Abstract
Bacterial indicators are used to indicate increased health risk from pathogens and to make beach closure and advisory decisions; however, beaches are seldom monitored for the pathogens themselves. Studies of sources and types of pathogens at beaches are needed to improve estimates of swimming-associated health risks. It would be advantageous and cost-effective, especially for studies conducted on a regional scale, to use a method that can simultaneously filter and concentrate all classes of pathogens from the large volumes of water needed to detect pathogens. In seven recovery experiments, stock cultures of viruses and protozoa were seeded into 10-liter lake water samples, and concentrations of naturally occurring bacterial indicators were used to determine recoveries. For the five filtration methods tested, the highest median recoveries were as follows: glass wool for adenovirus (4.7%); NanoCeram for enterovirus (14.5%) and MS2 coliphage (84%); continuous-flow centrifugation (CFC) plus Virocap (CFC+ViroCap) for Escherichia coli (68.3%) and Cryptosporidium (54%); automatic ultrafiltration (UF) for norovirus GII (2.4%); and dead-end UF for Enterococcus faecalis (80.5%), avian influenza virus (0.02%), and Giardia (57%). In evaluating filter performance in terms of both recovery and variability, the automatic UF resulted in the highest recovery while maintaining low variability for all nine microorganisms. The automatic UF was used to demonstrate that filtration can be scaled up to field deployment and the collection of 200-liter lake water samples.
INTRODUCTION
To protect beachgoers from illnesses associated with fecal contamination from sewage and other sources, officials generally rely on quantification of bacterial indicators to make decisions about beach closures and advisories. Concentrations above established standards indicate an unacceptable human health risk from the possible exposure to human or animal waste and pathogenic microorganisms. In 2007 and 2008, it was reported that E. coli O157:H7, Shigella, Cryptosporidium, and norovirus caused outbreaks of illness in the United States as a result of recreational exposure to contaminated waters (1). In recreational epidemiological studies, diarrhea and respiratory ailments are commonly reported health outcomes, and it is believed that these may be associated with a variety of unidentified enteric viruses (2).
There is no current legal requirement to examine recreational waters for pathogenic microorganisms. There is wide recognition, however, that bacterial indicators may not be adequate indicators of all types of pathogens. A few studies described concentrations of pathogens in recreational waters, but these were generally small-scale, local investigations (2, 3, 4, 5, 6). The exception was a 15-month study of pathogens and indicators at 25 freshwater recreational and water supply sites in New Zealand (7), the results of which were used to better understand pathogen presence and sources of fecal contamination in recreational waters, develop a quantitative microbial risk assessment for campylobacteriosis, and derive new national freshwater recreation guidelines for New Zealand. Studies of sources and types of potential pathogens present at beaches conducted on a regional scale in other areas can provide similar benefits. These types of studies can also provide data on the relationships of bacterial indicator concentrations and environmental and water-quality parameters that are used to provide estimates of swimming-associated health risks to pathogen concentrations.
Because pathogens are typically found in low numbers in environmental waters, it is necessary to concentrate relatively large sample volumes. It would be advantageous and cost-effective, especially for studies conducted on a regional scale, to use a method that can simultaneously target all classes of pathogens (viruses, bacteria, and protozoa), provided the method can provide acceptable and consistent recoveries. We evaluated two types of filtration approaches—virus adsorption-elution (VIRADEL) and ultrafiltration (UF). The VIRADEL filtration method is primarily used for recovering human enteric viruses from water matrices and concentrates viruses by charge interactions (8), with only limited testing of other microorganisms (9, 10). Ultrafiltration is a physical removal process and has been shown to effectively simultaneously concentrate viruses, bacteria, and protozoa (11).
This study was done to test and evaluate five filtration methods for recovery of seeded viruses and protozoa and naturally occurring bacterial indicators in lake water samples. One method was applied in a field setting. Although only lake water samples were tested during this study, the results may be applicable to other types of water samples.
MATERIALS AND METHODS
Sites and sampling methods.
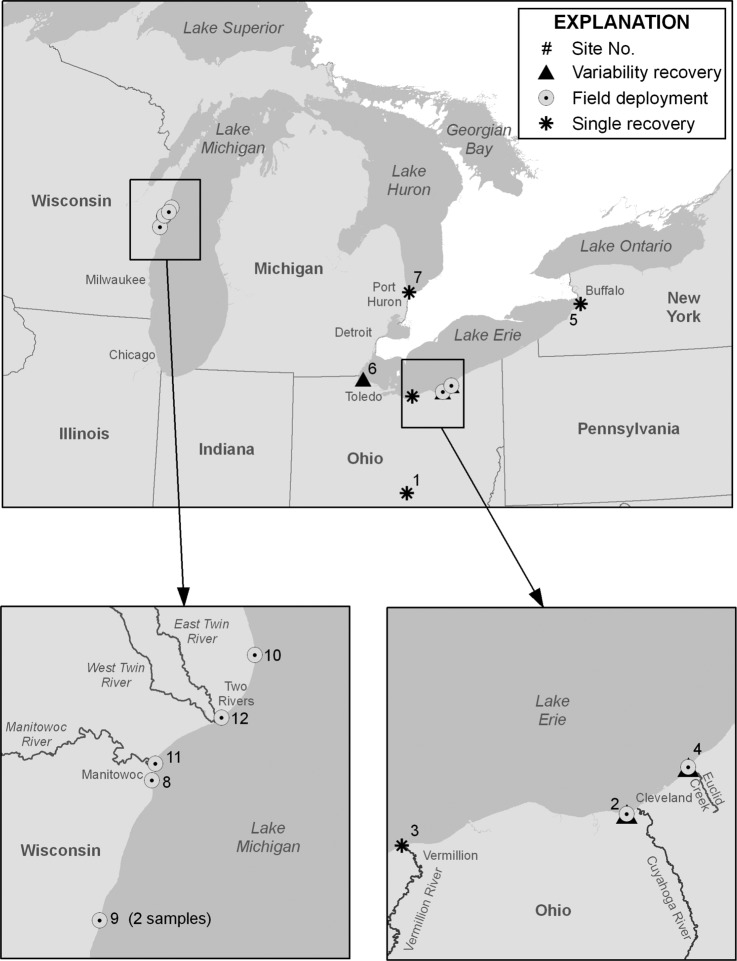
The study included seven recovery experiments with 10-liter lake water samples collected during July 2010 to August 2011 at one inland lake (site 1) and six Great Lakes beaches (Fig. 1). Field deployment studies (described below) were done during August to September 2010 at two sites in Ohio and at five sites in Wisconsin (Fig. 1). Site names, latitude and longitudes, study dates, and water-quality data are presented in the supplemental material (see Table S1 in the supplemental material).
Fig 1.
Sample sites for single and variability recovery experiments and field deployment.
For recovery experiments, 10 liters of lake water was collected into sterile carboys for seeding, and one 3-liter sample was collected for direct processing without filtration. Samples were collected by immersing carboys or bottles below the water surface at the center of the swimming area where water depths were 0.5 to 1.0 m. Lake samples were kept on ice and transported to the U.S. Geological Survey (USGS) Ohio Water Microbiology Laboratory in Columbus, OH (USGS Lab), for further processing. Two types of recovery experiments were designed—single and variability experiments. For single experiments, multiple carboys were collected, and one carboy was subsequently seeded for each filter method. For variability experiments, multiple carboys were collected, and triplicate filtrations for each filter method were run on the same day. Typically, 2 to 3 days were required to conduct all triplicate filtrations for a variability experiment. For all experiments, unseeded controls were collected, filtered, and analyzed within 24 h of collection. The unseeded controls added a duplicate (single studies) or quadruplicate (variability studies) sample for some organisms.
In the USGS Lab, duplicate measurements of turbidity were made with a portable turbidimeter (Hach Company, Loveland, CO) and pH was measured by established USGS methods (12), both from the 3-liter sample. If the pH of the sample was greater than 7.0, it was adjusted to pH 6.5 to 7.0 by adding 0.5 N HCl to the 10-liter sample before seeding and filtration. Reusable equipment was washed and sterilized as described elsewhere (13). To measure any potential for contamination, one equipment blank was processed for each filtration method. Equipment blanks were 10 liters of unseeded dechlorinated tap water that were filtered and processed in the same manner as regular samples.
Microorganisms and seeding.
Stock cultures of MS2 coliphage (an F-specific coliphage), enteric viruses (adenovirus, enterovirus, norovirus GII, and avian influenza virus), and protozoan pathogens (Cryptosporidium parvum and Giardia lamblia) were used to seed 10-liter water samples. Enterococci and Escherichia coli were not seeded so that recovery of these naturally occurring bacterial indicators (often abundant in lake water samples) could be determined. Wide ranges of seed amounts for coliphage and enteric viruses and of concentrations of naturally occurring bacterial indicators were included (Table 1). Concentrations for seeding were established to ensure that each microorganism was recovered after filtration. Seed amounts were representative of moderate to worst-case scenarios in natural settings.
Table 1.
Seed amounts and median percent recoveries of microorganisms in lake water samplesa
| Organism (unit of measurement)b | Avg (SD) seed amt (per 10 liters) | Median recovery (%) |
||||
|---|---|---|---|---|---|---|
| Glass wool (n = 14–20 trials) | NanoCeram (n = 8–13 trials) | CFC+ViroCap (n = 13 trials) | Automatic UF (n = 13–22 trials) | Dead-end UF (n = 13 trials) | ||
| E. coli (CFU) | 4,000 (4,000)c | 2.1 | 0.8 | 68.3 | 64.6 | 62.1 |
| Enterococci (CFU) | 5,000 (6,000)c | 0.9 | 0.4 | 27 | 45.8 | 80.5 |
| MS2 coliphage (PFU) | 460,000 (310,000) | 4.6 | 84 | 20.5 | 67.7 | 58.7 |
| Adenovirus (gc) | 590,000 (340,000) | 4.7 | 0.02 | 0.04 | 1.4 | 0.6 |
| Norovirus GII (gc) | 46,000 (81,000) | 2.0 | 0 | 0.06 | 2.4 | 2.1 |
| Enterovirus (gc) | 8.4E + 6 (1.5E + 7) | 10.5 | 14.5 | 11.3 | 3.5 | 0.9 |
| Avian influenza virus (gc) | 1.0E + 9 (9.3E + 8) | 0 | 0.007 | 0.002 | 0.01 | 0.02 |
| Cryptosporidium (no. of oocysts) | 100 (0)d | 10.7 | 0 | 54.0 | 35.3 | 41.0 |
| Giardia (no. of cysts) | 100 (0)d | 12.1 | 0 | 32.4 | 22.7 | 57.0 |
The results from the filtration method yielding the highest recovery for each organism are shaded. CFC, continuous-flow centrifugation.
gc, number of genomic copies.
Naturally occurring concentrations of microorganisms were used to determine recoveries.
500 oocysts and cysts were used for one glass wool and one NanoCeram filtration.
MS2 coliphage (ATCC 15597-B1) were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and maintained per ATCC instructions. Mahoney strain poliovirus (belongs to the enterovirus group and is referred to here as “enterovirus”) and adenovirus serotype 41 were made from pure culture inoculated cell lines, propagated and provided by other researchers (U.S. Environmental Protection Agency, Cincinnati, OH, and The University of North Carolina at Chapel Hill, Chapel Hill, NC). Norovirus GII stock culture was made by treating a norovirus GII-positive stool sample (obtained from the Ohio Department of Health) with Vertrel XF (Miller-Stephenson Chemical Company, Inc., Danbury, CT), a hydrocarbon degreasing compound. Avian influenza virus (AIV) strain A/turkey/Minnesota/3689-1551/1981 (H5N2) was propagated by inoculation into the allantoic sac of day 8 specific-pathogen-free embryonated chicken eggs and incubated at 37.2°C and 50% relative humidity for 3 days at the USGS National Wildlife Health Center (NWHC), Madison, WI. The titer of the AIV recovered from the allantoic fluid following incubation was quantified by inoculating serial dilutions, and the resultant 50% egg infectious dose (EID50) was calculated (14). Enterovirus, adenovirus, and norovirus GII were diluted in phosphate-buffered saline (Hardy Diagnostics, Santa Maria, CA), MS2 coliphage stocks were diluted in trypic soy broth (TSB), and AIV was diluted in Viral Transport Medium (VTM) (15) for seeding.
Parasitic cysts and oocysts were propagated and flow sorted at the U.S. Environmental Protection Agency (USEPA), National Exposure Research Laboratory (NERL), Cincinnati, OH. C. parvum oocysts (Iowa strain) were propagated and purified as previously described using sucrose and cesium chloride floatation (16). G. lamblia cysts (H3 strain) were propagated in Mongolian gerbils and purified using sucrose floatation followed by Percoll sedimentation (17, 18). Oocysts and cysts were used within 2 months and 3 weeks of purification, respectively. The parasite seeds of 100 or 500 cysts and oocysts were prepared and verified as previously described for C. parvum, except a FACSAria II cell sorter (BD Biosciences, San Jose, CA) and an Aqua-glo kit (Waterborne Inc., New Orleans, LA) were used for detection and sorting. Prepared seeds were stored and shipped overnight at 4°C to the USGS Lab and used within 10 days of preparation (19). Shipment temperature conditions and spike stability were verified with a Thermocron i-button tracking system (Maxim, Inc., Sunnyvale, CA) and trip controls (20). Stock tubes of protozoan oocysts and cysts were rinsed three times with 1 ml of 0.001% Tween 20 for seeding 10-liter water samples.
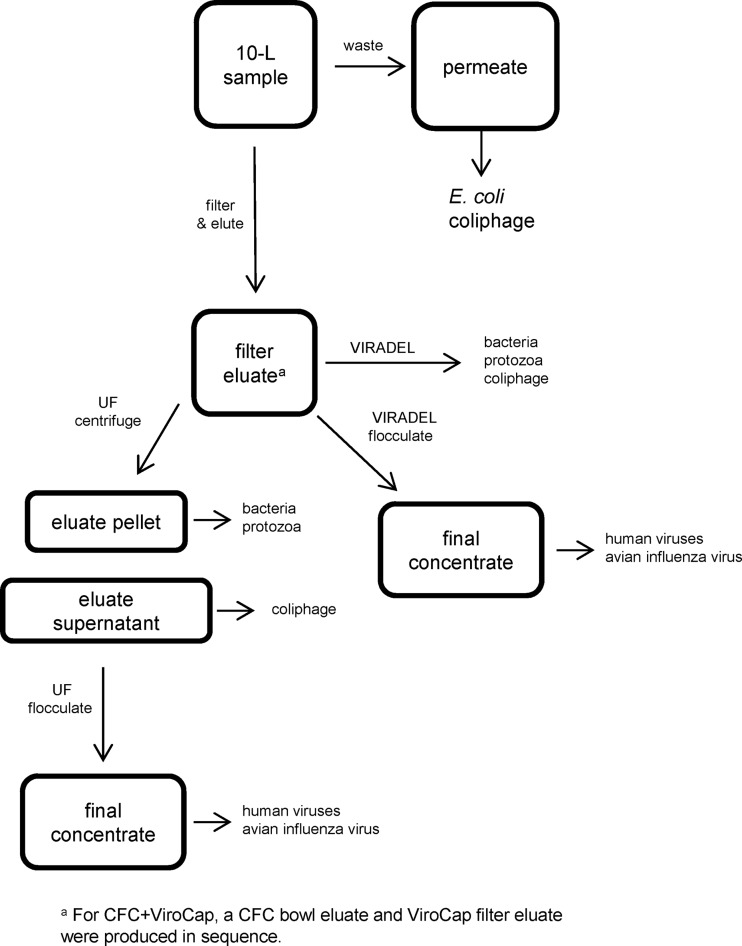
Filtration and postfiltration processing.
Each 10-liter carboy was filtered with a different filtration method, described in detail in the supplemental material (see Fig. S1 to S5 in the supplemental material). General steps are shown in Fig. 2 and described below. Three VIRADEL methods and two UF methods were tested. For most experiments, permeates (from the waste stream) from each filtration were analyzed for E. coli and F-specific coliphage to determine if these target organisms were lost as a result of crossing the filter medium and entering the waste stream. These microorganisms were chosen as representatives of filter compromise for bacteria and viruses.
Fig 2.
General steps in filtration and processing of lake water samples by virus adsorption-elution (VIRADEL) and ultrafiltration (UF) methods showing removals for different microorganisms.
VIRADEL filtration methods.
The VIRADEL filtration method concentrates microorganisms through charge interactions by use of an electropositive filter. These filters were designed specifically to concentrate negatively charged viruses. This study included the following three VIRADEL filters:
Glass wool fiber filter (special order from the U.S. Department of Agriculture [USDA] Agricultural Research Station, Marshfield, WI). For this method, the pH of the sample must be adjusted to pH 6.5 to 7.0 before filtration (21).
NanoCeram filter (Argonide, Inc., Sanford, FL), 2 μm average nominal pore size.
Continuous-flow centrifugation (CFC) with ViroCap capsule filter (CFC+ViroCap) (Scientific Methods, Granger, IN). The ViroCap contains a NanoCeram cartridge filter incorporated into a 12.7-cm-diameter disposable capsule.
The 10-liter sample was pumped through the filter, and microorganisms were eluted and concentrated for glass wool filtration (21, 22) (see Fig. S1 in the supplemental material) and NanoCeram filtration (23) (see Fig. S2 in the supplemental material). Aliquots for analysis of levels of bacteria, coliphage, and protozoa were removed from the filter eluate for glass wool-filtered samples collected at sites 1 to 4 (original procedure) and from all NanoCeram samples. For glass wool-filtered samples collected at sites 5 to 7, aliquots for analysis of levels of bacteria, coliphage, and protozoa were removed from the final concentrate (modified procedure) so that larger proportions were analyzed for these microorganisms (9). For CFC+Virocap filtration, the 10-liter samples were pumped through the CFC bowl and ViroCap filter in sequence and were eluted and concentrated from each (8) (see Fig. S3 in the supplemental material). Aliquots for analysis of levels of bacteria and protozoa were removed directly from the CFC bowl eluate for samples collected at sites 1 to 4 (original procedure). For samples collected at sites 5 to 7, the CFC bowl eluate was further concentrated by centrifugation and the resultant CFC bowl eluate concentrate was used for bacterial and protozoan analyses. For all CFC+Virocap samples, coliphage analyses were done from the filter eluates. For all VIRADEL methods, enteric virus analyses were done from the final concentrates (Fig. 2). Comparisons between the original and modified procedures for the glass wool and CFC+ViroCap filters showed no consistent differences in recoveries of affected microorganisms (data not shown).
Ultrafiltration methods.
Ultrafiltration (UF) methods rely on size exclusion and have pore sizes rated by molecular weight cutoffs which enable concentration by sieving, instead of adsorption or sedimentation (24). The ultrafilters used in this study were Rexbrane Membrane High-Flux, REXEED-25S (Asahi Kasei Kuraray Medical Co., Ltd., Japan), with a molecular weight cutoff of 29,000, surface area of 2.5 m2, and fiber inner diameter of 185 μm. Two UF methods were included in this study:
An automatic tangential-flow UF sampler (automatic UF) provided by USEPA, National Homeland Security Research Center (Teledyne Isco, Lincoln, NE).
Dead-end UF.
The 10-liter sample was pumped through and eluted from the automatic UF sampling device (25) (see Fig. S4 in the supplemental material) and the dead-end UF (26) (see Fig. S5 in the supplemental material). A computer-controlled system automates the process of concentrating microorganisms in the automatic method. The dead-end UF differs from automatic UF in that dead-end UF involves a single pass of water that is not recirculated. For both UF methods, the filter eluate was centrifuged and the resultant pellet was used for bacterial and protozoan analyses and the supernatant for coliphage analysis (Fig. 2). Enteric virus analyses were done from the final concentrates.
Bacterial indicator and coliphage quantification.
Enterococci and E. coli were enumerated by use of standard membrane filtration methods on membrane-enterococcus indoxyl-β-d-glucoside (mEI) agar (27) and modified membrane-thermotolerant Escherichia coli (mTEC) agar (28), respectively. F-specific coliphage were enumerated by use of the single-agar-layer procedure (29). This method detects any F-specific coliphage that is able to infect the host bacterium (E. coli Famp) and produce a circular lysis zone (plaque); MS2 is one strain of F-specific coliphage and was used to seed water samples in recovery experiments.
Protozoan quantification.
Cryptosporidium and Giardia were isolated and enumerated using USEPA method 1623 with heat dissociation (30, 31). Processed samples were shipped overnight at 4°C from the USGS Lab to the USEPA. One immunomagnetic separation (IMS) reaction was performed per sample. In highly turbid samples, an additional 10-ml deionized water rinse was added after the first IMS purification. The slides were stained with EasyStain G&C (BTF Pty Ltd., North Ryde, New South Wales, Australia) following the manufacturer's protocol, except steps 3, 6, and 7 were omitted.
Enteric virus quantification.
Viral RNA and DNA were extracted from 400 μl of the final concentrates using a QIAamp DNA Mini Extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, except the AL general lysis buffer was substituted for the AVL viral lysis buffer with the addition of carrier RNA (Qiagen, Valencia, CA). Of the 100 μl of extracted concentrate, 5 μl was analyzed by use of quantitative PCR (qPCR) for adenovirus (32) or quantitative reverse transcriptase PCR (qRT-PCR) for enterovirus (33) and norovirus GII (34). PCR inhibition was determined using matrix spikes by seeding the sample with an extracted positive-control virus in a duplicate qPCR or qRT-PCR. The concentration of target virus in the sample was then compared to the concentration of target virus in the clean matrix control that was seeded with the same extracted positive-control virus. Sample extracts were considered inhibited and were diluted if the seeded test sample was >2 threshold cycles (CT) higher than the seeded clean matrix control.
The standard curves for molecular detection of adenovirus, enterovirus, and norovirus GII were created using virus stocks treated with Benzonase (Novagen, Madison, WI), as described previously (21), except that the treated stocks were incubated overnight at 37°C, as recommended by Novagen, instead of for 30 min at 37°C and 2 days at 4°C. Treated stocks were extracted, the amount of virus RNA or DNA was measured by using RiboGreen or PicoGreen (Molecular Probes, Eugene, OR) and a spectrophotometer, and the number of genomic copies (gc) was calculated. After quantification, viral stocks were serially diluted using a 2% beef extract solution. Each standard point was extracted in duplicate and then tested by qPCR or qRT-PCR in duplicate on every plate.
Samples for AIV were quantified as described elsewhere (35). Briefly, avian influenza viral RNA was recovered from a 50-μl aliquot of the eluate using an Ambion MagMax AI/ND viral RNA kit according to the manufacturer's instructions. Viral RNA was eluted in a total of 50 μl, and 8 μl of the RNA was quantified to the matrix gene of the AIV genome in a qRT-PCR test. A standard curve was generated in each experiment using RNA extracted in a similar manner from AIV stocks of known concentration and the curve used to calculate the amount of AIV recovered. The qRT-PCR assay is capable of quantification of between 2.5 × 100 and 2.5 × 107 EID50 units of virus per reaction. Sample inhibition was detected by spiking with a heterologous AIV RNA (H7) to the recovered eluate in parallel qRT-PCR assays (35).
Field deployment experiments.
Eight samples were collected during field deployment experiments at seven sites (Fig. 1; see also Table S1 in the supplemental material). Sources of fecal contamination at sites 2 and 4 in Cleveland, OH, include urban storm water runoff and large populations of birds. An adjacent stream with combined sewer overflows and a wastewater pump station additionally affects water quality at site 4. The Wisconsin beaches are located in Manitowoc County and are potentially impacted by agricultural sources and storm water runoff. Water quality at sites 8 and 11, located within the city of Manitowoc, is affected by the Manitowoc River. Site 12 is located in the small community of Two Rivers, and potential sources include storm sewers and the Twin River. Site 10 is 11 miles north of Manitowoc in a recreational and agricultural area.
For field experiments, ambient concentrations of microorganisms were determined and samples were not seeded. The automatic UF method (24) was selected because a portable unit was available at the time, and this method provided acceptable recoveries in early experiments.
From the UF sampling unit, approximately 9 m of sterile inlet tubing was attached to the middle of a bar anchored to the lake bottom where water depths were 0.6 to 0.9 m. For some samples, a 100-mesh Alsco prefilter (Alsco Industrial Products, Lithia Springs, GA) with 150-μm openings was attached in-line outside the sampling unit to filter algae and other large particles and prevent filter clogging (see Fig. S6A in the supplemental material). As a substitute for or in addition to the prefilter for some samples, a 14-cm-long tubing “end connector” with irregular holes (approximately 2 mm in diameter) was attached to the end of the inlet tubing in the lake (see Fig. S6B in the supplemental material). As described above for recovery experiments, one 3-liter sample was collected for direct processing of bacterial indicators and coliphage and for measurements of turbidity.
Recovery calculations and statistical analysis.
For recovery and field deployment experiments, concentrations of each microorganism were determined by applicable analytical methods in the initial seed or in the grab sample for unseeded microorganisms (enterococci and E. coli) and from postfiltration processed samples specific to the target organism and filter.
The data from recovery experiments were analyzed to determine percent recoveries of each organism using different filtration methods. The recovery values, as calculated, do not distinguish the effects of the matrix from the effects of filtration, processing, and analysis. Recoveries for seeded microorganisms were not adjusted for concentrations found in unseeded lake water samples; in most cases, these concentrations were negligible compared to seeded amounts. Percent recoveries were calculated for each result, as follows:
| (1) |
Median percent recoveries were calculated by organism and by filter. A nonparametric analysis of variance (ANOVA), the rank transform test, and the Tukey-Kramer multiple-comparison test were used to compare median recoveries among all filter types for each organism.
A ranking system was developed to gain insight into which filter methods provided the highest recoveries of microorganisms in lake water while still maintaining low variability. The median percent recoveries were used to rank five filters for recovering each organism from 1 to 5, with the highest median recovery receiving the lowest rank (R). A variability rank score (RCV) was then calculated for each organism and filter method as follows:
| (2) |
where CV is the median coefficient of variation for the three variability recovery studies for each organism and filter.
The RCV was then ranked from lowest to highest (R′), with the lowest value representing the filter with the lowest RCV.
An average RCV was calculated for each filter for all microorganisms, giving equal weight to each type of organism (bacteria, viruses, and protozoa) as follows:
 |
RESULTS
Because turbidity was expected to affect recoveries, the samples collected represented a wide range of turbidities. Turbidities ranged from <5 to 280 nephelometric turbidity units (NTRU) in recovery and field deployment experiments (see Table S1 in the supplemental material), with an average of 58 NTRU (standard deviation [SD] = 87). Because pH adjustment to pH 6.5 to 7.0 is required for glass wool filtration, it was measured and adjusted for all filtered samples. Initial sample pH values ranged from 7.6 to 8.8 (see Table S1 in the supplemental material), with an average of pH 8.4 (SD = 0.2).
Recoveries in 10-liter seeded lake water samples.
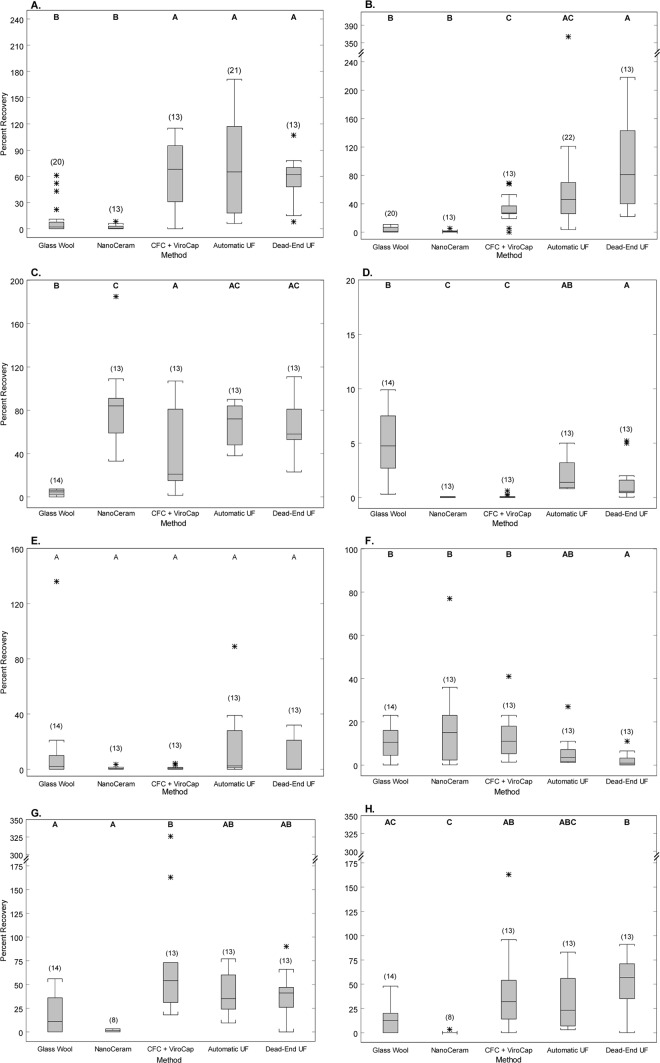
Median percent recoveries of microorganisms in lake water samples are presented in Table 1 with information on seed amounts and numbers of trials. Ranges for the numbers of trials were included because fewer protozoan analyses were done using the NanoCeram, and because unseeded controls for enterococci and E. coli were added in the sample counts for glass wool filtration and automatic UF. Box plots show the distributions of recoveries for each microorganism (Fig. 3); box plots for AIV recoveries were not included because recoveries were very low. Results from Tukey-Kramer multiple-comparison tests for median recoveries among filtration methods are presented as letters on the plots.
Fig 3.
Percent recoveries of viruses in lake water, 2010 to 2011. (A) E. coli, (B) enterococci, (C) MS2 coliphage, (D) adenovirus, (E) norovirus GII, (F) enterovirus, (G) Cryptosporidium, and (H) Giardia. The numbers of samples for each filtration method are indicated in parentheses. Each box includes the median concentration (center line) and the 25th and 75th percentiles (lower and upper box limits, respectively). Error bars represent data values less than or equal to 1.5 times the interquartile range outside the quartile. Asterisks indicate outlier values greater than 1.5 times the interquartile range outside the quartile. Results of the Tukey-Kramer multiple-comparison tests are presented as letters, and recoveries with at least one letter in common do not differ significantly. Note the different scales for percent recoveries for different microorganisms.
Recoveries for E. coli and enterococci were determined using concentrations from unseeded, naturally occurring bacteria in samples directly processed by membrane filtration (Table 1). Median recoveries of E. coli (Fig. 3A) and enterococci (Fig. 3B) for the CFC+ViroCap and UF methods were statistically higher than those for the glass wool and NanoCeram filtration methods. Concentrations of E. coli in permeates (waste stream) were below the limit of detection (<20 CFU/10 liters) for all filters except for the glass wool (data not shown). For the glass wool method, however, the average E. coli concentration in the permeates was 1,300 CFU/10 liters (SD = 1,800) (data not shown).
The analytical method for coliphage detects F-specific coliphages that are able to infect the host bacterium. In contrast, MS2 coliphage is one strain of F-specific coliphage and was used to seed water samples in recovery experiments. Percent recoveries of MS2 coliphage, therefore, were determined in seeded lake water samples, and concentrations of F-specific coliphage were determined in unseeded lake water samples. F-specific coliphage were detected in 53% of unseeded lake water samples; however, concentrations in unseeded samples (<4 to 170 PFU/10 liters; data not shown) were negligible compared to seed concentrations of MS2 coliphage (Table 1). Median recoveries of MS2 coliphage were >20% for all methods except for that for the glass wool filtration, which was statistically lower than those of all the other methods (Fig. 3C). Concentrations of MS2 coliphage in the permeates from the UF filters were all below detection (<100 PFU/10 liters; data not shown), and small amounts of MS2 coliphage were found in the permeates from the NanoCeram and CFC+ViroCap (averages of 2,300 and 900 PFU/10 liters, respectively; data not shown). From the glass wool, however, the average concentration in the permeates was 420,000 PFU/10 liters (SD = 320,000; data not shown), indicating poor trapping efficiency for coliphage.
Median recoveries of enteric viruses ranged from 0% to 14.5% (Table 1). For adenovirus in lake water, the highest median recovery was with the glass wool (4.7%), but this was not statistically higher than the recovery seen with the automatic UF method (Fig. 3D). For norovirus GII, the highest median recovery was for the automatic UF (2.4%), but this was not statistically higher than that for any other method (Fig. 3E). For enterovirus, the highest median recovery was for the NanoCeram (14.5%), but this was not statistically higher than that for any other method except for the dead-end UF (Fig. 3F). In unseeded lake water samples, norovirus GII and enterovirus were not detected. Adenovirus was detected in 47% of unseeded lake water samples in concentrations ranging from 3 to 170 gc/10 liters (data not shown), much lower than the seed amounts (Table 1). Avian influenza virus was not detected in any unseeded lake water samples. Recoveries of seeded AIV were very low (Table 1), with the highest median recovery for the dead-end UF (0.02%).
Aliquots of 100 or 500 Cryptosporidium oocysts and Giardia cysts were seeded into lake water samples. Fifteen unseeded lake water samples were analyzed, and background averages were generally low; eight of those had <1 cyst or oocyst per 10 liters, and four had either 1 oocyst or 1 cyst per 10 liters. The remaining three samples included results from glass wool and (or) automatic UF methods in two samples, with concentrations ranging from 1 to 52 oocysts or cysts per 10 liters. Only one sample had results with >16 oocysts and cysts per 10 liters, and these results were from a glass wool-filtered sample where considerable particulate clumping was observed. These results were considered an aberration due to particulate clumping and the fact that a small percentage of the sample was analyzed, both of which lead to higher variability. Consequently, seed values were not adjusted for background concentrations and recoveries may be slightly inflated. Median recoveries of Cryptosporidium and Giardia were >20% for the CFC+ViroCap and UF methods. Median recoveries were statistically higher for the CFC+Virocap for Cryptosporidium and the dead-end UF for Giardia than for the glass wool and NanoCeram (Fig. 3G and H).
Recovery and variability ranks for microorganisms.
In order to identify which filter performed best with lake water samples, median recoveries were ranked from 1 to 5 for each organism (R), with 1 representing the filter with the highest recovery (Table 2). Variability rank scores (RCV) and variability-weighted ranks (R′) based on the RCVs were calculated. The mean coefficient of variation, used as the variability measure for these calculations, was determined for each microorganism and filter method in the three lake water variability experiments. Different filters were ranked highest for different microorganisms (Table 2). When variability was considered along with recovery (R′), the number 1 rankings remained the same for all microorganisms as determined based on recovery alone (R); however, the number 2 rankings changed for enterovirus, AIV, Cryptosporidium, and Giardia. For recovery of all microorganisms, the automatic UF resulted in the lowest average RCV and R′ (Table 3).
Table 2.
Ranks of median percent recoveries, variability rank scores, and the ranks based on the RCV in lake water samples using five filtration systemsa
| Filtration system |
E. coli |
E. faecalis |
MS2 coliphage |
Adenovirus |
Norovirus GII |
Enterovirus |
Avian influenza virus |
Cryptosporidium |
Giardia |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | RCV | R′ | R | RCV | R′ | R | RCV | R′ | R | RCV | R′ | R | RCV | R′ | R | RCV | R′ | R | RCV | R′ | R | RCV | R′ | R | RCV | R′ | |
| Glass wool | 4 | 2.32 | 4 | 4 | 4.88 | 5 | 5 | 1.4 | 4 | 1 | 0.29 | 1 | 2 | 1.28 | 2 | 3 | 1.2 | 4 | 5 | 6.70 | 5 | 4 | 3.24 | 4 | 4 | 4.24 | 4 |
| NanoCeram | 5 | 2.8 | 5 | 5 | 4.05 | 4 | 1 | 0.16 | 1 | 5 | 4.1 | 5 | 5 | 5.15 | 5 | 1 | 0.67 | 1 | 3 | 2.25 | 4 | 5 | 4.35 | 5 | 5 | 8.65 | 5 |
| ViroCap | 1 | 0.15 | 1 | 3 | 1.29 | 3 | 4 | 2.32 | 5 | 4 | 4 | 4 | 4 | 3.88 | 4 | 2 | 1.14 | 3 | 4 | 1.68 | 2 | 1 | 0.43 | 1 | 2 | 1.24 | 3 |
| Automatic UF | 2 | 0.94 | 2 | 2 | 1.28 | 2 | 2 | 0.28 | 2 | 2 | 0.72 | 2 | 1 | 0.47 | 1 | 4 | 0.68 | 2 | 2 | 1.76 | 3 | 3 | 0.45 | 2 | 3 | 0.57 | 2 |
| Dead-end UF | 3 | 0.96 | 3 | 1 | 0.36 | 1 | 3 | 0.54 | 3 | 3 | 1.14 | 3 | 3 | 3.45 | 3 | 5 | 4.2 | 5 | 1 | 0.63 | 1 | 2 | 0.52 | 3 | 1 | 0.18 | 1 |
Ranks of 1 and 2 are shaded for each target microorganism. R is the rank of the median percentage of recoveries in all lake water samples, with the highest median recovery receiving the lowest rank. The lowest R indicates the highest recovery. RCV is the variability rank score, calculated by multiplying the rank (R) times the median coefficient of variation for the three multiple lake water samples. R′ is the rank of the RCVs, with the lowest value representing the filter with the lowest RCV.
Table 3.
Average variability rank scores and average ranks based on the RCVs for recovering all microorganisms from lake water samplesa
| Filtration system | Avg RCV | Avg R′ |
|---|---|---|
| Automatic UF | 0.79 | 2.00 |
| Dead-end UF | 1.33 | 2.56 |
| ViroCap | 1.79 | 2.89 |
| Glass wool | 2.84 | 3.67 |
| NanoCeram | 3.58 | 3.89 |
Avg RCV, average variability rank score; Avg R′, average rank based on the RCV. Filters are listed from lowest Avg RCV to highest.
Field deployment of automatic ultrafiltration.
To evaluate practicality and efficiency in using a filtration method at recreational beaches, eight samples were collected during field deployment experiments at seven sites (Fig. 1 and Table 4) using the automatic UF method. None of the samples were seeded with microorganisms. Ambient concentrations found by the use of direct plating ranged from 54 to 5,100 CFU/liter for bacterial indicators and <10 to 30 PFU/liter for F-specific coliphage (Table 4). Results for enteric virus analyses were inconclusive because all filtered samples showed inhibition due to matrix interference (data not shown). By use of automatic UF, two samples were positive for Cryptosporidium (0.06 oocysts/liter) and all were below detection for Giardia. Sample turbidities ranged from 2.9 to 280 NTRU (Table 4).
Table 4.
Recoveries of bacterial indicators and F-specific coliphage in unseeded lake water samples using automatic ultrafiltration during field deployment studies at seven sites in Ohio and Wisconsin in 2010
| Site no. | Ambient concn, per litera |
Turbidity (NTRU) | Filter information |
Time to filter (min) | Recovery after filtration (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli (CFU) | Enterococci (CFU) | F-specific coliphage (PFU) | Cryptosporidium (oocysts) | Giardia (cysts) | Type(s) of device(s) to reduce cloggingc | Vol filtered (liters) | E. coli (CFU) | Enterococci (CFU) | F-specific coliphage (PFU) | |||
| 2 | 2,300 | 1,500 | <10 | NRb | NR | 6.0 | Prefilter | 150 | 99 | 43.5 | 11.3 | NAd |
| 4 | 460 | 160 | <10 | NR | NR | 2.9 | Prefilter | 150 | 81 | 159 | 238 | NA |
| 10 | 5,100 | 2,700 | <10 | <0.1 | <0.1 | 100 | None | 29 | 272 | 4.5 | 10.4 | NA |
| 11 | 2,100 | 1,200 | <10 | <0.4 | <0.4 | 280 | Prefilter | 17 | 107 | 23.3 | 33.3 | NA |
| 11 | 4,500 | 4,200 | <10 | <0.5 | <0.5 | 220 | Prefilter and end | 41 | 211 | 35.6 | 38.1 | NA |
| 12 | 160 | 110 | 10 | 0.06 | <0.1 | 20 | End | 103 | 289 | 68.8 | 12.7 | 13.0 |
| 13 | 1,800 | 930 | 30 | 0.06 | <0.1 | 8.0 | End | 200 | 113 | 88.9 | 59.1 | 33.3 |
| 14 | 140 | 54 | 10 | <0.1 | <0.1 | 13 | End | 200 | 174 | 129 | 81.5 | 14 |
Determined by direct plating of the lake water sample for bacterial indicators or filtered samples for protozoan pathogens.
NR, not readable because of algae and debris.
Prefilter, 100 mesh, 150-mm diameter; End, end connector (tubing with irregular holes).
NA, not applicable because coliphage were not present in the direct-plating sample.
The automatic UF filtration method was relatively easy to deploy in the field. Although we attempted to filter approximately 150 to 200 liters, we were not able to filter the full volume in all samples due to filter clogging or system problems (Table 4). Prefilters were used in four of the early experiments; however, in later experiments, we learned that filtration worked more efficiently with attachment of a tubing end connector (see Fig. S6B in the supplemental material). With inclusion of the tubing end connector, filtration times at sites 13 and 14 (the last two sites sampled) were 113 and 174 min for ≥200 liters at a maximum of 2 to 3 liters/min. Median recoveries of E. coli and enterococci in field deployment experiments (56.2% and 35.7%, respectively) were slightly lower than those found for automatic UF in 10-liter recovery experiments (64.6% and 45.8%, respectively). The median recovery of coliphage in field deployment experiments (14.0%) was substantially lower than the median in 10-liter recovery experiments (67.7%).
DISCUSSION
This study was done to identify the filtration method(s) that could be used efficiently and simultaneously to sample for all classes of pathogens (bacteria, viruses, and protozoa) in recreational lake water studies. Even though the three virus adsorption-elution (VIRADEL) methods were designed specifically to concentrate viruses through charge interactions, we evaluated the use of VIRADEL for all microorganisms. Automatic and dead-end ultrafiltration (UF) methods were designed to concentrate all types of microorganisms. Stock cultures of MS2 coliphage, adenovirus, enterovirus, norovirus GII, avian influenza virus (AIV), C. parvum, and G. lamblia were used to seed 10-liter lake water samples. E. faecalis and E. coli were not seeded to determine recoveries of naturally occurring bacterial indicators.
There have been a few studies that presented data on recoveries of microorganisms in surface water that can be used as a comparison to recoveries of microorganisms in the present study. In the present study, using the automatic UF, median recoveries of naturally occurring E. coli and enterococci and seeded MS2 coliphage and Cryptosporidium in lake water samples were 64.6%, 45.8%, 67.7%, and 35.3%, respectively. Gibson and Schwab (36) reported recoveries of 68%, 56.4%, and 51.3% for E. coli, enterococci, and coliphage in seeded stream water samples by the use of tangential-flow UF. Using a similar UF method, Morales-Morales et al. (37) found an average recovery in seeded lake water samples for E. coli of 91.6% and for Cryptosporidium of 31.6%. Using dead-end ultrafiltration, Mull and Hill (38), recovered E. coli (81%), enterococci (85%), MS2 coliphage (66%), and Cryptosporidium (49%) from surface water samples. In the present study, using dead-end ultrafiltration, median recoveries were considerably lower for E. coli (62.1%) but only slightly lower for enterococci (80.5%), MS2 coliphage (58.7%), and Cryptosporidium (41.0%). Bennett et al. (8) found an average recovery for MS2 coliphage of 53% in surface water using the CFC+ViroCap filter; in the present study, median recovery of seeded MS2 coliphage by use of CFC+ViroCap filtration was 20.5%. Deboosere et al. (39) found that glass wool filtration could achieve an average recovery of 1% of H1N1 influenza virus—a higher recovery than they achieved using NanoCeram. In the present study, median recoveries of AIV were much lower—the median was 0% and the highest recovery of an individual sample was 0.009% (data not shown). Deboosere et al. (39) noted that the recovery was much more variable in lake water (0.01% to 7.89%) and that 50-liter samples had poor recovery due to interfering substances. These may be additional factors that resulted in poor AIV recovery in the current study. Millen et al. (9) used glass wool filtration to determine seeded enterovirus and Cryptosporidium recoveries in tap water amended with different amounts of silt loam soil and to determine AIV recoveries in seeded surface water samples. Enterovirus and AIV were quantified by qRT-PCR. Average recoveries of enterovirus, Cryptosporidium, and AIV ranged from 38% to 81%, 28% to 53%, and 7.8% to 42.9%, respectively, much higher than those in the current study (medians of 10.5%, 10.7%, and 0%). However, Millen et al. (9) calculated recoveries by setting the denominator equal to the seed concentration in a negative eluate (after filtration), therefore eliminating results from matrix differences. In the current study, recoveries were calculated by setting the denominator equal to the concentration in the seed before filtration. This enabled us to investigate how recoveries were affected by both the filtration method and the water matrix. To our knowledge, there are no other published studies of virus recovery from surface water samples using qPCR or qRT-PCR.
For E. coli, enterococci, and protozoa, median recoveries in lake water samples were >22% for the CFC+Virocap, automatic UF, and dead-end UF and were statistically higher than for the glass wool and NanoCeram. This is not surprising, in that the glass wool and NanoCeram filters were not originally designed for recovering bacteria and protozoa. Indeed, appreciable concentrations of E. coli were noted in the permeates (waste stream) from the glass wool but not from the other filters. For MS2 coliphage, median recoveries were >20% for all filters except for the glass wool. Similar to E. coli, lower recoveries of MS2 coliphage were attributed to poor trapping efficiency by glass wool filtration as shown by high concentrations in the permeates.
Variability-weighted ranks (R′) were calculated to combine both recovery and variability (from multiple lake water recovery measurements) into one value. Using this approach, the best filtration method would have a relatively high recovery while maintaining low variability. The five filters were ranked best (R′ = 1), as follows: the glass wool for adenovirus, the NanoCeram for MS2 coliphage and enterovirus, the CFC+ViroCap for E. coli and Cryptosporidium, the automatic UF for norovirus GII, and the dead-end UF for E. faecalis, AIV, and Giardia. The glass wool method performed consistently well in recovering the three targeted enteric viruses from lake water samples and ranked first for adenovirus, second for norovirus G11, and a close third for enterovirus. Although the glass wool method ranked low for the other microorganisms, it should not be discounted from inclusion in lake water pathogen studies when viruses are target microorganisms. Among the five filters, the automatic UF resulted in the best average R′ for all nine organisms.
Finally, we showed that the automatic UF method could be used efficiently for sampling 100-to-200-liter volumes of lake water for pathogens. During field sampling at seven Great Lakes beaches, the issue of filter clogging from high turbidity or algae was identified and reduced. The automatic UF method worked more efficiently with the attachment of a tubing end connector for removing large particles (see Fig. S5B in the supplemental material). To improve field filtration efficiency in future work, a polyvinyl chloride (PVC) end connector (40 cm long with 10 0.254-mm slotted openings) (see Fig. S5C in the supplemental material) was designed to replace the shorter tubing end connector (14 cm in length) used in the present study (see Fig. S5B in the supplemental material).
In addition to the automatic UF, other filtration methods have the potential to be practical and efficient for sampling large volumes of lake water. A dead-end concentrator was designed for sampling recreational waters (40) and is commercially available (IntelliSense Design, Inc., Tampa, FL). A manual UF system (41) that operates on the same principle as the automatic UF but without a computer-controlled system worked well in recovering pathogens from 100-liter samples during field studies at Great Lakes and inland lake beaches (data not shown). The glass wool filtration system, contained in a simple plastic tub, was used successfully to collect large volume samples at beaches in Wisconsin (Steve Corsi, USGS, written communication, 2011). In future studies, work is needed to identify appropriate sample volumes for detecting pathogens and to test the PVC end connector with 200-liter sample volumes, if larger volumes are needed. The volume identified would need to be a compromise between sampling as much water as practical and reducing the inhibition found in samples analyzed for viruses by qPCR and qRT-PCR.
To our knowledge, this was the first comprehensive evaluation for multipathogen sampling in recreational waters that included an evaluation of different filtration methods. Many previous studies were done with tap water or a relatively clean water matrix, whereas the present study was done with a variety of lake water samples. After comparison of filtration methods with 10-liter samples, we were able to scale up the methods to 150 to 200 liters and successfully deploy the technique to collect lake water samples from multiple public beaches in two states. The use of one or two filters to effectively concentrate all types of microorganisms makes filtration efficient for regional studies of public health risk at beaches.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vincent R. Hill and Bonnie Mull (Centers for Disease Control and Prevention) for help with the dead-end UF, Fu-Chi Hsu (Scientific Methods Inc.) for help with the CFC+ViroCap, Mark A. Borchardt and Susan K. Spencer (USDA ARS Laboratory) for help with glass wool filtration, and Brian Morris (Pegasus Technical Services) for help with the automated UF device. We also thank David O. Erisman for technical support. We thank Elizabeth Bohuski and other members of the National Wildlife Health Center Diagnostic Virology Laboratory for their capable assistance in AIV analysis.
Support for this study was provided by the U.S. Geological Survey, Coastal Marine Program, and by the U.S. Environmental Protection Agency through its Office of Research and Development.
This publication has been reviewed by the U.S. Environmental Protection Agency but does not necessarily reflect Agency views. No official endorsement by the U.S. Environmental Protection Agency should be inferred. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. government.
Footnotes
Published ahead of print 21 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03117-12.
REFERENCES
- 1. Hlavsa MC, Roberts VA, Anderson AR, Hill VR, Kahler AM, Orr M, Garrison LE, Hicks LA, Newton A, Hilborn ED, Wade TJ, Beach MJ, Yoder JS. 2011. Surveillance for waterborne disease outbreaks and other health events associated with recreational water—United States, 2007–08. MMWR Surveill. Summ. 60:1–32 [PubMed] [Google Scholar]
- 2. Wong M, Kumar L, Jenkins TM, Xagoraraki I, Phanikumar MS, Rose JB. 2009. Evaluation of public health risks at recreational beaches in Lake Michigan via detection of enteric viruses and human-specific bacteriological marker. Water Res. 43:1137–1149 [DOI] [PubMed] [Google Scholar]
- 3. Mocé-Llivina L, Lucena F, Jofre J. 2005. Enteroviruses and bacteriophages in bathing waters. 2005. Appl. Environ. Microbiol. 71:6838–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xagoraraki I, Kuo DHW, Wong K, Wong M, Rose JB. 2007. Occurrence of human adenoviruses at two recreational beaches of the Great Lakes. Appl. Environ. Microbiol. 73:7874–7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schets FM, van Wijnen JH, Schijven JF, Schoon H, de Roda Husman AM. 2008. Monitoring of waterborne pathogens in surface waters in Amsterdam, The Netherlands, and the potential health risk associated with exposure to Cryptosporidium and Giardia in these waters. Appl. Environ. Microbiol. 74:2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abdelzaher AM, Wright ME, Ortega C, Solo-Gabriele HM, Miller G, Elmir S, Newman X, Shih P, Bonilla Alfredo J, Bonilla TD, Palmer CJ, Scott T, Lukasik J, Harwood VJ, McQuaig S, Sinigalliano C, Gidley M, Plano LRW, Zhu X, Wang JD, Fleming LE. 2010. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Appl. Environ. Microbiol. 76:724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Till D, McBride G, Ball A, Taylor K, Pyle E. 2008. Large-scale freshwater microbiological study: rationale, results and risks. J. Water Health 6:443–460 [DOI] [PubMed] [Google Scholar]
- 8. Bennett HB, O'Dell HD, Norton G, Shin G, Hsu F-C, Meschke JS. 2010. Evaluation of a novel electropositive filter for the concentration of viruses from diverse water matrices. Water Sci. Tech. 61:317–322 [DOI] [PubMed] [Google Scholar]
- 9. Millen HT, Gonnering JC, Berg RK, Spencer SK, Jokela WE, Pearce JM, Borchardt JS, Borchardt MA. 2012. Glass wool filters for concentrating waterborne viruses and agricultural zoonotic pathogens. J. Vis. Exp. 3:e3930 doi:10.3791/3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Polaczyk AL, Roberts JM, Hill VR. 2007. Evaluation of 1MDS electropositive microfilters for simultaneous recovery of multiple microbe classes from tap water. J. Microbiol. Methods 68:260–266 [DOI] [PubMed] [Google Scholar]
- 11. Holowecky PM, Jamer RR, Lorch DP, Straka SE, Lindquist HDA. 2009. Evaluation of ultrafiltration cartridges for a water sampling apparatus. J. Appl. Microbiol. 106:738–747 [DOI] [PubMed] [Google Scholar]
- 12. Wilde FD. (ed). 2011. Field measurements—U.S. geological survey techniques of water-resources investigations, book 9, chapter A6, sections 6.1, 6.3, and 6.7. U.S. Geological Survey, Reston, VA: http://pubs.water.usgs.gov/twri9A6/ Accessed October 2011 [Google Scholar]
- 13. Myers DN, Stoeckel DM, Bushon RN, Francy DS, Brady AMG. 2007. Fecal indicator bacteria. U.S. geological survey techniques of water-resources investigations, book 9, chapter A7, section 7.1. U.S. Geological Survey, Reston, VA: http://pubs.water.usgs.gov/twri9A/ [Google Scholar]
- 14. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. J. Hyg. (Lond) 27:493–497 [Google Scholar]
- 15. Docherty DE, Slota PG. 1988. Use of Muscovy duck embryo fibroblasts for the isolation of viruses from wild birds. J. Tiss. Cult. Methods 11:165–170 [Google Scholar]
- 16. Ware MW, Villegas EN. 2010. Improved Cryptosporidium parvum oocyst propagation using dexamethasone suppressed CF-1 mice. Vet. Parasitol. 168:329–331 [DOI] [PubMed] [Google Scholar]
- 17. Belosevic M, Faubert GM, MacLean JD, Law C, Croll NA. 1983. Giardia lamblia infections in Mongolian gerbils: an animal model. J. Infect. Dis. 147:222–226 [DOI] [PubMed] [Google Scholar]
- 18. Sauch JF. 1984. Purification of Giardia muris cysts by velocity sedimentation. Appl. Environ. Microbiol. 48:454–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ware MW, Schaefer FW., III 2005. The effects of time and temperature on flow cytometry enumerated live Cryptosporidium parvum oocysts. Lett. Appl. Microbiol. 41:385–389 [DOI] [PubMed] [Google Scholar]
- 20. Francy DS, Simmons OD, III, Ware MW, Granger EJ, Sobsey MD, Schaefer FW., III 2004. Effects of seeding procedures and water quality on recovery of Cryptosporidium oocysts from stream water by using U.S. Environmental Protection Agency method 1623. Appl. Environ. Microbiol. 70:4118–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambertini E, Spencer SK, Bertz PD, Loge FJ, Kiek BA, Borchardt MA. 2008. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 74:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francy DS, Stelzer EA, Bushon RN, Brady AMG, Mailot BE, Spencer SK, Borchardt MA, Elber AG, Riddell KR, Gellner TM. 2011. Quantifying viruses and bacteria in wastewater—results, interpretation methods, and quality control. U.S. Geological Survey Scientific Investigations Report 2011-5150. U.S. Geological Survey, Columbus, OH [Google Scholar]
- 23. Karim MK, Rhodes ER, Brinkman N, Wymer L, Fout GS. 2009. New electropositive filter for concentrating enterovirus and norovirus from large volumes of water. Appl. Environ. Microbiol. 75:2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu P, Hill VR, Hahn D, Johnson TB, Pan Y, Jothikumar N, Moe CL. 2012. Hollow-fiber ultrafiltration for simultaneous recovery of viruses, bacteria, and parasites from reclaimed water. J. Microbiol. Methods 88:155–161 [DOI] [PubMed] [Google Scholar]
- 25. U. S. Environmental Protection Agency and Centers for Disease Control and Prevention 2011. Comparison of ultrafiltration techniques for recovering biothreat agents in water. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 26. Smith CM, Hill VR. 2009. Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl. Environ. Microbiol. 75:5284–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. U. S. Environmental Protection Agency 2006. Method 1600—enterococci in water by membrane filtration using membrane-enterococcus indoxyl-β-D-glucoside agar (mEI). EPA/821/R-06/009. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 28. U. S. Environmental Protection Agency 2006. Method 1603—Escherichia coli in water by membrane filtration using modified membrane-thermotolerant Escherichia coli aga. EPA 821-R-06-011. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 29. U. S. Environmental Protection Agency 2001. Method 1602—Male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. EPA 821-R-01-029. Office of Water, U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 30. Ware MW, Wymer L, Lindquist HD, Schaefer FW., III 2003. Evaluation of an alternative IMS dissociation procedure for use with method 1622—detection of Cryptosporidium in water. J. Microbiol. Methods 55:575–583 [DOI] [PubMed] [Google Scholar]
- 31. U. S. Environmental Protection Agency 2005. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA-815-R-05-002. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 32. Jothikumar N, Cromeans TL, Hill VR, Lu X, Sobsey MD, Erdman DD. 2005. Quantitative real-time PCR assays for detection of human adenovirus and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gregory JB, Litaker RW, Noble RT. 2006. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal positive control for detection of enteroviruses in environmental samples. Appl. Environ. Microbiol. 72:3960–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jothikumar N, Lowther JA, Henshilwood K, Lees DN, Hill VR, Vinje J. 2005b. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl. Environ. Microbiol. 71:1870–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gibson KE, Schwab KJ. 2011. Tangential-flow ultrafiltration with integrated inhibition detection for recovery of surrogates and human pathogens from large-volume source water and finished drinking water. Appl. Environ. Microbiol. 77:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morales-Morales HA, Vidal G, Olszewski J, Rock CM, Dasgupta D, Oshim KH, Smith GB. 2003. Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl. Environ. Microbiol. 69:4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mull B, Hill VR. 2012. Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J. Microbiol. Methods 91:429–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deboosere N, Horm SV, Pinon A, Gachet J, Coldefy C, Buchy P, Vialette M. 2011. Development and validation of a concentration method for the detection of influenza A viruses from large volumes of surface water. Appl. Environ. Microbiol. 77:3802–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leskinen SD, Kearns EA, Jones WL, Miller RS, Bevitas CR, Kingsley MT, Brigmon RL, Lim DV. 2012. Automated dead-end ultrafiltration of large volume water samples to enable detection of low-level targets and reduce sample variability. J. Appl. Microbiol. 113:351–360 [DOI] [PubMed] [Google Scholar]
- 41. Francy DS, Bushon RN, Brady AMG, Bertke EE, Kephart CM, Likirdopulos CA, Mailot BE, Schaefer FW, III, Lindquist HDA. 2009. Comparison of traditional and molecular analytical methods for detecting biological agents in raw and drinking water following ultrafiltration. J. Appl. Microbiol. 107:1479–1491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.