RT-PCR parameters were as follows: 60 min at 42°C; 7 min at 95°C; 40 cycles of 1 min at 95°C, 2 min at 45°C, and 3 min at 72°C; and a final incubation at 72°C for 10 min.
PCR products were detected by transferring a 10-μl PCR sample to a Hybond N+ membrane (Amersham Pharmacia Biotech Benelux, Roosendaal, The Netherlands) and then hybridizing the sample with a biotin-labeled probe (5′-CTGTTAATATCCCACACCAGTGGCATGC-3′) as described before (7). DNA fragments were visualized by exposing the blot to X-ray film.
RNA isolation and generation of cDNA.
RNA was isolated with a High Pure RNA isolation kit (Roche Diagnostics, Almere, The Netherlands) according to the manufacturer's instructions. A 5-μl RNA sample was used for the generation of cDNA with specific primers and Superscript III RT enzyme (Invitrogen, Breda, The Netherlands) in a final volume of 20 μl at 55°C according to the manufacturer's instructions. For specificity tests with RNA from other respiratory viral agents, cDNA was generated with random-hexamer primers (Promega) at 50°C. Aliquots of 5 μl of cDNA were used for each real-time PCR. All real-time RT-PCR assays were performed by using two-step reactions, except for experiments in which the limits of detection of the NL-N and ALT-N assays were compared by using runoff transcripts (see below).
Generation of RNA runoff transcripts.
RNA runoff transcripts were made to generate a standard curve for the quantitation of RNA copy numbers. The nucleoprotein sequences of the prototype hMPV strains were cloned into pCITE (Novagen, Madison, Wis.) under the control of a T7 promoter. Runoff transcripts were generated by using the Riboprobe T7 system (Promega, Leiden, The Netherlands) according to the manufacturer's instructions. RNA runoff transcripts were checked by gel electrophoresis and quantitated by measuring the A260 with a spectrophotometer.
Design of oligonucleotides and probes.
Two sets of Taqman primers and probes (NL-N and ALT-N assays) were designed to identify all four genetic lineages of hMPV on the basis of sequence information for the nucleoprotein gene from 53 hMPV-positive clinical specimens. All four genetic lineages of hMPV were present within the panel of isolates. To identify conserved regions in the hMPV nucleoprotein gene and to identify the mismatches of the tested primers and probes with their respective target sequences, entropy plots were created with the BioEdit software package (available through http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The selected primers and probes were located within the most conserved regions of the nucleoprotein gene in all sequences.
The sequences of the primers and probe used for the ALT-N assay were as follows: primer ALT-N-forward (5′-CAACAACATAATGCTAGGACATGTATC-3′), primer ALT-N-reverse (5′-CCGAGAACAACACTAGCAAAGTTG-3′), and probe ALT-N-probe (5′-FAM-TGGTGCGAGAAATGGGTCCTGAATCTGG-3BHQ_1-3′). The sequences for the NL-N assay were as follows: primer NL-N-forward (5′-CATATAAGCATGCTATATTAAAAGAGTCTC-3′), primerNL-N-reverse (5′-CCTATTTCTGCAGCATATTTGTAATCAG-3′), and probe NL-N-probe (5′-FAM-TGYAATGATGAGGGTGTCACTGCGGTTG-TAMRA-3′, in which Y is either a C or a T residue).
Real-time RT-PCR assays.
The real-time RT-PCR assays with viral RNA runoff transcripts to compare the NL-N and ALT-N assays were performed as one-step reactions with a single tube and a final volume of 50 μl containing 10 μl of RNA, 300 nM each primer, 200 nM probe, 5 mM MgCl2, 10 U of StrataScript RT, and 2.5 U of SureStart Taq DNA polymerase. Cycling parameters were as follows: 30 min at 48°C, 5 min at 95°C, and 45 cycles of 30 s at 95°C and 1 min at 60°C. Amplification and detection of RNA from virus isolates or clinical specimens were performed with an ABI Prism 7000 Taqman machine (Applied Biosystems) but with a separate RT step (see above). Each PCR mixture (total volume, 25 μl) contained Taqman universal PCR master mix (Applied Biosystems), 5 μl of cDNA, forward primer (500 nM NL-N-forward or 300 nM ALT-N-forward), reverse primer (250 nM NL-N-reverse or 300 nM ALT-N-reverse), and probe (500 nM NL-N-probe or 200 nM ALT-N-probe). Amplification parameters were 5 min of denaturation and activation at 95°C and 45 cycles of 30 s at 95°C and 1 min at 60°C.
Nucleotide sequence accession numbers.
The nucleoprotein sequences determined in this study can be obtained from the GenBank database as follows: for lineage A2, strain NL/17/00, GenBank accession no. AY355324; for lineage B1, strain NL/1/99, GenBank accession no. AY355328; and for lineage B2, strain NL/1/94, GenBank accession no. AY355335.
RESULTS
Detection of hMPV by end-point RT-PCR and DNA blotting.
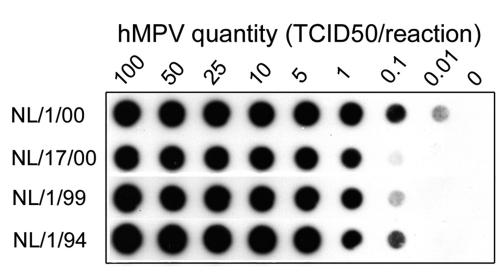
Upon the initial identification of hMPV, an assay including a conventional end-point RT-PCR and hybridization with a biotin-labeled probe was designed for hMPV detection on the basis of the L gene of the hMPV genome (13). Figure 1 demonstrates that the limits of detection of this assay were 0.01 50% tissue culture infective doses (TCID50s) per reaction for the A1 prototype virus strain (NL/1/00), 0.1 TCID50 per reaction for the B prototype virus strains (NL/1/99 and NL/1/94), and 1 TCID50 per reaction for the A2 prototype virus strain (NL/17/00) (Fig. 1). These results illustrate the variable sensitivity of this assay with end-point RT-PCR and DNA blotting for the detection of the four different lineages of hMPV. Recent and more detailed information on the nucleotide sequences of the four known genetic lineages of hMPV indicates that the primer and probe sequences used in this assay contain a significant number of mismatches compared to their target sequences (Fig. 2). These mismatches pose a risk for a low sensitivity of this assay in detecting hMPV strains from certain genetic lineages and may cause false-negative results. Furthermore, this assay is rather time-consuming and not amenable to high-throughput sample processing. Therefore, real-time RT-PCR was applied to improve the sensitivity, reliability, and throughput for the detection of hMPV in a large number of clinical samples.
FIG. 1.
Hybridization blot showing detection of serial dilutions of viral RNAs of the prototype hMPV strains from genetic lineages A1 (NL/1/00), A2 (NL/17/00), B1 (NL/1/99), and B2 (NL/1/94) by using the conventional end-point RT-PCR assay targeting the hMPV L gene (15). Values above the lanes indicate the amounts of virus (in TCID50) from which RNA used in each reaction was obtained.
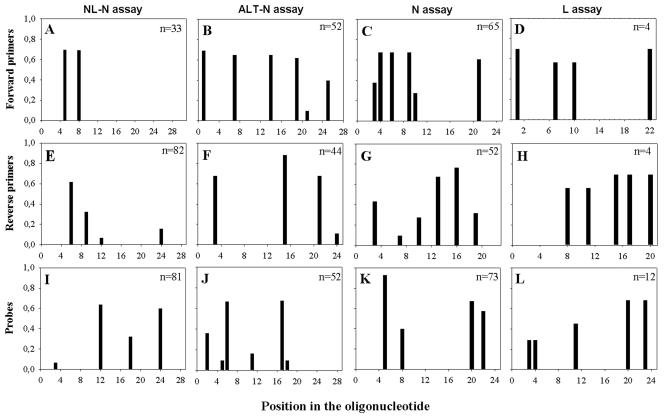
FIG. 2.
Entropy plots of oligonucleotide-annealing sites in hMPV sequences available from the GenBank database. The sequences recognized by the individual oligonucleotides were compared to all available hMPV sequences, and their heterogeneities are displayed as entropy values on the x axis. A higher entropy value indicates mismatches at a particular position in the oligonucleotide with a larger number of target sequences analyzed. The number of sequences upon which each plot was based is given in the upper right corner of each plot. Oligonucleotide positions are given in the 5′-3′ direction, with position 1 being the extreme 5′ nucleotide.
Design of primers and probes for real-time RT-PCR.
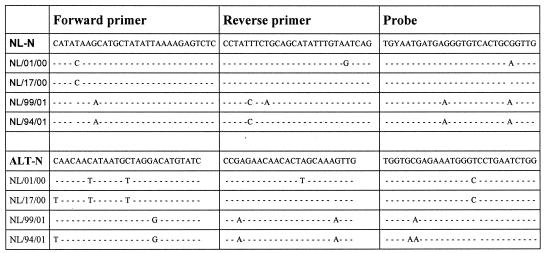
Two sets of primers and probes (for the ALT-N and NL-N assays) were designed to detect virus strains from all known genetic lineages of hMPV. Primers and probes were targeted at the most conserved sequences within the nucleoprotein gene of hMPV and were designed to match all four genetic lineages of hMPV. Figure 3 shows an alignment of the primer and probe sequences with the target sequences of the prototype viruses from all four genetic lineages of hMPV. Previously described primer sets targeting the N (10) or L (15) gene sequences of hMPV were based on the limited sequence information available at that time. The sequences of these primer sets and our newly designed primer sets were compared to the hMPV sequences from the GenBank database and from several strains sequenced in our laboratory. The variability between the viral sequences and each oligonucleotide sequence was calculated by using the entropy algorithm available from the BioEdit software package and revealed a considerable degree of heterogeneity for most of the primer sets (Fig. 2). For successful detection of viral RNA and to minimize the risk of false-negative results, a good match of the 3′ end of the primers is of utmost importance. Only the sequence of the primer set for the NL-N assay showed a good match with the sequences from all available hMPV isolates. In real-time RT-PCR, a good match of the 5′ end of the probe with its target sequence is critical for detecting the presence of viral RNA, as this probe is degraded from the 5′ end onward during amplification of the viral target sequences. The entropy plots in Fig. 2 indicate that the probe for the NL-N amplicon has the fewest number of mismatches and therefore is expected to perform best for all available amplicons.
FIG. 3.
Alignment of sequences of primers and probes for the NL-N and ALT-N assays with the target sequences of the four prototype hMPV strains. Residue Y in the third position of the NL-N probe represents either a C or a T.
Detection of hMPV by real-time RT-PCR with previously described primer sets.
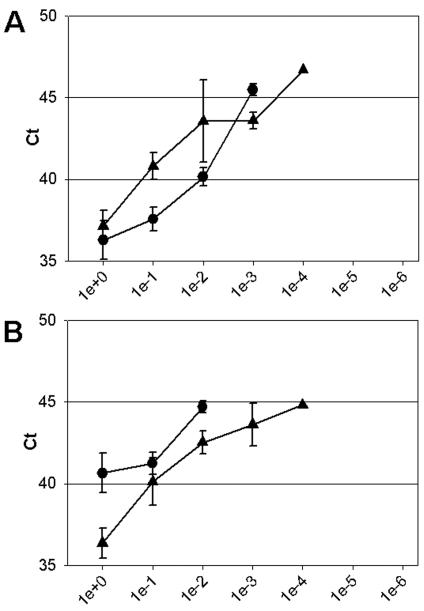
To examine the influence of mismatches in the previously described primers and probes in a real-time RT-PCR setting, we tested whether they were able to detect the four genetic lineages of hMPV. The results obtained with serial dilutions of RNAs from the prototype viruses showed that the primers and probes for the N (10) and L (15) gene assays did amplify target sequences from the hMPV A lineage in our real-time RT-PCR setting but not from viruses of the B lineage (Fig. 4). Hence, the L and N gene assay primers and probes may be of use for conventional RT-PCR (Fig. 1) and for the LightCycler platform (10), respectively, but not for Taqman real-time RT-PCR.
FIG. 4.
Real-Time RT-PCR amplification in the N (A) and L (B) gene assays of serially diluted RNAs of prototype hMPV strains. The values on the x axes indicate the amounts of RNA per reaction, ranging from undiluted (1e+0) to millionfold diluted (1e−6) RNAs. Ct values on the y axes indicate the first PCR cycle at which a positive signal was detected for each sample. Black circles and triangles represent the lineage A1 (NL/1/00) and A2 (NL/17/00) prototype virus strains, respectively. No amplification of viral RNA from lineage B viruses was detected in these experiments. Error bars indicate standard deviations for triplicate samples.
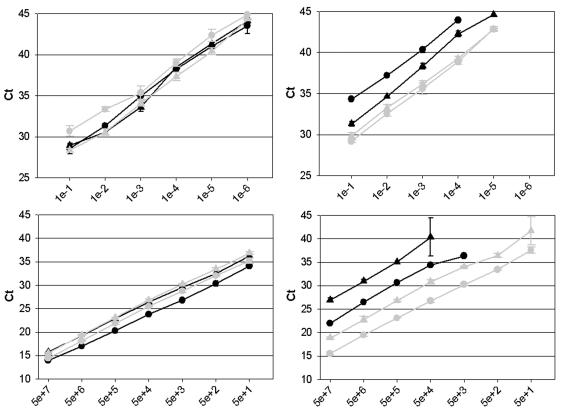
Comparison of NL-N and ALT-N assays.
The newly designed assays (NL-N and ALT-N assays) were tested for their sensitivities as real-time RT-PCR assays by using serially diluted RNAs obtained from the four hMPV prototype strains (Fig. 5, top panels). Identical RNA aliquots were used in both the NL-N and the ALT-N assays. With both assays, we were able to detect viral RNAs from all four prototype strains, but the ALT-N assay showed more variation in the efficiency of detection of the four prototype strains. The NL-N assay, on the other hand, proved to be equally efficient in detecting all four prototype strains. The detection of RNAs from the hMPV prototype strains was maintained up to a dilution factor of 106-fold by the NL-N assay (Fig. 5, top left panel). Further dilution did not result in a positive signal for any of the strains. The approximate limits of detection of the NL-N assay were calculated to be at least 0.006 TCID50 for the A2 prototype hMPV and 0.01 TCID50 for the other prototype viruses. For the ALT-N assay, viral RNAs could be detected up to a dilution of 104-fold for the A1 prototype virus and up to a dilution of 105-fold for the other three prototype viruses (Fig. 5, top right panel).
FIG. 5.
Real-time RT-PCR amplification of serially diluted RNAs from virus stocks (top panels) and RNA runoff transcripts from the nucleoprotein gene (bottom panels) of four prototype hMPV strains inthe NL-N assay (left panels) and the ALT-N assay (right panels). Black symbols are the same as those defined in the legend to Fig. 4. Grey circles and triangles represent the lineage B1 (NL/1/99) and B2 (NL/1/94) prototype virus strains, respectively. The values on the x axes in the top panels indicate the serial dilutions of RNA per reaction, ranging from 0-fold (1e−) to millionfold (1e−) dilutions. The values on the x axes in the bottom panels indicate the numbers of RNA molecules per reaction, ranging from 50 million (5e+7) to 50 (5e+1). Error bars indicate standard deviations. Ct, threshold cycle.
Quantitated RNA runoff transcripts from the nucleoprotein genes of the prototype strains were also used to determine the sensitivities and limits of detection of both assays (Fig. 5, bottom panels). A comparison confirmed that both assays were able to detect target sequences from all four genetic lineages of hMPV, although serial dilutions of the RNA runoff transcripts showed that the ALT-N assay was less sensitive for detecting sequences of the A1 and A2 prototype virus strains (Fig. 5, bottom right panel). The NL-N assay could easily detect 50 RNA copies from each of the four prototype hMPV strains with equal efficiencies (as indicated by the similar Ct values [first PCR cycle at which a positive signal was detected for each sample] for each dilution of the different RNA samples). Further dilution of the target RNAs from the A1 and B1 prototype virus strains to five copies still yielded a positive signal in most experiments (data not shown). Considering errors in quantitation of the RNA runoff transcript copy numbers and pipetting errors in making serial dilutions, we estimate the limit of detection of the NL-N assay to be about 5 to 10 copies per reaction. The ALT-N assay could detect 50 RNA copies from the B1 and B2 prototype virus strains but was much less efficient in detecting RNA copies from the A1 and A2 prototype virus strains (as indicated by the higher Ct values for each dilution and the lower sensitivity in detecting diluted RNA).
Specificity of the NL-N assay.
To test whether the primers and probe designed for the NL-N assay were specific for hMPV, we used RNAs from 15 other common respiratory viral agents as templates for real-time RT-PCR analysis. These templates were isolated from stocks of measles virus, mumps virus, simian virus 5, Newcastle disease virus, respiratory syncytial viruses A and B, avian pneumoviruses A, B, and C, human parainfluenza viruses 1, 2, 3, and 4, and influenza viruses A and B. The hMPV A1 prototype strain was used as a positive control sample. For this experiment, cDNAs from all samples were generated with random-hexamer primers in a separate RT step. None of the RNA templates isolated from the non-hMPV strains resulted in a positive signal, confirming that the designed primers and probe were specific for hMPV (data not shown).
We next used the newly developed real-time RT-PCR NL-N assay to detect hMPV RNA in clinical samples that had previously been found to be either positive or negative for hMPV by the conventional RT-PCR specific for the L gene (15). A panel of 38 clinical samples that had previously been found to be positive were all confirmed to be positive by our real-time RT-PCR NL-N assay. An additional panel of 54 samples that had previously been found to be negative did not give a positive signal in our new assay (data not shown). These results indicate that our new assay is at least as sensitive as conventional RT-PCR targeting of the L gene of hMPV and does not produce false-negative results.
DISCUSSION
The implications of the discovery of hMPV for respiratory tract disease, especially in young children and immunocompromised individuals, emphasize the need for a reliable and sensitive assay for detecting this virus. Recently, it became clear, on the basis of sequence analyses of the hMPV genome, that hMPV strains circulating around the world can be divided into two main genetic lineages (A and B) representing two serotypes and each comprising two sublineages (A1, A2, B1, and B2) (1, 16). Upon the initial identification of hMPV, a set of primers and a probe targeting the L gene sequence of hMPV were designed and shown to be able to amplify and detect hMPV strains from all four genetic lineages in a conventional end-point RT-PCR setting. This assay, however, was not equally sensitive for virus strains from all four known genetic lineages. Furthermore, because the primers and probe used in this assay were designed on the basis of limited sequence data, the oligonucleotide sequences showed a considerable number of mismatches compared to their target sequences, posing a risk of obtaining false-negative results.
Here we describe the development and evaluation of a real-time RT-PCR assay (NL-N assay) that can detect the four genetic lineages of hMPV described to date with comparable sensitivities. Primers and a probe were designed on the basis of a wide range of hMPV sequences available from public databases with a minimum number of mismatches. Figures 2 and 3 show that for the NL-N assay primers, a few mismatches do exist, but since these are mainly in the 5′ end, they probably have little impact on PCR amplification. The probe was chosen such that any mismatches were at the 3′ end and not at the 5′ end of the probe. This choice ensures that the 5′ end of the probe is firmly attached to its target sequence during PCR and is thus efficiently hydrolyzed by the endonuclease activity of the DNA polymerase.
The entropy plots showing the mismatches of the primer and probe sequences with their target sequences (Fig. 2) also indicate why the previously described primers and probes targeting the N (10) and L (15) gene sequences of hMPV were not successful in efficiently amplifying RNAs from all four prototype hMPV strains in our experimental real-time RT-PCR setup. As both of these assays were developed on the basis of limited sequence information primarily available for lineage A strains of hMPV, mismatches between primer and probe sequences and their respective target sequences were most severe for lineage B strains. These multiple mismatches are most likely the reason why we were not able to amplify RNAs from lineage B prototype virus strains (NL/1/99 and NL/1/94) with these primers in our Taqman setup (Fig. 4). The inefficiency of both assays is also illustrated by the high Ct values (>35 cycles) and the high number of cycles needed to produce a clear signal. It should be noted that the N gene primers described by Mackay et al. (10) were originally designed for use with a LightCycler platform. We cannot rule out the possibility that these primers in LightCycler assays can detect all four genetic lineages of hMPV. With a Taqman platform, the relatively low annealing temperature of the probe (<60°C) may result in inefficient hydrolysis of the probe and poor detection of the amplified product. Although the L gene assay did not perform well in the Taqman real-time RT-PCR setting, Fig. 1 shows that the primers and probe used for this assay are capable of detecting target sequences for all prototype hMPV strains in a conventional end-point RT-PCR setting. This result suggests that the mismatches in the L gene primers had much less of an effect in the conventional RT-PCR assay than in the real-time RT-PCR assay. This discrepancy is probably the result of the lower annealing temperature (45°C) used in the conventional RT-PCR setting, where the level of binding of the primers to the target sequence was apparently sufficient for PCR amplification.
In general, real-time RT-PCR assays have a very high level of specificity. Apart from the primers, the specificity of the reaction is also determined by binding of the probe to its specific target sequence. The specificity of the assay is especially important, as clinical samples from patients with symptoms of respiratory disease may contain any of a number of different respiratory viral agents. The NL-N assay did not give any positive signal when RNAs from other respiratory viruses were used as templates, indicating that the assay is indeed specific for hMPV. The assay performed well with clinical samples from patients with symptoms of respiratory disease. From a panel of 92 clinical samples, the assay could identify all samples that had been shown to be hMPV positive by previously conducted conventional RT-PCR analysis or virus isolation. Samples that had been shown to be negative by these previous tests did not yield a positive signal in our real-time RT-PCR assay. These results indicate that the risk of misinterpretating clinical samples, both as false-negative results and as false-positive results, is very limited with this real-time RT-PCR assay.
The main reason for developing this new assay was to develop a single test that can identify virus strains from all four hMPV genetic lineages with similar sensitivities. By testing serial dilutions of RNAs from four prototype hMPV strains, we concluded that the assay worked equally well for all four genetic lineages of hMPV, with a limit of detection of about 0.01 TCID50. Whereas TCID50 is an indication of the number of infectious viral particles, it may be a poor measure of viral genomic RNA copy numbers. To obtain a more accurate indication of the limit of detection of our NL-N assay, we generated RNA runoff transcripts from the nucleoprotein gene of the A1 and B1 prototype hMPV strains. Upon quantitation, serial dilutions of the RNA runoff transcripts tested in our assay indicated that the limit of detection of our assay was about 5 to 10 copies of the RNA target sequence.
In conclusion, we have developed a real-time RT-PCR assay for the amplification and detection of RNA of hMPV from all four genetic lineages. The results show that the sensitivities of the assay for strains of hMPV from all four genetic lineages are similar and that the assay is approximately 10-fold more sensitive than the previously described conventional RT-PCR assay targeting the hMPV L gene sequence (15). Our assay also proved to be faster and highly specific and can be used for future implementation in a diagnostic setting.
REFERENCES
- 1.Bastien, N., S. Normand, T. Taylor, D. Ward, T. C. T. Peret, G. Boivin, L. J. Anderson, and Y. Li. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., G. De Serres, S. Cote, R. Gilca, Y. Abed, L. Rochette, M. G. Bergeron, and P. Dery. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domachowske, J. B., and H. F. Rosenberg. 1999. Respiratory syncytial virus infection: immune response, immunopathogenesis, and treatment. Clin. Microbiol. Rev. 12:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esper, F., D. Boucher, C. Weibel, R. A. Martinello, and J. S. Kahn. 2003. Human metapneumovirus infection in the Unites States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111:1407-1410. [DOI] [PubMed] [Google Scholar]
- 6.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., T. M. Bestebroer, S. Herfst, L. Van der Kemp, G. F. Rimmelzwaan, and A. D. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freymuth, F., A. Vabret, L. LeGrand, N. Eterradossi, F. Lafay-Delaire, J. Brouard, and B. Guillois. 2003. Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr. Infect. Dis. J. 22:92-94. [DOI] [PubMed] [Google Scholar]
- 9.Jartti, T., B. van den Hoogen, R. P. Garofalo, A. D. Osterhaus, and O. Ruuskanen. 2002. Metapneumovirus and acute wheezing in children. Lancet 360:1393-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackay, I. M., K. C. Jacob, D. Woolhouse, K. Waller, M. W. Syrmis, D. M. Whiley, D. J. Siebert, M. Nissen, and T. P. Sloots. 2003. Molecular assays for detection of human metapneumovirus. J. Clin. Microbiol. 41:100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nissen, M. D., D. J. Siebert, I. M. Mackay, T. P. Sloots, and S. J. Withers. 2002. Evidence of human metapneumovirus in Australian children. Med. J. Aust. 176:188-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peret, T. C., G. Boivin, Y. Li, M. Couillard, C. Humphrey, A. D. Osterhaus, D. D. Erdman, and L. J. Anderson. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 185:1660-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van den Hoogen, B. G., J. C. De Jong, J. Groen, T. Kuiken, R. De Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van den Hoogen, B. G., T. M. Bestebroer, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 15.Van den Hoogen, B. G., G. J. J. van Doornum, J. Fockens, J. J. Cornelissen, W. E. P. Beyer, R. De Groot, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalised patients. J. Infect. Dis. 188:1571-1577. [DOI] [PubMed]
- 16.Van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo-Neto, R. L. de Swart, A. D. M. E. Osterhaus, and R. A. M. Fouchier. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 17.Viazov, S., F. Ratjen, R. Scheidhauer, M. Fiedler, and M. Roggendorf. 2003. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 41:3043-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]