Abstract
Future rates of anthropogenic N deposition can slow the cycling and enhance the storage of C in forest ecosystems. In a northern hardwood forest ecosystem, experimental N deposition has decreased the extent of forest floor decay, leading to increased soil C storage. To better understand the microbial mechanisms mediating this response, we examined the functional genes derived from communities of actinobacteria and fungi present in the forest floor using GeoChip 4.0, a high-throughput functional-gene microarray. The compositions of functional genes derived from actinobacterial and fungal communities was significantly altered by experimental nitrogen deposition, with more heterogeneity detected in both groups. Experimental N deposition significantly decreased the richness and diversity of genes involved in the depolymerization of starch (∼12%), hemicellulose (∼16%), cellulose (∼16%), chitin (∼15%), and lignin (∼16%). The decrease in richness occurred across all taxonomic groupings detected by the microarray. The compositions of genes encoding oxidoreductases, which plausibly mediate lignin decay, were responsible for much of the observed dissimilarity between actinobacterial communities under ambient and experimental N deposition. This shift in composition and decrease in richness and diversity of genes encoding enzymes that mediate the decay process has occurred in parallel with a reduction in the extent of decay and accumulation of soil organic matter. Our observations indicate that compositional changes in actinobacterial and fungal communities elicited by experimental N deposition have functional implications for the cycling and storage of carbon in forest ecosystems.
INTRODUCTION
The extent to which terrestrial ecosystems will function as a future sink for anthropogenic CO2 in the atmosphere will be modified by current and future rates of anthropogenic nitrogen (N) deposition from the atmosphere (1). In addition to increasing ecosystem carbon (C) storage by fostering higher rates of net primary production (NPP), anthropogenic N deposition may reduce the decomposition of plant detritus, thereby increasing the storage of C in long-lived soil organic matter. This response has been documented in a wide range of ecosystems (2), but such a response is not universal. We have studied the impact of elevated atmospheric N deposition on forest C cycling in a long-term replicated field experiment in Michigan (Fig. 1; see Table S1 in the supplemental material). We have provided evidence that experimental N deposition, at a rate expected by midcentury, has decreased litter decay and increased soil C storage (3). Although experimental N deposition has increased NPP (+10%), it has not altered the amount of leaf or fine root litter entering the forest floor (Table 1) (4). Meanwhile, the leaf litter N concentration has increased by 25%, and inorganic N concentrations in the forest floor have increased by 288% (3, 5). With the use of field observations and a biogeochemical simulation model, we further determined that chronic N deposition has decreased the extent of microbial decay, thereby leading to an accumulation of soil organic matter (6). In combination, these observations indicate that a microbial mechanism underlies a decline in litter decay under experimental N deposition, which has enhanced soil C storage (+10%) (3, 7).
Fig 1.
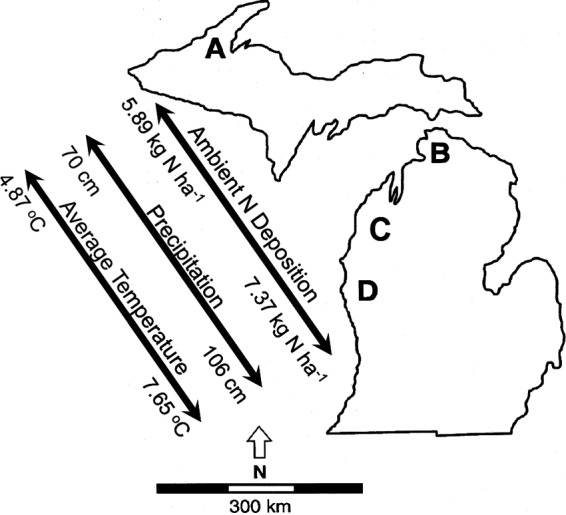
Approximate locations and climatic information for four replicate sugar maple-dominated northern hardwood forest stands, A to D, across the upper and lower peninsulas of Michigan.
Table 1.
Microbial responses summarized for forest floor (Oe/Oa)a
| Response to chronic N deposition | % change | Reference(s) |
|---|---|---|
| Plant | ||
| NPP | +10 | 7 |
| Leaf litter production | 0 | 7 |
| Leaf litter N concn | +25 | 3 |
| Fine root litter | 0 | 4 |
| Fine root N concn | 0 | 4 |
| Biogeochemical | ||
| Forest floor mass | +51 | 3 |
| Forest floor turnover | −60 | 3 |
| Soil organic matter content | +18 | 3 |
| Soil solution NO3− concn | +288 | 3 |
| NO3− leaching | +680 | 5 |
| Dissolved organic matter leaching | +26 | 5 |
| Microbial | ||
| Soil respiration | −15 | 4 |
| Active microbial biomass (PLFAb) | +23 | 36, 59 |
| Phenol oxidase activity | −33 | 36, 59 |
| Peroxidase activity | −30 | 36, 59 |
| Basidiomycete laccase copy no. | −5 to −8 | 16 |
Eighteen years of experimental N deposition have increased the storage and slowed the cycling of C in forest floor and surface mineral soil. With the exception of the laccase copy number, all of these responses are statistically significant.
PLFA, phospholipid fatty acid analysis.
Saprotrophic basidiomycete and ascomycete fungi are the primary agents of litter decay, especially during the later stages dominated by degrading plant cell wall polymers (8). These fungi facilitate the rate-limiting step of litter decay in which cell wall lignin is depolymerized. Laboratory studies have found that increased concentrations of inorganic N can suppress the transcription of fungal genes required for the metabolism of lignin and lignocellulose (9). In our long-term field study, experimental N deposition has also decreased the transcription of fungal genes encoding ligninolytic enzymes and induced a shift in the composition of basidiomycete fungi (10). Some actinobacteria may also be able to solubilize and modify lignin and lignocellulose, but not as completely as basidiomycete fungi, which mineralize lignin to CO2 (11–14). Rather, the end products of actinobacterial lignin metabolism are dissolved organic compounds and soil organic matter. In our study, experimental N deposition has also altered the composition of actinobacteria, which occurred in concert with reduced decay and increased production of phenolic dissolved organic matter (15).
Here, we focused on functional genes from the Dikarya fungi (basidiomycetes and ascomycetes), as well as actinobacteria involved in plant and fungal cell wall decay, to help elucidate the potential microbial mechanisms mediating reduced decay under experimental N deposition. We quantified the abundance, diversity, and composition of genes involved in degrading four components of plant detritus: starch, hemicellulose, cellulose, and lignin. We also quantified genes encoding enzymes that metabolize chitin, a major component of the fungal cell wall. Previously, we explored the extent to which experimental N deposition has altered the cellulolytic and lignolytic capabilities of basidiomycete and ascomycete fungi (3, 10, 16); here, we examine a broader suite of Dikarya fungal and actinobacterial genes mediating the decay of detritus to better understand how this agent of global change has reduced the extent of plant litter decay under experimental N deposition. We hypothesized that experimental N deposition has altered the functional capability, and therefore diminished the gene content, of forest floor microbial communities. To test our hypothesis, we used GeoChip 4.0 (17), a closed-format metagenomic approach to compare communities of actinobacteria and fungi growing under ambient and experimental N deposition.
MATERIALS AND METHODS
Study sites.
The impact of chronic atmospheric N deposition on northern hardwood forest ecosystems has been documented in a long-term experiment spanning the upper and lower peninsulas of Michigan (18). Four replicate sugar maple (Acer saccharum) forest stands similar in age, plant community composition, and edaphic properties span a 500-km climatic and ambient N deposition gradient (Fig. 1; see Table S1 in the supplemental material). There are six 30-m by 30-m plots within each site; three plots receive ambient N deposition, and three have received an additional 3 g NO3−-N m−2 year−1 over the growing season. These treatments were initiated in 1994 and have been maintained to the present.
Sample collection and DNA extraction.
Forest floor samples were collected in early October 2009; samples from all four sites were gathered over a 4-day period. A 10-cm by 10-cm frame was randomly placed at 10 locations within each experimental plot. From the center of the frame, the portion of the organic soil horizon that consisted of freshly fallen intact leaf litter (Oi horizon) was removed by hand; the intermediately to moderately decomposed (Oe/Oa) horizon was collected down to the mineral soil. Material from all 10 random locations within each plot was composited and homogenized in the field with sterilized scissors; a 5-g subsample was removed and frozen in liquid N2. Samples were then stored at −80°C prior to DNA extraction. DNA was extracted from approximately 2.5 g of forest floor using the MoBio Power Soil DNA Extraction Kit (MoBio Laboratories, Carlsbad, CA) following the manufacturer's instructions. Total microbial DNA was quantified with Quant-iT PicoGreen (Invitrogen) according to the manufacturer's instructions.
Target preparation and GeoChip hybridization.
GeoChip 4.0 was used to quantify a broad suite of key fungal and actinobacterial functional genes encoding enzymes that disassemble plant and fungal cell walls; hybridization of DNA and preprocessing of data were described by Lu et al. (17). Briefly, 1 μg of genomic DNA (gDNA) from each plot was purified by the Genomic DNA Clean and Concentrator kit (Zymo Research, Irvine, CA) and labeled with the fluorescent dye Cy3 using random primers (19). The labeled gDNA was dried and rehydrated with 2.7 μl of a sample-tracking control, followed by incubation at 50°C for 5 min. This DNA solution was then mixed with 7.3 μl of hybridization buffer containing a universal standard DNA labeled with the fluorescent dye Cy5; it was denatured at 95°C for 5 min and maintained at 42°C until the DNA was loaded onto GeoChip 4.0 microarrays (NimbleGen, Madison, WI). The hybridization was performed on a Hybridization Station (Maui; Roche, CA) at 42°C for 16 h with agitation. After washings, the arrays were scanned using an MS 200 Microarray Scanner (NimbleGen) at laser power of 100% photomultiplier tube (PMT). Each biological replicate was run once on the microarray.
GeoChip data preprocessing.
The signal intensities of GeoChip hybridization spots were normalized by the Cy5-labeled universal standard DNA across samples and by dividing the signal intensity of each spot by the average intensity of all positive spots within each sample. After normalization, unreliable spots were removed if their original signal intensities were below the noise level (<2,000), the signal-to-noise ratio (SNR) was ≤2.0, or the coefficient of variation of the background was >0.8 (17). All signal intensities were log-normal transformed. Details of further normalization steps can be found in the supplemental material of Lu et al. (17).
Statistical analysis.
To examine the effect of experimental N deposition on functional-gene composition, a subset of normalized GeoChip 4.0 signal intensity data was analyzed for diversity, functional richness, and composition. The set of genes considered encode enzymes involved in the depolymerization of the biochemical components of plant and fungal litter: starch, hemicellulose, cellulose, chitin, and lignin. We chose to specifically analyze all variants of these genes belonging to the Actinobacteria and Dikarya fungi (hereafter referred to as fungi). A full list of genes, the substrate category in which they belong as designated by He et al. (20), and the number of their variants (hybridization spots on the microarray) included in these analyses can be found in Table S2 in the supplemental material. Actinobacterial and fungal data were analyzed separately.
The diversity of actinobacterial and fungal functional genes in each sample was calculated with the Shannon diversity index in PRIMER v6 (21) using the normalized signal intensity of all relevant gene variants. The total functional richness of each sample was calculated as the sum of the presence-absence data of all gene variants across all genes considered sensu stricto (22). We calculated the functional richness of the genes in each substrate category (e.g., lignin) by summing the presence of all gene variants for genes that code for enzymes acting upon that substrate. Differences in diversity and functional richness due to experimental N deposition and site were examined with two-way analysis of variance (ANOVA). If an interaction effect was present, it was further elucidated with the protected Fisher's least significant difference (LSD) test. Univariate statistics were conducted in SPSS (IBM Statistics version 20; IBM Corp., Armonk, NY).
To assess the responses of actinobacterial and fungal taxonomic groups (class, order, and family) to experimental N deposition, we reorganized the data to sum the signal intensities for all genes within each taxonomic group under ambient and experimental N deposition. The response ratio of each taxonomic group was then calculated by taking the natural log of the ratio of the mean summed signal intensity of experimental N deposition samples to the mean summed signal intensity of ambient N deposition samples sensu lato (23, 24). We then calculated the variance and 90, 95, and 99% confidence intervals (CIs) for each response ratio to assess the significance of the response ratios (24).
Multivariate statistics were conducted in PRIMER v6 (21). Similarity matrices for the Actinobacteria and Dikarya fungi were calculated with the Bray-Curtis similarity metric on presence-absence transformed data for all relevant gene variants. Principal-coordinate analyses (PCOs) were created to visualize the differences in composition between ambient and elevated N deposition functional-gene assemblages. The correlation of each gene with the primary PCO axis was then calculated with the vector overlay function in PRIMER, averaging the Pearson correlation metrics of all gene variants showing a correlation with the first PCO axis. We used the multivariate community similarity analyses ANOSIM (25), PERMANOVA (26), and PERMDISP (27) to analyze whether experimental N deposition induced compositional dissimilarities in functional-gene variants present in the forest floor. To examine which genes contributed most to the difference in the compositions of functional-gene assemblages, we used the SIMPER tool in PRIMER v6 to calculate the contribution of each gene to the average Bray-Curtis dissimilarity between treatments. All genes were given equal weight for this analysis, to account for the bias inherent in having a wide range of gene variants for each gene on the array.
RESULTS
We examined 20 actinobacterial functional genes and 26 ascomycete and basidiomycete functional genes encoding enzymes involved in degrading starch, hemicellulose, cellulose, chitin, and lignin, all common biochemical constituents of plant and fungal detritus. Experimental N deposition consistently and significantly altered the composition of actinobacterial and fungal genes mediating plant and fungal cell wall depolymerization, as we explain below.
Diversity and richness.
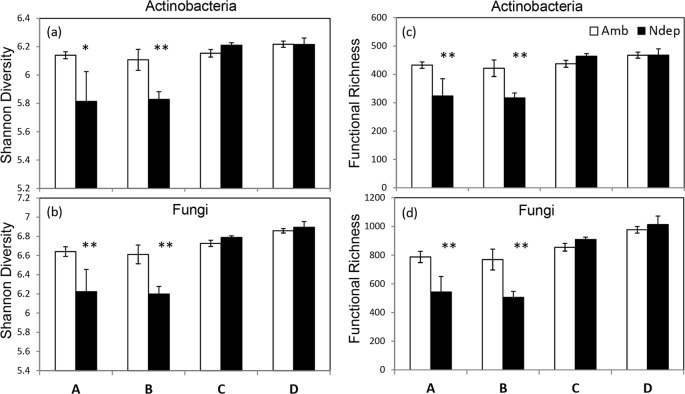
The 20 actinobacterial genes of interest were represented by 635 gene variants (probes on the GeoChip) from 25 actinobacterial families on GeoChip 4.0 (see Table S2 in the supplemental material). The mean Shannon diversity index (H) of these actinobacterial genes declined under experimental N deposition (H = 6.01) relative to ambient N deposition (H = 6.51; P = 0.035). Additionally functional richness also declined under experimental N deposition (richness = 393) relative to ambient N deposition (richness = 440; P = 0.026). There were significant site differences for both actinobacterial diversity (P = 0.013) and functional richness (P = 0.003). The overall decrease in actinobacterial diversity and richness was driven by the 25 to 35% decrease in gene variants detected in the northern two sites, A and B (Fisher's protected LSD; P < 0.035), and no response in the southern sites, C and D (Fig. 2a and c). This led to an interaction between site and treatment in diversity (P = 0.08) and richness (P = 0.043), as shown by two-way ANOVA.
Fig 2.
(a and b) Shannon diversity indexes of actinobacterial (a) and fungal (b) functional genes of soil microbial communities at each site, A to D. (c and d) Functional richness, determined by the average sum of the gene variants detected in actinobacteria (c) and fungi (d). Open bars represent ambient N deposition (Amb); closed bars represent experimental N deposition (Ndep). The diversity of functional genes decreased under experimental deposition in actinobacteria (P = 0.035) and fungi (P = 0.017). Richness declined under experimental N deposition in actinobacteria (P = 0.026) and fungi (P = 0.018). Site effects and site × treatment interactions were present in all analyses. **, significant protected Fisher's LSD, P < 0.015; *, P < 0.035. The error bars indicate standard error.
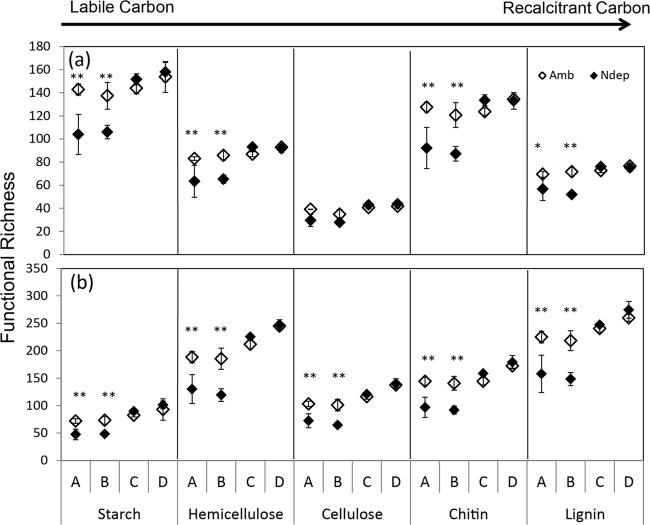
Actinobacterial functional genes encoding enzymes involved in degrading starch, hemicellulose, chitin, and lignin decreased in richness under experimental N deposition at sites A and B, but not C and D (Fig. 3a). Genes encoding enzymes for the breakdown of cellulose did not respond to the treatment. There was a disproportionately greater decrease in the richness of actinobacterial genes involved in chitin degradation compared to the other substrates (P = 0.037).
Fig 3.
Functional richness at each site, A to D, as the sum of all detected genes within each C substrate category for actinobacteria (a) and fungi (b). The error bars represent standard errors. N deposition decreased the total signal intensity for each substrate, except for actinobacterial cellulose-degrading genes, across all sites with a treatment effect (P < 0.04), site effect (P < 0.005), and interaction (P = 0.07 to 0.006). **, protected Fisher's LSD, P < 0.025; *, P < 0.05.
Twenty-six fungal functional genes involved in the depolymerization of the biochemical components of plant and fungal cell walls were also measured on GeoChip. There were 1,294 variants of these genes from 56 fungal families within the Dikarya represented on the microarray. The fungal functional genes responded to experimental N deposition in a manner nearly identical to that of the actinobacterial genes. Shannon's diversity of fungal functional genes declined significantly under experimental N deposition (H = 6 0.71) relative to the ambient treatment (H = 6.52; P = 0.017), and the functional richness declined from 824 to 723 under ambient and experimental N deposition, respectively (P = 0.018). There was a significant site effect for diversity and richness (P < 0.001). The overall decrease in diversity and richness was again driven by an ∼30% decrease in the gene variants detected in the northern two sites (P < 0.001); there was no change in diversity or richness in the southern sites, C and D (Fig. 2). The interaction effect was significant for both diversity (P = 0.032) and functional richness (P = 0.014). Unlike the actinobacterial genes, the decrease in richness of fungal functional genes occurred equally in all five C substrates (−14%). Significantly, the decrease occurred in sites A and B (P < 0.001), but not in C or D (Fig. 3b).
RRs of taxonomic groups.
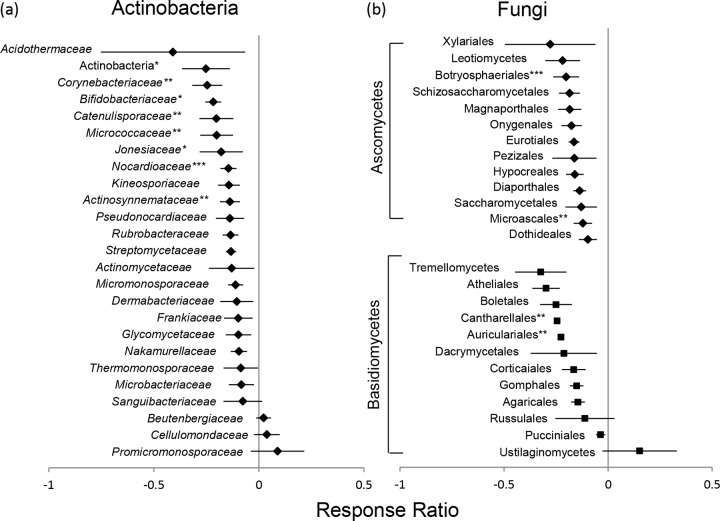
The response ratios (RRs) of actinobacterial families to experimental N deposition were consistently negative (Fig. 4a). A negative response ratio indicates a decrease in gene abundance under experimental N deposition relative to the set of genes exposed to ambient N deposition, whereas a positive response ratio indicates the opposite. The Corynebacteriaceae, Bifidobacteriaceae, Catenulisporaceae, Micrococcaceae, Jonesiaceae, and Actinosynnemataceae all significantly decreased under experimental N deposition (Fig. 4a). The families Beutenbergaceae, Cellulomondaceae, and Promicromonosporaceae have slightly positive response ratios (<0.15); however, with the standard error considered, these values cannot be distinguished from a null response.
Fig 4.
Response ratios of actinobacterial (a) and Dikarya fungal (b) taxa in soil microbial communities under experimental N deposition relative to ambient N deposition. Fungal ascomycete taxa are represented by diamonds; basidiomycetes are represented by squares. The lines represent standard error. ***, 99%; **, 95%; *, 90% of CIs did not cross zero.
The response ratios of fungal ascomycete and basidiomycete groups to experimental N deposition were also mostly negative (Fig. 4b), with the exception of members of the Ustilaginomycetes, which responded positively (RR = 0.15), although not significantly. Members of the Cantharellales, Auricularales, Botryoosphaeriles, and Microscales all had a significant negative response to experimental N deposition, where the 90%, 95%, and 99% CIs did not cross zero.
Multivariate analysis.
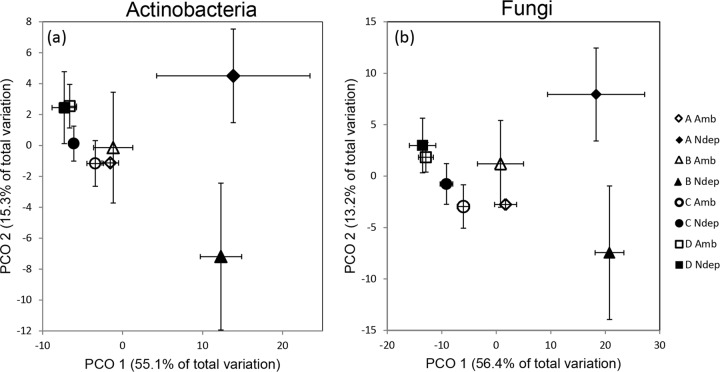
Compositional shifts in actinobacterial functional-gene assemblages were visualized using PCOs (Fig. 5a). The first PCO axis accounted for 55.1% of the total variation, and the second PCO axis accounted for 15.3%. Gene assemblages under ambient and experimental N deposition at sites A and B separated on the first PCO axis. Experimental N deposition gene assemblages at sites C and D shifted slightly in the opposite direction along PCO axis 1. The genes for phenol oxidase, glyoxal oxidase (glx), and endoglucanase had the strongest correlations to the first PCO axis with Pearson's r correlation values of −0.68, −0.56, and −0.65, respectively. All other genes had correlation values between −0.18 and −0.46 (see Table S3 in the supplemental material). Functional genes exposed to experimental N deposition were more dispersed in multivariate space than those occurring under ambient N deposition (Fig. 5a); the dispersion effect of experimental N was confirmed with a significant PERMDISP (P = 0.004).
Fig 5.
PCO displaying the effect of N deposition on the sets of actinobacterial (a) and fungal (b) functional genes involved in the decomposition of forest floor material. Dissimilarity matrices were constructed from the Bray-Curtis similarity metric based on the presence/absence of all relevant gene variants. For actinobacterial and fungal functional genes, there were significant treatment (P = 0.022,0.013), site (P = 0.001, 0.001), and interaction (P = 0.005,0.001) effects, respectively. Pairwise comparisons with Monte Carlo simulations found significant treatment effects in sites B and C (P < 0.05) for both actinobacteria and fungi. Sites A and D did not have a significant response to treatment. The error bars indicate standard error.
ANOSIM (analysis of similarity) revealed that there were significant differences between actinobacterial gene assemblages under ambient and experimental N deposition (R = 0.658; P = 0.01). Large R values (close to 1) indicate complete separation between sets of genes, whereas small R values (close to 0) imply completely random grouping. The assemblages of functional genes within each site were distinct; regardless of treatment, actinobacterial genes within a site were more similar to themselves than to those in other sites (R = 0.613; P = 0.01).
The differences in the compositions of functional genes were further elucidated with PERMANOVA, the multivariate adaptation of ANOVA. There were significant treatment (P = 0.022), site (P = 0.001), and interaction (P = 0.005) effects for actinobacterial functional genes. Pairwise comparisons with Monte Carlo simulations revealed significant treatment effects on functional-gene composition at sites B (P = 0.036) and C (P = 0.026). The composition of functional genes at site D did not significantly differ under experimental N deposition.
Within the bounds of the incomplete coverage of the complete microbial community due to the nature of the microarray, we observed an 18% difference in the compositions of ambient and experimental N deposition actinobacterial functional-gene variants based on the Bray-Curtis similarity metric with SIMPER analysis. When all genes were given equal weight, regardless of gene variant number, the composition of phenol oxidase genes, which may be involved in lignin degradation, contributed the most (10%) to compositional differences. When summed, the composition of genes possibly involved in lignin degradation contributed 27% of the difference in these functional-gene assemblages. The overall influence of these oxidoreductase-encoding genes was a negative response; however, actinobacterial peroxidase contributed 6.3% of the difference, which increased under experimental N deposition. Differences in the composition of genes encoding enzymes involved in decomposing starch contributed 19% of the dissimilarity, followed by hemicellulose (17%), cellulose (16%), and chitin (13%) (see Table S4 in the supplemental material).
Differences in fungal functional-gene compositions between ambient and experimental N deposition were visualized with a PCO (Fig. 5b). The first PCO axis accounted for 56.4% of the total variation in fungal gene assemblages, and the second PCO axis accounted for 13.2%. The gene encoding exochitinase had the strongest correlation (Pearson's r = −0.61) with the first axis; the remaining genes all had correlation values between −0.33 and −0.49 (see Table S3 in the supplemental material). The separation of experimental N deposition and ambient gene assemblages in sites A and B primarily occurred on PCO axis 1, as well. The set of fungal genes under experimental N deposition had more variation between samples than the ambient gene assemblages (PERMDISP; P = 0.001).
Similar to the actinobacterial set of functional genes, the ANOSIM found fungal functional-gene assemblages under experimental N deposition to be significantly different from those under ambient N deposition (R = 0.72; P = 0.01). PERMANOVA results supported this; there were significant treatment (P = 0.013), site (P = 0.001), and interaction (P = 0.001) effects within the fungal functional-gene assemblages. Pairwise comparisons with Monte Carlo simulations revealed significant treatment effects on functional genes at sites B (P = 0.047) and C (P = 0.027).
The composition of fungal functional gene variants under elevated N deposition was 25% different from that of the ambient functional genes. According to the SIMPER analysis, differences in the composition of exochitinase, which encodes a chitin-depolymerizing enzyme, contributed the most (8%) to the difference between ambient and experimental N deposition. All other genes contributed less than 5% when considered individually. When pooled by their substrate categories, genes encoding enzymes for the depolymerization of hemicellulose contributed 25% of the difference in ambient and N deposition fungal functional-gene assemblages, followed by chitin (21%), cellulose (19%), lignin (16%), and starch (11%) (see Table S4 in the supplemental material).
DISCUSSION
Experimental N deposition has elicited changes in the diversity, richness, and composition of both actinobacterial and fungal functional-gene assemblages involved in the decomposition of plant detritus in our long-term field experiment. Specifically, experimental N deposition decreased the richness and diversity of genes at the northern two sites, A and B; altered the composition of functional genes at sites B and C; and increased the heterogeneity of functional-gene assemblages at all sites. These changes occurred in concert with the previously documented decreased extent of forest floor decay, which has increased the accumulation of organic matter in forest floor and surface mineral soil (Table 1). The work we report here supports the idea that compositional shifts in the forest floor decomposer community have functional implications for C cycling and C storage in forest ecosystems under future rates of anthropogenic N deposition from the atmosphere.
In the northern two sites, the decreases in functional richness occurred for both actinobacterial and fungal gene assemblages under experimental N deposition, indicating there were fewer genes detected that mediate the process of plant and fungal cell wall decay. The suppression of functional diversity by enhanced levels of N has been documented in boreal forest floor fungal communities (28) and arctic soil bacterial communities (29). Several studies have further linked decreases in the diversity of microbial communities to a decline in function; for example, Salonius (30) correlated a decline in the metabolic capabilities of soil bacterial communities with diminished diversity; this response did not result from decreased cell counts. Decreased soil saprotrophic fungal richness has also been found to lower respiration in species-poor communities (31).
In our study, decreased functional richness and diversity of saprotrophic microorganisms under experimentally elevated N deposition may be related to the decreased extent of forest floor decay and increased C storage in soil organic matter. As explained by Hanson et al. (32), the decomposition of decay-resistant biopolymers, such as lignocellulose, requires a more diverse assemblage of fungi than is required for other components of plant litter. This is due to the complexity of the lignocellulose molecule and the multiple enzymes needed for depolymerization, many of which are produced by different species and act upon different parts of the molecule (33–35). Therefore, the decrease in the alpha diversity of functional genes under experimental N deposition is one plausible mechanism underlying the decreased extent of decay and organic matter accumulation in our experiment.
The decrease in fungal functional richness by ∼14% occurred across all gene categories (Fig. 3b). The response of the actinobacteria was similar, with a widespread decrease in functional genes, except there was no decrease in the richness of genes encoding enzymes responsible for the decomposition of cellulose. The lack of response from cellulolytic genes is not surprising in that previous studies examining the impact of experimental N deposition on the later stages of forest floor decay found no effect of N on either the activity of cellobiohydrolase enzymes (36) or the transcription of fungal cellobiohydrolase genes (10). It has been suggested that increased N availability is associated with higher cellulolytic activities and initial rates of mass loss (37); however, high N availability neither promoted nor suppressed the breakdown of cellulose in the later stages of leaf decay in our experiment (10).
We also observed a disproportionately large (P = 0.037) decrease in actinobacterial genes encoding chitin-degrading enzymes compared to the decrease in other gene categories. Actinobacteria typically produce chitin-degrading enzymes to requisition the energy (e.g., C) and N in fungal cell walls (38, 39). In N-enriched ecosystems, the decomposition of chitin for N may not be as important. Furthermore, the decrease in genes encoding chitin-degrading enzymes is intriguing when considering competitive interactions between actinobacteria and fungi (40). High N concentrations decreased fungal biomass (36) and competitive ability (41); this altered environment may be selecting for actinobacteria less dependent on decomposing fungal litter. However, previous studies have not detected any response of the enzyme activity of the chitinase enzyme N-acetyl-glucosaminidase to experimental N deposition (36).
Experimental N deposition not only decreased the alpha diversity of fungal and actinobacterial functional genes, it increased the beta diversity between the ambient and experimental N deposition forest floor functional-gene assemblages. This is evidenced by significant PERDISP (P < 0.004) results; functional-gene assemblages under experimental N deposition were more dispersed in multivariate space than ambient genes (Fig. 5). The incomplete depolymerization of lignin under experimental N deposition may feed back to increase heterogeneity in the decomposer community, as well as suppress the diversity and function of saprotrophic microorganisms, and could likely contribute to the variation in site-to-site responses to experimental N deposition.
Lignins are polyphenolic compounds with low solubility; as they are depolymerized by saprotrophic organisms, soluble polyphenols are released into the environment (42). The accumulation of soluble polyphenols can either stimulate or inhibit fungal spore germination and hyphal growth (43). Basidiomycete fungi tend to depolymerize lignin completely to CO2; however, ascomycetes and actinobacteria are limited in their ability to mineralize lignin and therefore incompletely degrade lignin compounds, resulting in the variable production of such soluble polyphenolics (11, 44, 45). There are many catabolic pathways that require a diverse set of enzymes for the complete depolymerization of lignin and its components (33), so it stands to reason that saprotrophic communities that differ in species composition, as our study sites do (46), may change the rate and extent of lignin decomposition in response to experimental N deposition. This would result in a different composition of soluble polyphenolics in the forest floor and therefore different potential feedback on the microbial communities. To understand more about this possible mechanism regulating soil C storage, further insight into the composition of polyphenolic compounds in partially decomposed litter under experimental N deposition is necessary.
Fungal and actinobacterial functional-gene assemblages responded differently to experimental N deposition at each site, yet they all had the same functional response: decreased litter decay. This disconnect between the measured physiological response of decreased litter decay and the inconsistent shift in functional-gene composition between sites could result from the dispersion effect of experimental N deposition on the sapotrophic communities. While these forest floor communities likely contain functional redundancy (47), it seems that both a suppression of richness and diversity (sites A and B) and a shift in overall community composition (sites B and C) lead to similar functional outcomes (i.e., reduced decay and organic matter accumulation). Seasonal differences between sites at the time of sampling could further contribute to the site-specific responses. All sites were sampled within 4 days of each other; however, due to the climatic gradient on which the sample sites lie, each forest site was at a different seasonal phase. It is well established that there is strong temporal variation in forest floor microbial communities (48); the site-to-site differences seen in this study could be attributed to the communities being in different stages of autumnal forest floor decomposition.
This study revealed that even small changes in community composition (25% difference in fungi; 18% in actinobacteria) could lead to important biogeochemical implications. Given the high diversity of soil microbial communities and the limited coverage of this diversity by GeoChip, we are unable to draw conclusions regarding the generality of this response in nature. Nonetheless, a link between composition and function is supported by a review linking soil microbial diversity and composition to C-cycling dynamics, which concluded that changes in microbial community composition resulting from global change are more likely to be relevant to ecosystem function than changes in species richness alone (47). The driving force leading to changes in actinobacterial functional-gene assemblages under experimental N deposition was the response of genes encoding oxidoreductases possibly involved in lignin degradation. Our SIMPER results revealed that the composition of genes possibly related to lignin degradation accounts for 27% of the difference in actinobacterial gene assemblages. Furthermore, the genes encoding putative phenol oxidase and glyoxal oxidase (glx) were two of the genes most strongly correlated with the first axis of the PCO (Fig. 5a); this axis accounted for 55.1% of the variation in assemblages.
The importance of genes possibly involved in ligninolysis in structuring the actinobacterial functional community is further indicated by the significant negative response of the family Micrococcaceae to experimental N deposition. The Micrococcaceae are represented on the microarray by genes encoding muconate-lactonizing enzymes that are involved in the breakdown of lignin-derived aromatics in soil. Taylor et al. (49) isolated two members of the Micrococcaceae able to grow on minimal medium containing lignocellulose, and they metabolized Kraft lignin into lower-molecular-weight products. Even though actinobacteria are not the primary agents of the later stages of plant litter decay, the changes in actinobacterial functional-gene composition observed here, occurring concomitantly with a decreased fungal presence, could be a mechanism responsible for lower decay under experimental N deposition.
Most other taxonomic groups of actinobacteria responded negatively to experimental N deposition (Fig. 4a); the Corynebacteriaceae, Bifidobacteriaceae, Catenulisporaceae, Jonesaceae, and Actinosynnemataceae all had significantly negative response ratios. The lignolytic capabilities of members of these families are either nonexistent or unknown; nonetheless, the Catenulisporaceae and Actinosynnemataceae are known to have vegetative and areal mycelia and have been isolated from fallen leaves and soil (50, 51). The Bifidobacteriaceae are able to break down complex carbohydrates, and their genomes contain genes for complex sugar metabolism; however, these enzymes are not suspected to be extracellular (52).
The actinobacterial families that responded positively are the Beutenbergiaceae, Cellulomonadaceae, and Promicromonosporaceae, all of which belong to the suborder Micrococcineae. The Beutenbergiaceae have been found in cave ecosystems and are known to use glucose and cellobiose as growth substrates; 12% of their genome is dedicated to carbohydrate transport and metabolism (53). The members of the suborder Micrococcineae exist in a wide variety of ecosystems and have been found to be positively correlated with the decomposition of Microcystis blooms in aquatic ecosystems (54), although their autoecology in forest floor ecosystems is not well understood.
The composition of fungal functional-gene assemblages under experimental N deposition was found to be 25% different from that of ambient assemblages, and the gene encoding exochitinase contributed most to the difference between ambient and experimental N deposition communities. The composition of this gene also had the strongest correlation of all genes to the primary PCO axis along which the functional community split. The altered composition of genes encoding the degradation of chitin could be indicative of a different saprotrophic competitive ability of fungi exposed to experimental N deposition (41).
The functional genes from the fungal orders Auriculariales, Botryosphaeriales, Cantharellales, and Microscales all significantly declined under experimental N deposition. The orders Cantharallales and Microscales were primarily represented by genes encoding lignolytic enzymes on the microarray. The Cantharellales are ectomycorrhizal fungi from whose fruiting bodies distinctive laccases have been isolated (55); these fungi are active decomposers in our study sites (unpublished data). The Microscales lignolytic genes represented on the microarray are from the family Halosphaeriaceae of the Sordariomycetes. These are primarily aquatic fungi (56); however, the negative response of the genes encoding lignolytic enzymes provides evidence for experimental N deposition inhibiting the growth of lignolytic fungi. Even though changes in gene composition and the richness of genes encoding lignolytic enzymes has not been detected as a driving force of changes in fungal gene composition, the decrease in fungal taxa known to be active lignin decomposers supports our findings of decreased fungal lignolytic activity in forest floor ecosystems (10). Similar to our findings in Michigan, experimental N deposition decreases the lignolytic capability of microorganisms across nine biomes around the world (57).
In summary, 18 years of experimental atmospheric N deposition have reduced the functional capability of sapotrophic actinobacteria, basidiomycetes, and ascomycetes in the forest floor, consistent with a decreased extent of plant litter decay. Experimental N deposition has led to smaller and less diverse sapotrophic functional-gene assemblages and increased the amount of variation between them. Changes in the actinobacterial functional-gene composition were driven by changes in genes encoding enzymes possibly involved in lignin degradation. This supports the notion of Bugg et al. (58) that the bacterial metabolism of lignin may be more important to decomposition than previously thought. The observations we report here, in combination with a documented reduction in litter decay, provide a plausible mechanism by which chronic experimental deposition has slowed decay and increased soil C storage, as well as enhanced production of phenolic dissolved organic C.
Supplementary Material
ACKNOWLEDGMENTS
This research was made possible by support from the National Science Foundation's Long-Term Research in Environmental Biology program and the Department of Energy's Biological and Environmental Research Division.
We thank Rima A. Upchurch and Lauren Cline from Don Zak's soil ecology laboratory for their valuable contributions to the analysis of the data presented here and Tong Yuan from Jizhong Zhou's laboratory for her work in running the microarray.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03156-12.
REFERENCES
- 1. Reay DS, Dentener F, Smith P, Grace J, Feely RA. 2008. Global nitrogen deposition and carbon sinks. Nat. Geosci. 1:430–437 [Google Scholar]
- 2. Liu LL, Greaver TL. 2010. A global perspective on belowground carbon dynamics under nitrogen enrichment. Ecol. Lett. 13:819–828 [DOI] [PubMed] [Google Scholar]
- 3. Zak DR, Holmes WE, Burton AJ, Pregitzer KS, Talhelm AF. 2008. Simulated atmospheric NO3− deposition increases soil organic matter by slowing decomposition. Ecol. Appl. 18:2016–2027 [DOI] [PubMed] [Google Scholar]
- 4. Burton AJ, Pregitzer KS, Crawford JN, Zogg GP, Zak DR. 2004. Simulated chronic NO3− deposition reduces soil respiration in northern hardwood forests. Glob. Change Biol. 10:1080–1091 [Google Scholar]
- 5. Pregitzer KS, Zak DR, Burton AJ, Ashby JA, MacDonald NW. 2004. Chronic nitrate additions dramatically increase the export of carbon and nitrogen from northern hardwood ecosystems. Biogeochemistry 68:179–197 [Google Scholar]
- 6. Whittinghill KA, Currie WS, Zak DR, Burton AJ, Pregitzer KS. 2012. Anthropogenic N deposition increases soil C storage by decreasing the extent of litter decay: analysis of field observations with an ecosystem model. Ecosystems 15:450–461 [Google Scholar]
- 7. Pregitzer KS, Burton AJ, Zak DR, Talhelm AF. 2008. Simulated chronic nitrogen deposition increases carbon storage in northern temperate forests. Glob. Change Biol. 14:142–153 [Google Scholar]
- 8. Frankland JC. 1998. Fungal succession—unravelling the unpredictable. Mycol. Res. 102:1–15 [Google Scholar]
- 9. Li D, Alic M, Gold MH. 1994. Nitrogen regulation of lignin peroxidase gene tanscription. Appl. Environ. Microbiol. 60:3447–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards IP, Zak DR, Kellner H, Eisenlord SD, Pregitzer KS. 2011. Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PLoS One 6:e20421 doi:10.1371/journal.pone.0020421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tuomela M, Vikman M, Hatakka A, Itavaara M. 2000. Biodegradation of lignin in a compost environment: a review. Bioresour. Technol. 72:169–183 [Google Scholar]
- 12. le Roes-Hill M, Khan N, Burton SG. 2011. Actinobacterial peroxidases: an unexplored resource for biocatalysis. Appl. Biochem. Biotechnol. 164:681–713 [DOI] [PubMed] [Google Scholar]
- 13. Kirby R. 2006. Actinomycetes and lignin degradation. Adv. Appl. Microbiol. 58:125–169 [PubMed] [Google Scholar]
- 14. Godden B, Ball AS, Helvenstein P, McCarthy AJ, Penninckx MJ. 1992. Towards elucidation of the lignin degradation pathway in actinomycetes. J. Gen. Microbiol. 138:2441–2448 [Google Scholar]
- 15. Eisenlord SD, Zak DR. 2010. Simulated atmospheric nitrogen deposition alters actinobacterial community composition in forest soils. Soil Biol. Biochem. 74:1157–1166 [Google Scholar]
- 16. Hassett JE, Zak DR, Blackwood CB, Pregitzer KS. 2009. Are basidiomycete laccase gene abundance and composition related to reduced lignolytic activity under elevated atmospheric NO3− deposition in a northern hardwood forest? Microb. Ecol. 57:728–739 [DOI] [PubMed] [Google Scholar]
- 17. Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, Lee YJ, Mason OU, Dubinsky EA, Chavarria KL, Tom LM, Fortney JL, Lamendella R, Jansson JK, D'Haeseleer P, Hazen TC, Zhou J. 2012. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J. 6:451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zak DR, Pregitzer KS, Burton AJ, Edwards IP, Kellner H. 2011. Microbial responses to a changing environment: implications for the future functioning of terrestrial ecosystems. Fung. Ecol. 4:386–395 [Google Scholar]
- 19. Wu L, Liu X, Schadt CW, Zhou J. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He ZL, Deng Y, Van Nostrand JD, Tu QC, Xu MY, Hemme CL, Li XY, Wu LY, Gentry TJ, Yin YF, Liebich J, Hazen TC, Zhou JZ. 2010. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 4:1167–1179 [DOI] [PubMed] [Google Scholar]
- 21. Clarke KR, Gorley RN. 2006. PRIMER v6: user manual/tutorial. PRIMER-E, Plymouth, United Kingdom [Google Scholar]
- 22. Yergeau E, Bokhorst S, Kang S, Zhou JZ, Greer CW, Aerts R, Kowalchuk GA. 2012. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J. 6:692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo Y, Hui D, Zhang D. 2006. Elevated CO2 stimulates net accumulations of carbon and nitrogen in land ecosystems: a meta-analysis. Ecology 87:53–63 [DOI] [PubMed] [Google Scholar]
- 24. Liang Y, Li G, Van Nostrand JD, He Z, Wu L, Deng Y, Zhang X, Zhou J. 2009. Microarray-based analysis of microbial functional diversity along an oil contamination gradient in oil field. FEMS Microbiol. Ecol. 70:324–333 [DOI] [PubMed] [Google Scholar]
- 25. Clarke KR. 1993. Nonparametric multivariate analyses of changes in community structure. Austral. J. Ecol. 18:117–143 [Google Scholar]
- 26. Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26:32–46 [Google Scholar]
- 27. Anderson MJ. 2006. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253 [DOI] [PubMed] [Google Scholar]
- 28. Allison SD, Hanson CA, Treseder KK. 2007. Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol. Biochem. 39:1878–1887 [Google Scholar]
- 29. Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EA. 2010. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ. Microbiol. 12:1842–1854 [DOI] [PubMed] [Google Scholar]
- 30. Salonius PO. 1981. Metabolic capabilities of forest soil microbial populations with reduced species diversity. Soil Biol. Biochem. 13:1–10 [Google Scholar]
- 31. Setala H, McLean MA. 2004. Decomposition rate of organic substrates in relation to the species diversity of soil saprophytic fungi. Oecologia 139:98–107 [DOI] [PubMed] [Google Scholar]
- 32. Hanson CA, Allison SD, Bradford MA, Wallenstein MD, Treseder KK. 2008. Fungal taxa target different carbon sources in forest soil. Ecosystems 11:1157–1167 [Google Scholar]
- 33. Bugg TD, Ahmad M, Hardiman EM, Rahmanpour R. 2011. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 28:1883–1896 [DOI] [PubMed] [Google Scholar]
- 34. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kirk TK, Farrell RL. 1987. Enzymatic combustion: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465–505 [DOI] [PubMed] [Google Scholar]
- 36. DeForest JL, Zak DR, Pregitzer KS, Burton AJ. 2004. Atmospheric nitrate deposition, microbial community composition, and enzyme activity in northern hardwood forests. Soil Sci. Soc. Am. J. 68:132–138 [Google Scholar]
- 37. Waldrop MP, Zak DR, Sinsabaugh RL, Gallo M, Lauber C. 2004. Nitrogen deposition modifies soil carbon storage through changes in microbial enzymatic activity. Ecol. Appl. 14:1172–1177 [Google Scholar]
- 38. Bhattacharya D, Nagpure A, Gupta RK. 2007. Bacterial chitinases: properties and potential. Crit. Rev. Biotechnol. 27:21–28 [DOI] [PubMed] [Google Scholar]
- 39. Gooday GW. 1990. Physiology of microbial degradation of chitin and chitosan. Biodegradation 1:177–190 [Google Scholar]
- 40. Jayasinghe BATD, Parkinson D. 2008. Actinomycetes as antagonists of litter decomposer fungi. Appl. Soil Ecol. 38:109–118 [Google Scholar]
- 41. Fog K. 1988. The effect of added nitrogen on the rate of decomposition of organic matter. Biol. Rev. Camb. Philos. Soc. 63:433–462 [Google Scholar]
- 42. Hattenschwiler S, Vitousek PM. 2000. The role of polyphenols in terrestrial ecosystem nutrient cycling. Trends Ecol. Evol. 15:238–243 [DOI] [PubMed] [Google Scholar]
- 43. Kuiters AT. 1990. Role of phenolic substances from decomposing forest litter in plant-soil interactions. Bot. Acta 39:329–348 [Google Scholar]
- 44. Trigo C, Ball AS. 1994. Is the solubilized product from the degradation of lignocellulose by Actinomycetes a precursor of humic substances. Microbiology 140:3145–3152 [DOI] [PubMed] [Google Scholar]
- 45. Baldrian P, Voriskova J, Dobiasova P, Merhautova V, Lisa L, Valaskova V. 2011. Production of extracellular enzymes and degradation of biopolymers by saprotrophic microfungi from the upper layers of forest soil. Plant Soil 338:111–125 [Google Scholar]
- 46. Eisenlord SD, Zak DR, Upchurch RA. 2012. Dispersal limitation and the assembly of soil Actinobacteria communities in a long-term chronosequence. Ecol. Evol. 2:538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nielsen UN, Ayres E, Wall DH, Bardgett RD. 2011. Soil biodiversity and carbon cycling: a review and synthesis of studies examining diversity-function relationships. Eur. J. Soil Sci. 62:105–116 [Google Scholar]
- 48. Voriskova J, Baldrian P. 2012. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. doi:10.1038/ismej.2012.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taylor CR, Hardiman EM, Ahmad M, Sainsbury PD, Norris PR, Bugg TDH. 2012. Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J. Appl. Microbiol. 113:521–530 [DOI] [PubMed] [Google Scholar]
- 50. Busti E, Cavaletti L, Monciardini P, Schumann P, Rohde M, Sosio M, Donadio S. 2006. Catenulispora acidiphila gen. nov., sp. nov., a novel, mycelium-forming actinomycete, and proposal of Catenulisporaceae fam. nov. Int. J. Syst. Evol. Microbiol. 56:1741–1746 [DOI] [PubMed] [Google Scholar]
- 51. Tamura T, Hayakawa M, Nonomura H, Yokota A, Hatano K. 1995. 4 new species of the genus Actinokineospora: Actinokineospora inagensis sp nov, Actinokineospora globicatena sp nov, Actinokineospora terrae sp nov, and Actinokineospora diospyrosa sp nov. Int. J. Syst. Bacteriol. 45:371–378 [Google Scholar]
- 52. Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylura. Microbiol. Mol. Biol. Rev. 71:495–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Land M, Pukall R, Abt B, Goker M, Rohde M, Del Rio TG, Tice H, Copeland A, Cheng JF, Lucas S, Chen F, Nolan M, Bruce D, Goodwin L, Pitluck S, Ivanova N, Mavromatis K, Ovchinnikova G, Pati A, Chen A, Palaniappan K, Hauser L, Chang YJ, Jefferies CC, Saunders E, Brettin T, Detter JC, Han C, Chain P, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lapidus A. 2009. Complete genome sequence of Beutenbergia cavernae type strain (HKI 0122(T)). Stand. Genomic Sci. 1:21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li HB, Xing P, Chen MJ, Bian YQ, Wu QLL. 2011. Short-term bacterial community composition dynamics in response to accumulation and breakdown of Microcystis blooms. Water Res. 45:1702–1710 [DOI] [PubMed] [Google Scholar]
- 55. Ng TB, Wang HX. 2004. A homodimeric laccase with unique characteristics from the yellow mushroom Cantharellus cibariusi. Biochem. Biophys. Res. Commun. 313:37–41 [DOI] [PubMed] [Google Scholar]
- 56. Hawksworth DL. 2001. Fungal ecology. eLS doi:10.1038/npg.els.0000356 [Google Scholar]
- 57. Ramirez KS, Craine JM, Fierer N. 2012. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Change Biol. 18:1918–1927 [Google Scholar]
- 58. Bugg TD, Ahmad M, Hardiman EM, Singh R. 2011. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22:394–400 [DOI] [PubMed] [Google Scholar]
- 59. DeForest JL, Zak DR, Pregitzer KS, Burton AJ. 2005. Atmospheric nitrate deposition, declines in decomposition and increases in DOC: test of a potential mechanism. Soil Sci. Soc. Am. J. 69:1233–1237 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.