Abstract
Edible brown algae are used as major food material in Far East Asian countries, particularly in South Korea and Japan. They contain fermentable dietary fibers, alginic acid (uronic acid polymer) and laminaran (β-1,3-glucan), that are fermented into organic acids by intestinal bacteria. To clarify the effect of edible algae on the intestinal environment, the cecal microbiotas of rats fed diets containing no dietary fiber (control) or 2% (wt/wt) sodium alginate or laminaran for 2 weeks were analyzed using FLX amplicon pyrosequencing with bar-coded primers targeting the bacterial 16S rRNA gene. The most abundant phylum in all groups was Firmicutes. Specifically, Allobaculum was dominant in all diet groups. In addition, Bacteroides capillosus (37.1%) was abundant in the alginate group, while Clostridium ramosum (3.14%) and Parabacteroides distasonis (1.36%) were only detected in the laminaran group. Furthermore, rats fed alginate showed simplified microbiota phylotypes compared with others. With respect to cecal chemical compounds, laminaran increased cecal organic acid levels, particularly propionic acid. Alginate increased total cecal organic acids. Cecal putrefactive compounds, such as indole, H2S, and phenol, were decreased by both alginate and laminaran. These results indicate that edible brown algae can alter the intestinal environment, with fermentation by intestinal microbiota.
INTRODUCTION
The adult human intestine contains 1013 to 1014 bacteria, involving at least 500 different species or strains (1), which make up the gut microbiota. While up to 9 different bacterial phyla typically comprise the microbiota, Firmicutes and Bacteroidetes account for >90% of microbiota (2). The intestinal microbiota plays an important role in host health (3). The role of bacteria can be seen as 2-fold, encompassing both beneficial and harmful effects on the host. Beneficial effects include prevention of pathogen colonization and stimulation of immune responses (4, 5), assistance in digestion and absorption (6), and vitamin synthesis (7). Harmful effects include the production of intestinal putrefactive compounds, such as ammonia, H2S, amines, phenols, and indoles (8). These putrefactive compounds are regarded as putative carcinogens and toxins (9). The intestinal microbiota depends on various factors, such as aging, stress, climate, infectants, disease, drugs, and diet (10). Moreover, diet composition is dependent on geographic location and culture. Such differences can also affect the intestinal microbiota. For example, De Filippo et al. (11) reported that the microbiota of breast-fed babies in Europe and Burkina Faso were identical, irrespective of location and culture. However, when breast-fed children were exposed to the local diet microbiota, alterations were observed. For example, levels of Firmicutes fecal bacteria increased in European children, while levels of Bacteroidetes fecal bacteria increased in Burkina Faso children.
In Far East Asian countries, particularly in South Korea and Japan, various marine algae (seaweeds) are used as food material. In particular, many kinds of edible brown algae, Phaeophyceae, such as kombu (Laminaria japonica), wakame (Undaria pinnatifida), and hijiki (Hijikia fusiforme), are consumed (12). Recently, it was proposed that marine red algae, Rhodophyta (such as nori Porphyra species), and associated marine bacteria might have been the route by which novel carbohydrate active enzymes, capable of degrading the red-algal polysaccharide porphyran, were acquired by the intestinal bacteria of Japanese individuals (13). This knowledge has had an impact on the research area. However, since ancient times, the supply of brown algae has actually been greater than that of red algae in Japan (14). Brown algae contain water-soluble polysaccharides (dietary fibers), such as alginate, fucoidan, and laminaran (12, 15). Alginates are viscous compounds found in the cell wall matrix and are polymers of glucuronic and mannuronic acids. Fucoidans are sulfated fucans, which are also located in the cell wall matrix. On the other hand, laminaran is a β-1,3-glucan (terminating with glucose) that is contained in all cells as a storage polysaccharide. These polysaccharides are not digested by human intestinal enzymes but are instead broken down and fermented into propionic and butyric acids by the intestinal microbiota, similar to the effects of prebiotics (16, 17). Furthermore, these fermentable fibers were observed to suppress intestinal putrefactive compounds, such as indole and phenol (8). Several intestinal bacteria, such as Bacteroides distasonis, Bacteroides ovatus, and Clostridium ramosum, are known as alginic acid and/or laminaran fermentable intestinal bacteria (18). Kuda et al. (19) reported that oligosaccharides derived from laminaran by C. ramosum promoted the growth of Bifidobacterium strains. Moreover, increased levels of bifidobacteria in the rat cecum by laminaran diet administration have been reported (17). However, these results were determined by culture-dependent methods able to detect only 1% of microorganisms from most environments and capable of cultivating only 20 to 40% of intestinal microbiota, due to culture media limitations and the existence of very-oxygen-sensitive bacteria (20).
In the last decade, molecular methods such as denaturing gradient gel electrophoresis (DGGE) (21) and terminal restriction fragment length polymorphism (T-RFLP) (22) have been employed in the analysis of microbiota from various environments, including the intestine. Recently, FLX amplicon pyrosequencing was used in the identification of microbial communities in the human intestinal tract (23). This method can clearly and minutely identify the microbiota in one run.
To clarify the effect of edible algae on the intestinal environment, cecal microbiotas of rats fed diets containing no dietary fiber (control), 2% (wt/wt) sodium alginate, or 2% (wt/wt) laminaran for 2 weeks were analyzed using FLX amplicon pyrosequencing with bar-coded primers targeting the bacterial 16S rRNA gene. Furthermore, effects on cecal organic acid composition and putrefactive compounds were also determined.
MATERIALS AND METHODS
Animal experiment.
The animal experiment was performed in compliance with the fundamental guidelines for proper conduct of animal experiments and related activities in academic research institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan and approved by the animal experiment committee of Tokyo University of Marine Science and Technology (approval no. 2011-10). Four-week-old male Wistar rats (Clea Japan Inc., Tokyo, Japan) were housed separately in metal wire cages and allowed free access to water and food. After acclimation with a control diet (AIN-76-based diet) (Table 1) for 7 days, the animals were divided into three groups (n = 6) and given a control diet or experimental diet containing 2% (wt/wt) of either sodium alginate (Wako Pure Chemical Industries, Osaka, Japan) or laminaran (Tokyo Kasei, Tokyo, Japan) for 2 weeks. Body weight, fecal frequency, and fecal amount were determined at the same time daily. Next, rats were anesthetized with diethyl ether and exsanguinated from the abdominal aorta. The cecum was excised and weighed. A portion of the cecal content was used for direct total cell counts using the Gram stain method (24). The remaining cecal content was stored at −80°C and used for subsequent experiments.
Table 1.
Composition of test diets
| Diet type | Composition (grams/100 g) of test diet |
||
|---|---|---|---|
| Control | Alginate | Laminaran | |
| Sucrose | 50.0 | 50.0 | 50.0 |
| Casein | 20.0 | 20.0 | 20.0 |
| Corn starch | 20.0 | 18.0 | 18.0 |
| Sodium alginate | 2.0 | ||
| Laminaran | 2.0 | ||
| Corn oil | 5.0 | 5.0 | 5.0 |
| AIN-76 mixed minerals | 3.5 | 3.5 | 3.5 |
| AIN-76 mixed vitamins | 1.0 | 1.0 | 1.0 |
| dl-Methionine | 0.3 | 0.3 | 0.3 |
| Choline bitartrate | 0.2 | 0.2 | 0.2 |
Cecal chemical compounds.
Cecal contents were diluted with four volumes of distilled water, and levels of organic acids, cecal polysaccharides, and low-molecular-weight saccharides were determined.
Organic acids (lactic acid, acetic acid, propionic acid, and n-butyric acid) were determined by high-pressure liquid chromatography (HPLC) procedures according to our previous report (8). Briefly, 200 μl of cecal suspension was acidified with 50 μl of 1 mol/liter sulfuric acid and centrifuged at 15,000 × g for 3 min at 4°C. After centrifugation, the supernatant was passed through a 0.45-μm-pore-size filter and injected into the HPLC instrument using an ICSep ICE-ORH-801 column (Tokyo Kasei), operated at 35°C, and eluted with 0.005 mol/liter of H2SO4 at a flow rate of 0.8 ml/min. Eluted compounds were detected by a refractive index detector.
The cecal polysaccharide and low-molecular-weight saccharide contents were determined by the phenol-sulfuric method (25) as mentioned in a previous report (15). Briefly, saccharide in the aqueous solution was determined as the total saccharide. Polysaccharide in the solution was precipitated by the addition of ethanol. Next, the saccharide content in the supernatant was determined as the low-molecular-weight saccharide. Polysaccharide content was calculated by the subtraction of the low-molecular-weight saccharide from the total water-soluble saccharide.
Levels of ammonia, phenol, and sulfide compounds were determined using reagent sets for water analysis (no. 7, 17, and 53; Kyoritsu Chemical-Check Lab. Co., Tokyo, Japan) after dilution with four volumes of distilled water (8). The level of indole was measured using Kovac's reagent (26).
Pyrosequencing of bacterial 16S rRNA fragments.
Bacterial DNA from each cecal content sample was extracted using a Fast Pure DNA kit (TaKaRa Bio Inc., Shiga, Japan) (27). Purified DNA was dissolved in Tris-EDTA buffer and used as the DNA template in pyrosequencing.
DNA sequences of cecal contents were amplified individually with primer pairs for the 16S rRNA gene. The region of the 16S rRNA gene was amplified by PCR using bar-coded primers targeting 27 to 338 bp. The primers contained five-base sample-specific bar code sequences denoted as “X” and common linker (AC) sequences in the 5′ end (23). The forward primer was 5′-ctatgcgccttgccagcccgctcag NNNNN AGAGTTTGATCCTGGCTCAG-3′, where the sequence of the A adapter is shown in lowercase letters and N represents a 5-bp bar code that is unique for each sample. The reverse primer was 5′-ctatgcgccttgccagcccgctcag TGCTGCCTCCCGTAGGAGT-3′, where the sequence of the A adapter is shown in lowercase letters. PCR amplification was performed in 100-μl reaction mixtures composed of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 50 pmol of each primer, 0.2 mM (each) 4 deoxynucleoside triphosphates (dNTPs), 2.5 U of TaKaRa Ex Taq DNA polymerase (TaKaRa Bio), and 50 ng of template DNA. PCRs were performed with an initial annealing temperature set at 7.8°C above the expected annealing temperature, decreasing by 0.6°C every second cycle until the expected annealing temperature (60.2°C) was reached (total of 26 cycles), followed by 5 additional cycles at the expected annealing temperature. Amplification was carried out using the following cycle: denaturation was carried out at 94°C for 30 s, the annealing time was 30 s, and primer extension was performed at 72°C for 10 s using a GeneAmp 9700 thermal cycler (Applied Biosystems). Aliquots (5 μl) of the PCR products were analyzed first by electrophoresis in 1% (wt/vol) agarose gels. The PCR products were purified using the MinElute PCR purification kit (Qiagen). An equal quantity (100 ng) of each PCR amplicon, tagged with the sample-specific bar code sequences, was pooled to give a total amount of 3.6 μg.
The pooled DNA samples were adapter ligated with beads and amplified by emulsion PCR using a GS FLX Titanium SV emPCR kit (Roche, CT). Beads were counted after emulsion PCR, and the same amount of beads was put in a PicoTiterPlate. Pyrosequencing was performed using the genome sequencer FLX system (Roche, CT). Sequences obtained from pyrosequencing were analyzed with 454 BaseCaller 2.3 in genome sequencer FLX system software (Roche, CT). FLX pyrosequencing and analysis were carried out at TaKaRa Bio.
Taxonomy-based analysis at the phylum, family, genus, and species levels were performed by assigning taxonomic status to each sequence using BLAST analysis with the DNA Data Bank of Japan (DDBJ).
Statistical analysis.
Data for body, cecal, and fecal weights and cecal chemical compounds were expressed as means ± standard deviations (SD) or standard errors of the means (SEM). Statistical analysis for the animal experiment was performed using EXCEL Statistic 5.0 (Esumi Co., Ltd., Tokyo, Japan). One-way analysis of variance (ANOVA) was used to assess the effects of treatments. Significant differences were accepted at P values of <0.05. When significant variation was found, Dunnett's test was used to determine differences between the control and test diet groups.
RESULTS
Body, cecal, and fecal weights.
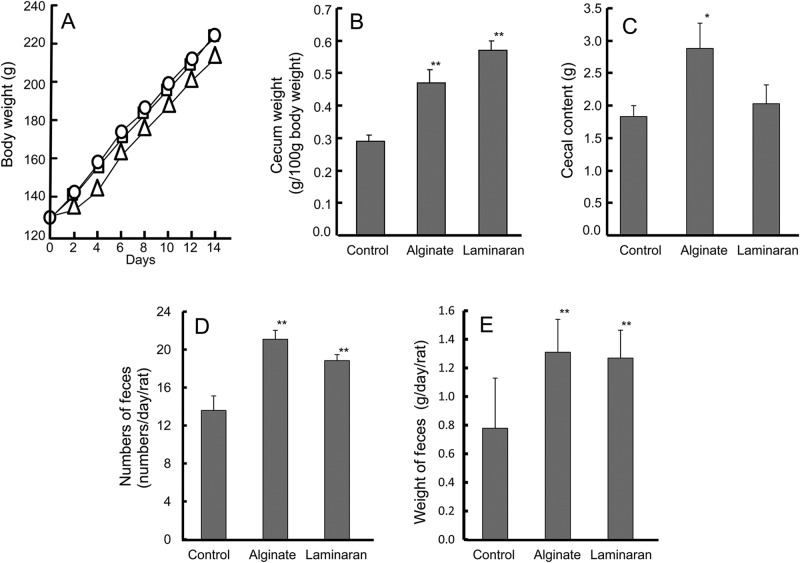
Body, cecal, and fecal weights of rats fed AIN-76-based diets containing 2% (wt/wt) of either alginate or laminaran are shown in Fig. 1. There were no significant differences among the three groups in final body weight gain, ranging from 214 g/rat to 252 g/rat (Fig. 1A), although the increases in rats fed alginate tended to be smaller than in rats fed the other diets. Cecal weight was increased by both alginate and laminaran (Fig. 1B); in particular, the laminaran group cecal weight was 2-fold greater than the control group. The amounts of cecal contents in the control, alginate, and laminaran groups were 1.8 g, 2.8 g, and 2.0 g, respectively (Fig. 1C). The average fecal frequencies were about 7.0, 12.1, and 8.5 times/day/rat, while the average fecal amounts were about 0.77, 1.31, and 1.27 g/day/rat in rats fed control, alginate, and laminaran diets, respectively (Fig. 1D and E).
Fig 1.
Body, cecal, and fecal weights of rats fed diets containing no dietary fiber (control), 2% (wt/wt) Na-alginate, or 2% laminaran. (A) Changes in rat body weight. (B) Endpoint cecal weights. (C) Endpoint cecal content. (D) Numbers of feces per day. (E) Amount of feces per day. Symbols: control (circles), alginate (triangles), or laminaran (squares). Values are expressed as means and SEM. *, P < 0.05; **, P < 0.01 (n = 6).
Cecal carbohydrate, organic acid, and putrefactive compound contents.
As shown in Fig. 2, the levels of soluble polysaccharides were 1.6, 4.4, and 2.9 mg/g and those of low-molecular-weight saccharides were 0.7, 2.8, and 1.4 mg/g cecal content in rats fed control, alginate, and laminaran diets, respectively. Alginate administration increased soluble polysaccharide and low-molecular-weight saccharide levels in the cecum; >2-fold increases were observed. In the laminaran group, these levels showed a tendency to be elevated.
Fig 2.
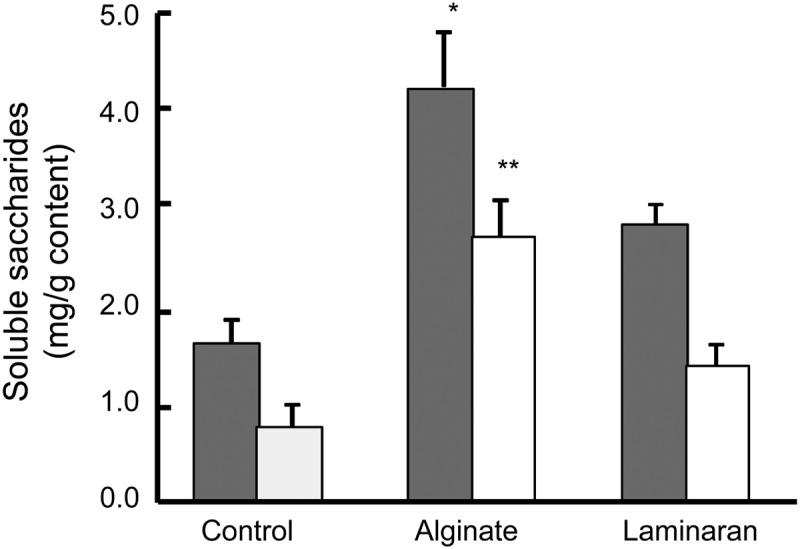
Cecal carbohydrates in rats fed diets containing no dietary fiber (control), 2% (wt/wt) Na-alginate, or 2% laminaran. Cecal polysaccharides (gray bars) and low-molecular-weight saccharides (white bars). Error bars indicate SEM. *, P < 0.05; **, P < 0.01 (n = 6).
Cecal organic acids are summarized in Fig. 3. Laminaran supplementation increased total organic acids; notably, propionic acid was significantly increased from 6.9 mmol/kg (control) to 12.5 mmol/kg. Furthermore, other organic acids and total organic acids in the cecal contents showed a tendency to increase with laminaran administration. On the other hand, total cecal organic acids were the highest in rats fed alginate (Fig. 3B).
Fig 3.
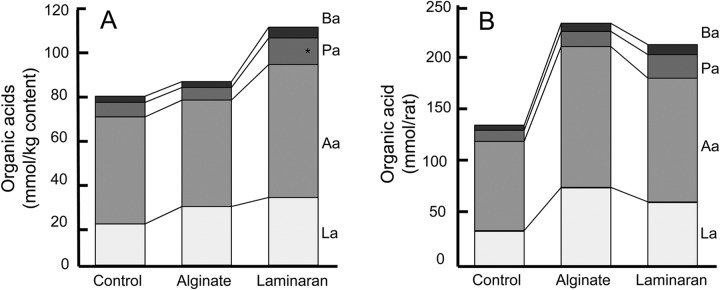
Cecal organic acid content of rats fed diets containing no dietary fiber (control), 2% (wt/wt) Na-alginate, or 2% laminaran. (A and B) Cecal organic acid composition per gram content (A) and whole content (B). Ba, butyric acid; Pa, propionic acid; Aa, acetic acid; La, lactic acid. *, P < 0.05 (n = 6).
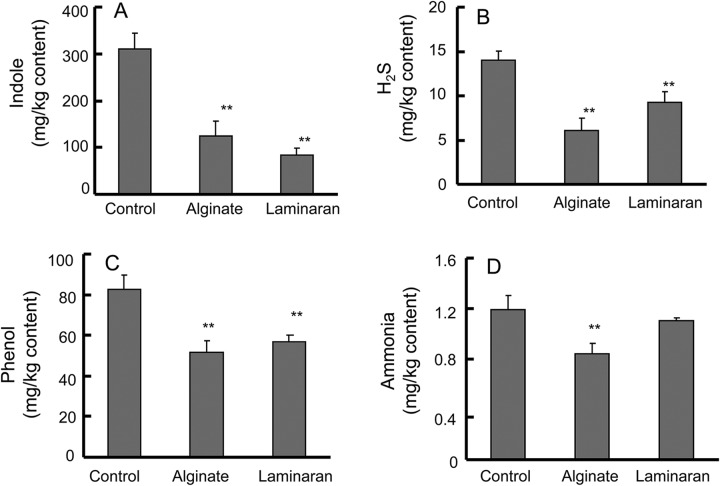
Production of intestinal putrefactive compounds was notably suppressed by the alginate and laminaran diets (Fig. 4A to D). Indole, H2S, and phenol were significantly decreased by alginate, as well as laminaran, compared with the control; indole was suppressed from 311.9 to 123.6 and 83.5 mg/kg cecal content by alginate and laminaran, respectively. H2S levels were 14.0, 6.1, and 9.3 mg/kg cecal content in rats fed control, alginate, and laminaran, respectively. Phenol levels decreased from 82.8 to 51.7 and 56.7 mg/kg cecal content in rats fed alginate and laminaran, respectively. However, ammonia levels were decreased by alginate administration alone.
Fig 4.
Cecal putrefactive compound levels in rats fed diets containing no dietary fiber (control), 2% (wt/wt) Na-alginate, or 2% laminaran. (A to D) Putrefactive compounds indole (A), H2S (B), phenol (C), and ammonia (D). Error bars indicate SEM. **, P < 0.01 (n = 6).
Microbiota in cecal contents.
From direct counts using Gram staining, the total bacterial counts were 10.38, 10.52, and 10.61 log cells/gram cecal content in rats fed control, alginate, and laminaran, respectively. The intestinal microbiota was examined using pyrosequencing with bar-coded primers targeting the bacterial 16S rRNA gene. Total reads of each sample were 11,400 ± 349, 13,217 ± 516, and 12,053 ± 1,362 in rats fed control, alginate, and laminaran diets, respectively (Table 2). Additionally, the average sequence length was around 346 bp and matched the targeted region.
Table 2.
Characteristics of pyrosequencing qualitya
| Characteristic | Pyrosequencing quality of test diets |
||
|---|---|---|---|
| Control | Alginate | Laminaran | |
| No. of sequences | 11,400 ± 349 | 13,217 ± 516* | 12,053 ± 1,362 |
| Sequence length (bp) | 350 ± 2 | 349 ± 0 | 344 ± 3 |
| No. of phylotypes | 152 ± 9 | 112 ± 18** | 162 ± 6 |
Values are means and SD.
, P < 0.05;
, P < 0.01 (n = 6).
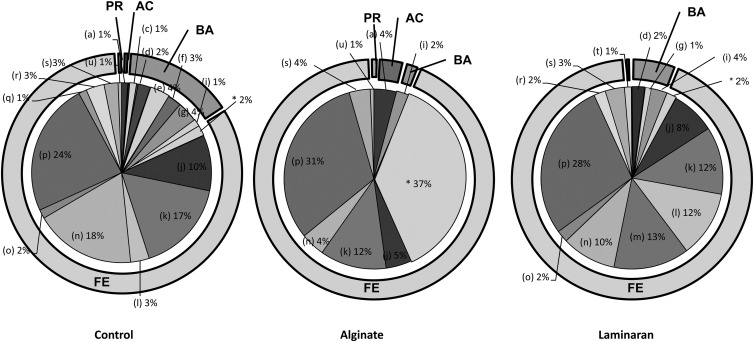
The microbiota at the phylum level is shown in Fig. 5. The most represented phylum in the three diet groups was Firmicutes, accounting for about 81.1 to 93.4% of genes. Compared with the control diet (81.1%), the ratio of Firmicutes genes to total 16S rRNA genes increased in the alginate (93.4%) and laminaran (90.6%) groups. In addition, the ratio of Actinobacteria genes to total 16S rRNA genes was increased by alginate administration, from 1.2% (control) to 4.2%, while the ratio of Proteobacteria was increased in the laminaran group (1.3%).
Fig 5.
Characterization of the intestinal microbiota in rats fed diets containing no dietary fiber (control), 2% (wt/wt) Na-alginate, or 2% laminaran using pyrosequencing of bacterial 16S rRNA genes. The composition of intestinal microbiota was detected at the family level. The phylum percentages are shown in each outer circle. FE, Firmicutes; BA, Bacteroidetes; AC, Actinobacteria; and PR, Proteobacteria. Additionally, each circle is separated by family level: Coriobacteriaceae (a), other Actinobacteria (b), Bacteroidaceae (c), Bacteroidales bacterium (d), Cytophagaceae (e), Marinilabiaceae (f), Prevotellaceae (g), Porphyromonadaceae (h), other Bacteroidetes (i), Clostridiaceae (j), Clostridiales bacterium (k), Eubacteriaceae (l), Lachnospiraceae (m), Lactobacillaceae (n), Lactobacillales bacterium (o), Erysipelotrichaceae (p), Oscillospiraceae (q), Ruminococcaceae (r), other Firmicutes (s), Alcaligenaceae (t), other Proteobacteria (u), and B. capillosus (*).
At the family level, Clostridiaceae, Erysipelotrichaceae, and Lactobacillaceae were detected in all diet groups at similar levels across groups. Specifically, Erysipelotrichaceae dominated in the control and laminaran groups (control, 23.9%; alginate, 31.4%; and laminaran, 28.2%).
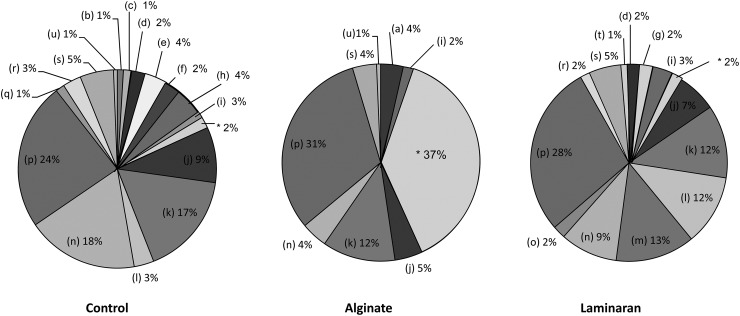
Figure 6 shows the cecal microbiota at the genus level. Allobaculum dominated in all groups (control, 24.0%; alginate, 31.4%; and laminaran, 28.2%); however, Bacteroides was observed in the alginate group (37.6%), with Bacteroides capillosus as the majority. In the control group, Lactobacillus (18.2%), Clostridiales bacterium (16.8%), and Clostridium (9.1%) followed Allobaculum, whereas Pontibacter, Anaerophaga, Prevotella, and Oscillibacter were only detected in the control group. Enterorhabdus was only confirmed in the alginate group, whereas Eubacterium and Ruminococcus were not detected. In addition, only 6 genus types constituting >1% were observed in the alginate group, with low phylotype diversity compared with others. The actual numbers of phylotypes were 152, 112, and 162 in rats fed control, alginate, and laminaran diets, respectively (Table 3). In the laminaran group, a number of genera were detected that were not present in the other groups. For example, Parabacteroides, Lachnospiracea bacterium, and Parasutterella accounted for 1.4, 13.0, and 1.2% in the laminaran group.
Fig 6.
Characterization of the intestinal microbiota of rats fed diets containing no dietary fiber (control), 2% (wt/wt) Na-alginate, or 2% laminaran using pyrosequencing of bacterial 16S rRNA genes. The composition of intestinal microbiota was detected at the genus level: Enterorhabdus (a), other Actinobacteria (b), Bacteroides (c), Bacteroidales bacterium (d), Pontibacter (e), Anaerophaga (f), Parabacteroides (g), Prevotella (h), other Bacteroidetes (i), Clostridium (j), Clostridiales bacterium (k), Eubacterium (l), Lachnospiraceae bacterium (m), Lactobacillus (n), Lactobacillales bacterium (o), Allobaculum (p), Oscillibacter (q), Ruminococcus (r), other Firmicutes (s), Parasutterella (t), other Proteobacteria (u), and B. capillosus (*).
Table 3.
Predominant bacteria in the cecal content of rats fed test diets, analyzed by FLX pyrosequencing and BLAST
| Expt | Predominant bacterium in cecal content of rats fed test diets |
|||||
|---|---|---|---|---|---|---|
| Control | Ratio of identified clones to total detected clones (%) | Alginate | Ratio of identified clones to total detected clones (%) | Laminaran | Ratio of identified clones to total detected clones (%) | |
| 1 | Allobaculum stercoricanis | 22.98 | Bacteroides capillosus | 37.10 | Allobaculum stercoricanis | 28.42 |
| 2 | Clostridiales bacterium | 16.87 | Allobaculum stercoricanis | 31.42 | Lachnospiraceae bacterium | 13.20 |
| 3 | Lactobacillus intestinalis | 11.77 | Clostridiales bacterium | 11.96 | Clostridiales bacterium | 12.16 |
| 4 | Clostridium sp. | 7.12 | Enterorhabdus caecimuris | 3.85 | Eubacterium rectale | 7.23 |
| 5 | Lactobacillus reuteri | 5.42 | Lactobacillus intestinalis | 3.50 | Lactobacillus reuteri | 3.62 |
| 6 | Pontibacter sp. | 3.84 | Clostridium sp. | 1.90 | Lactobacillus intestinalis | 3.28 |
| 7 | Prevotella sp. | 3.52 | Clostridium islandicum | 0.69 | Clostridium ramosum | 3.14 |
| 8 | Anaerophaga sp. | 2.44 | Clostridium orbiscindens | 0.68 | Lactobacillus johnsonii | 2.38 |
| 9 | Eubacterium coprostanoligenes | 2.43 | Turicibacter sanguinis | 0.66 | Eubacterium cylindroides | 2.34 |
| 10 | Ruminococcus sp. | 2.31 | Parasutterella excrementihominis | 0.65 | Lactobacillales bacterium | 2.14 |
| 11 | Bacteroidetes bacterium | 2.27 | Lactobacillus johnsonii | 0.60 | Bacteroidetes bacterium | 1.65 |
| 12 | Bacteroides capillosus | 2.24 | Ruminococcus sp. | 0.49 | Bacteroides capillosus | 1.64 |
| 13 | Lactobacillales bacterium | 1.60 | Clostridium bolteae | 0.44 | Clostridium orbiscindens | 1.44 |
| 14 | Allobaculum sp. | 1.10 | Prevotella sp. | 0.40 | Parabacteroides distasonis | 1.36 |
| 15 | Oscillibacter sp. | 1.10 | Bacteroidetes bacterium | 0.37 | Clostridium sp. | 1.28 |
In this study, pyrosequencing data were also analyzed at the species level (Table 3). Among them, 3.14% of all sequence copies from the laminaran group were C. ramosum. In addition, Parabacteroides distasonis was detected at 1.36% in the laminaran group. These were not detected in the control and alginate groups. Furthermore, B. capillosus comprised 37.1% of all sequence copies detected in the alginate group; levels detected in the control and laminaran groups were <2.5%.
DISCUSSION
Although seaweeds have become increasingly popular as a health food (marine vegetables) outside Japan and other Far East Asian countries (28), the consumption of edible algae in Japanese adults remains high (approximately 14.3 g/day) (29). Recently, a study characterized the first porphyranases from a member of the marine Bacteroidetes, Zobellia galactanivoran (13). These porphyranases were active on the sulfated polysaccharide porphyran from the marine red algae of the genus Porphyra, and genes encoding these porphyranases, agarases, and associated proteins have been transferred to the intestinal bacterium Bacteroidetes plebeius, isolated from Japanese individuals (13). However, the consumption of brown algae such as kombu and wakame exceeds that of red algae in Japan (14).
Low-molecular-weight polysaccharides and oligosaccharides derived from brown algae have been reported to exhibit prebiotic activity (30, 31). In this study, increases in soluble polysaccharides and mono- and oligosaccharides were observed in the cecal contents of rats fed alginate and laminaran. This result suggests that dietary fiber from the small intestine was the source of sugars in the cecum and/or was liberated from polysaccharides by cecal microbiota.
Levels of cecal organic acids were increased, while the levels of putrefactive compounds were decreased, by alginate and laminaran administration (Fig. 3), results that confirm our previous study (8). Decreases in intestinal putrefactive compounds are thought to reduce the risk of cancer (9). In this study, butyrate tended to be high in the cecal contents of rats fed laminaran (Fig. 3B). Interestingly, butyrate is the preferred energy source for colonocytes and has a protective effect against colon disease (32). Propionic acid is an important anti-inflammatory agent, influencing adipokine secretion by stimulating leptin and reducing resistin expression, as well as having an anabolic effect by stimulating two important metabolic pathways that are also stimulated by insulin (33). Some have reported that the production of putrefactive compounds is regulated by poly- and oligosaccharides. For example, lactulose has been used as medicine for hyperammonemia (34). Laminaran is a highly fermentable polysaccharide of low viscosity, thereby capable of regulating levels of indole, H2S, and phenol; paradoxically, the effects of the highly viscous alginate were largely the same. Potential toxic substances, such as ammonia, indole, and phenols, which arise from intestinal putrefaction, are also thought to be important in the development of colorectal cancer, ageing, etc. (35). Furthermore, putrefactive fermentation products were used for the growth of some saccharolytic species, such as Clostridium sp. and Bacteroides sp. For example, when human feces are incubated with lactulose or other fermentable substrate, such as glucose or mannitol, ammonia is utilized by the microbiota (36).
We employed FLX pyrosequencing with bar-coded primers targeting the bacterial 16S rRNA gene, a method capable of detecting bacterial changes undetectable by culture-dependent methods. Actually, the intestine hosts 1013 to 1014 bacteria belonging to at least 500 different species or strains in adult human intestine contents (1), with the majority unculturable. Until recently, the study of microbiota has employed molecular methods such as DGGE and T-RFLP. It has been reported that while the procedure was sensitively performed, DGGE analysis could not differentiate between all species (37). Furthermore, results obtained by T-RFLP underestimated the number of strains or species in the bacterial community (38). However, we detected 110 to 160 phylotypes in each diet group using our primer sets and pyrosequencing.
At the family level, Lachnospiraceae were abundant in the laminaran group. Lachnospiraceae are known as pectin and cellulose degraders that are important in colonic fermentation of dietary fibers (39). While we used different dietary fibers, alginate and laminaran, the Lachnospiraceae observed in the laminaran group might play the same role in the intestine following consumption of pectin and cellulose. Furthermore, Bacteroidaceae were dominant in the alginate group. Among these, the greatest proportion was composed of B. capillosus. B. capillosus is usually found in the intestinal tract of humans (40) and has the ability to ferment 1,3-1,4-β-glucanase (27) and pectin (41). However, in recent reports, B. capillosus was reclassified using genotyping to Firmicutes and was found to be closely related to Clostridium or Ruminococcus (40, 42, 43).
At the genus level, Allobaculum was dominant in all diet groups and was increased by supplementation with algal dietary fibers. Allobaculum, a Gram-positive non-spore-forming anaerobic rod, was isolated from dog feces (44), the intestines of mice fed diets containing 50% ground beef (45), and the feces of commercial pigs (46); however, it was not dominant in these samples. On the other hand, Allobaculum was detected as dominant in the intestine of hamsters fed an AIN-93 M diet and was increased by grain sorghum lipid extract supplementation, which was correlated with cholesterol metabolic improvement (24). The blood cholesterol-lowering effect of alginate has been reported, which might be associated with its fermentation properties (47).
C. ramosum was identified as predominant in the laminaran group, although this species was not detected in the control and alginate groups. C. ramosum is a normal bacterium of the human intestine and is known to ferment laminaran (25). The degradation products produced from laminaran by C. ramosum, such as glucose, laminaribiose, and laminaritriose, might promote other bacteria, including Bifidobacterium, in the intestine (19). In addition, it was reported that Bacteroides thetaiotaomicron and Parabacteroides distasonis were laminaran-fermenting bacteria (21). P. distasonis was also increased in the laminaran group, indicating that it might play a similar role to C. ramosum in the intestine. While B. ovatus is known as an alginate-fermenting bacteria, we were unable to detect it. However, another Bacteroides bacterium was abundantly found in the alginate group and might be involved in alginate fermentation.
In conclusion, changes in intestinal microbiota by alginate and laminaran administration were clarified using FLX pyrosequencing. We confirmed that dietary fiber was fermented by intestinal bacteria and polysaccharide-fermenting bacteria increased with the ingestion of dietary fiber. This is the first time that we have detected increases in levels of C. ramosum, which was reported as a habitat for laminaran-fermenting bacteria in normal human intestine, in a rat model following laminaran administration. Rat models have been widely employed for extrapolating results to humans; we expect that our results can be similarly applied. The results also indicate that edible brown algae can alter and possibly improve the intestinal environment. However, currently, numerous alginate- and/or laminaran-related bacterial genes have not been classified to the species or genus level. Further studies regarding isolation and identification of novel intestinal fermentative bacteria, as well as metagenomic analysis of intestinal content, are now in progress.
ACKNOWLEDGMENT
The present study was supported in part by the Ministry of Education, Science, Sports and Culture, Japan, Grant-in-Aid for Scientific Research (C), 22580221, 2010–2012.
Footnotes
Published ahead of print 26 November 2012
REFERENCES
- 1. Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280 doi:10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sears CL. 2005. A dynamic partnership: celebrating our intestinal flora. Anaerobe 11:247–251 [DOI] [PubMed] [Google Scholar]
- 4. Nakamura S, Kuta T, An C, Kanno T, Takahashi H, Kimura B. 2012. Inhibitory effects of Leuconostoc mesenteroides 1RM3 isolated from narezushi, a fermented fish with rice, on Listeria monocytogenes infection to Caco-2 cells and A/J. mice. Anaerobe 18:19–24 [DOI] [PubMed] [Google Scholar]
- 5. Willing BP, Russell SL, Finlay BB. 2011. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9:233–243 [DOI] [PubMed] [Google Scholar]
- 6. Willing BP, Kassei V. 2010. Host pathways for recognition: establishing gastrointestinal microbiota as relevant in animal health and nutrition. Livestock Sci. 133:82–91 [Google Scholar]
- 7. Rossi M, Amaretti A, Raimondi S. 2011. Folate production by probiotic bacteria. Nutrients 3:118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuda T, Yano T, Matsuda N, Nishizawa M. 2005. Inhibitory effects of laminaran and low molecular alginate against the putrefactive compounds produced by intestinal microbiota in vitro and in rats. Food Chem. 91:745–749 [Google Scholar]
- 9. Windey K, De Preter V, Verbeke K. 2012. Relevance of protein fermentation to intestinal health. Mol. Nutr. Food Res. 56:184–196 [DOI] [PubMed] [Google Scholar]
- 10. Sekirov I, Russell SL, Antunes CM, Finlay BB. 2010. Intestinal microbiota in health and disease. Physiol. Rev. 90:859–904 [DOI] [PubMed] [Google Scholar]
- 11. De Filippo C, Cavalieri D, Paola MD, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping intestinal microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A. 107:14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuda T, Goto H, Yokoyama M, Fujii T. 1998. Fermentable dietary fiber in dried products of brown algae and their effects on cecal microbiota and levels of plasma lipid in rats. Fisheries Sci. 64:582–588 [Google Scholar]
- 13. Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michelet G. 2010. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese intestinal microbiota. Nature 464:908–912 [DOI] [PubMed] [Google Scholar]
- 14. Ohno M. 2004. Biology and technology of economic seaweeds, p 333–474 Uchidarokakuho, Tokyo, Japan: (In Japanese.) [Google Scholar]
- 15. Kuda T, Taniguchi E, Nishizawa M, Yokoyama M. 2002. Fate of water-soluble polysaccharides in dried Chorda filum a brown alga during water washing. J. Food Comp. Anal. 15:3–9 [Google Scholar]
- 16. Kuda T, Enomoto T, Yano T. 2009. Effects of two storage β-1,3-glucans, laminaran from Eicenia bicyclis and paramylon from Euglena gracili, on cecal environment and plasma lipid levels in rats. J. Funct. Food. 1:399–404 [Google Scholar]
- 17. Kuda T, Goto H, Yokoyama M, Fujii T. 1998. Effects of dietary concentration of laminaran and depolymerised alginate on rat cecal microbiota and plasma lipids. Fisheries Sci. 64:589–593 [Google Scholar]
- 18. Fujii T, Kuda T, Saheki K, Masayo O. 1992. Fermentation of water-soluble polysaccharides of brown algae by human intestinal bacteria in vitro. Nippon Suisan Gakkaishi 58:147–152 [Google Scholar]
- 19. Kuda T, Fujii T, Hasegawa A, Okuzumi M. 1992. Effect of degraded products of laminaran by Clostridium ramosum on the growth of intestinal bacteria. Nippon Suisan Gakkaishi 58:1207–1311 [Google Scholar]
- 20. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satokari RM, Vaughan EE, Akkermans AD, Saarela M, Willem MV. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piacentini G, Peroni D, Bessi E, Morelli L. 2010. Molecular characterization of intestinal microbiota in infants fed with soymilk. J. Pediatr. Gastroenterol. Nutr. 51:71–76 [DOI] [PubMed] [Google Scholar]
- 23. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core intestinal microbiome in obese and lean twins. Nature 457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miriam B, Buenviaje MD. 1989. Quantitative sputum culture and gram stain: pulmonary infection vs. colonization. Philipp. J. Microbiol. Infect. Dis. 18:28–35 [Google Scholar]
- 25. Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350–356 [Google Scholar]
- 26. Lombard GL, Dowell VR. 1983. Comparison of three reagents for detecting indole production by anaerobic bacteria in microtest systems. J. Clin. Microbiol. 18:609–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. An C, Takahashi H, Kimura B, Kuda T. 2010. Comparison of PCR-DGGE and PCR-SSCP analysis for bacterial flora of Japanese traditional fermented fish products, aji-narezushi and iwashi-nukazuke. J. Sci. Food Agric. 90:1796–1801 [DOI] [PubMed] [Google Scholar]
- 28. O'Sullivan L, Murphy B, McLoughlin P, Duggan P, Lawlor PG, Hughes H, Gardiner GE. 2010. Prebiotics from marine macroalgae for human and animal health applications. Mar. Drugs 8:2038–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iso H, Date C, Noda H, Yoshimura T, Tamakoshi A. 2005. Frequency of food intake and estimated nutrient intake among men and women: the JACC Study. J. Epidemiol. 15:S24–S42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Deville C, Gharbi M, Dandrifosse G, Peulen O. 2007. Study on the effects of laminarin, a polysaccharide from seaweed, on gut characteristics. J. Sci. Food Agric. 87:1717–1725 [Google Scholar]
- 31. Warrand J. 2006. Healthy polysaccharides. Food Technol. Biotechnol. 44:355–370 [Google Scholar]
- 32. Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133–139 [DOI] [PubMed] [Google Scholar]
- 33. Al-Lahham S, Roelofsen H, Rezaee F, Weening D, Hoek A, Vonk R, Venema K. 2011. Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur. J. Clin. Invest. doi:10.1111/j.1365-2362.2011.02590.x [DOI] [PubMed] [Google Scholar]
- 34. Bircher J, Guggenheim P, Haemmerli UP. 1966. Treatment of chronic portal-systemic encephalopathy with lactulose. Lancet 287:890–893 [DOI] [PubMed] [Google Scholar]
- 35. Silverman SJ, Andrews AW. 1977. Bile acids: co-mutagenic activity in the Salmonella-mammalian-microsome mutagenicity test. J. Natl. Cancer Inst. 59:1557–1559 [DOI] [PubMed] [Google Scholar]
- 36. Wrong O. 1978. Nitrogen metabolism in the gut. Am. J. Clin. Nutr. 31:1587–1593 [DOI] [PubMed] [Google Scholar]
- 37. Nikolcheva LG, Cockshutt AM, Bärlocher F. 2003. Determining diversity of freshwater fungi on decaying leaves: comparison of traditional and molecular approaches. Appl. Environ. Microbiol. 69:2548–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu WT, Marsh TL, Cheng H, Forney LJ. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rode LM, Genthner BR, Bryant MP. 1981. Syntrophic association by cocultures of the methanol- and CO(2)-H(2)-utilizing species Eubacterium limosum and pectin-fermenting Lachnospira multiparus during growth in a pectin medium. Appl. Environ. Microbiol. 42:20–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carlier JP, Bedora-Faure M, K'ouas G, Alauzet C, Mory F. 2010. Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Séguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 60:585–590 [DOI] [PubMed] [Google Scholar]
- 41. Sirotek K, Marounek M, Rada V, Benda V. 2001. Isolation and characterization of rabbit caecal pectinolytic bacteria. Folia Microbiol. (Praha) 46:79–82 [DOI] [PubMed] [Google Scholar]
- 42. Gloux K, Berteau O, El oumami H, Béguet F, Leclerc M, Doré J. 2011. A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. U. S. A. 108:4539–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karlsson FH, Ussery DW, Nielsen J, Nookaew I. 2011. A closer look at Bacteroides: phylogenetic relationship and genomic implications of a life in the human gut. Microb. Ecol. 61:473–485 [DOI] [PubMed] [Google Scholar]
- 44. Greetham HL, Gibson GR, Giffard C, Hippe H, Merkhofferc B, Steinerc U, Falsend E, Collinsa MD. 2004. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe 10:301–307 [DOI] [PubMed] [Google Scholar]
- 45. Li W, Dowd SE, Scurlock B, Acosta-Martinez V. 2009. Memory and learning behavior in mice is temporally associated with diet-induced alterations in intestinal bacteria. Physiol. Behav. 96:557–567 [DOI] [PubMed] [Google Scholar]
- 46. Kim HB, Borewicz K, White BA, Singer RS. 2011. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 153:124–133 [DOI] [PubMed] [Google Scholar]
- 47. Kuda T, Yokoyama M, Fujii T. 1997. Effect of high and low viscous sodium alginates on levels of serum lipids and cecal microbiota in rats. Shokuhin Kagaku Kogaku Kaishi 44:226–229 [Google Scholar]