Abstract
Orange, white, and yellow vacuolated Beggiatoaceae filaments are visually dominant members of microbial mats found near sea floor hydrothermal vents and cold seeps, with orange filaments typically concentrated toward the mat centers. No marine vacuolate Beggiatoaceae are yet in pure culture, but evidence to date suggests they are nitrate-reducing, sulfide-oxidizing bacteria. The nearly complete genome sequence of a single orange Beggiatoa (“Candidatus Maribeggiatoa”) filament from a microbial mat sample collected in 2008 at a hydrothermal site in Guaymas Basin (Gulf of California, Mexico) was recently obtained. From this sequence, the gene encoding an abundant soluble orange-pigmented protein in Guaymas Basin mat samples (collected in 2009) was identified by microcapillary reverse-phase high-performance liquid chromatography (HPLC) nano-electrospray tandem mass spectrometry (μLC–MS-MS) of a pigmented band excised from a denaturing polyacrylamide gel. The predicted protein sequence is related to a large group of octaheme cytochromes whose few characterized representatives are hydroxylamine or hydrazine oxidases. The protein was partially purified and shown by in vitro assays to have hydroxylamine oxidase, hydrazine oxidase, and nitrite reductase activities. From what is known of Beggiatoaceae physiology, nitrite reduction is the most likely in vivo role of the octaheme protein, but future experiments are required to confirm this tentative conclusion. Thus, while present-day genomic and proteomic techniques have allowed precise identification of an abundant mat protein, and its potential activities could be assayed, proof of its physiological role remains elusive in the absence of a pure culture that can be genetically manipulated.
INTRODUCTION
Guaymas Basin is a midocean spreading center located at about 2,000 m depth in the Gulf of California (1), where hydrothermal fluid percolates to the surface through a complex system of basaltic sills and dikes, which are buried in organic-rich sediments derived from terrestrial runoff and the highly productive overlying waters (2, 3). Orange and white Beggiatoaceae filaments are typically found together in the central portion of Guaymas Basin sediment-surface microbial mats, where subsurface temperature gradients are steepest, surrounded by a mat dominated by white filaments (4). The results of physiological studies of orange Beggiatoa (“Candidatus Maribeggiatoa” [5]) filaments collected from mat material have been consistent with a nitrate-reducing, sulfur-oxidizing metabolism (6), although pure cultures have not yet been obtained. Orange Guaymas Beggiatoa (“Candidatus Maribeggiatoa”) species accumulate nitrate (4, 7), likely within their vacuoles, as has been observed or postulated for other, larger-diameter Beggiatoaceae (8, 9). In order to better understand the physiology of these uncultured bacteria, a single orange Guaymas Basin filament was physically isolated and purified of epibionts for genomic sequencing (B. J. MacGregor, J. F. Biddle, C. Harbort, A. G. Matthysse, and A. Teske, submitted for publication).
Orange filaments examined by epifluorescence microscopy during our 2008 and 2009 Guaymas Basin cruises had a diameter of ∼35 μm, corresponding to the 25- to 35-μm size class reported from repeated earlier sampling (10). Prince et al. (11) identified proteins extracted from a mixed natural population of orange Guaymas Beggiatoaceae (average cell width, 30 μm; sample 1615) as c-type cytochromes. Pigmented (yellow and orange) and nonpigmented Beggiatoaceae are also found in the neighborhood of cold seeps in the Gulf of Mexico, and pigmented cells collected there contained a soluble protein with peak absorbance at ∼390 nm (12).
In centrifuging Guaymas Basin mat samples that had been stored at −80°C for RNA extraction, we noted that the supernatant from orange mats was deep orange, suggesting release of some soluble material from the cells. Denaturing protein gel electrophoresis of this supernatant revealed a single pigmented band, which comigrated with the predominant Coomassie-stained band; no corresponding strong band was detected in white-mat supernatants. The identification, preliminary characterization, and possible physiological role of the protein are described here, in the context of the single-filament genome sequence.
MATERIALS AND METHODS
Genome sequencing and annotation.
An orange tuft retrieved from core 4489-10 (Fig. 1A) from the RV Atlantis/HOV Alvin cruise AT15-40 (13 December 2008) at the UNC Gradient Mat site in Guaymas Basin, Gulf of California, Mexico (latitude 27°0.450300′N, longitude 111°24.532320′W; depth, 2,001 m) was purified of epibionts, and its genome was amplified, sequenced, assembled, and annotated as described elsewhere (MacGregor et al., submitted). A total of 99.3% of the sequence was assembled into 822 contigs, suggesting good coverage was achieved. A total of 4.7 Mb of sequence was recovered, with 80% of the sequence forming large (≥15-kb) contigs. Throughout this paper, annotated sequences are referred to by 5-digit contig and 4-digit open reading frame (ORF) numbers, e.g., 00024_0691, and the genome is called the BOGUAY genome (the IMG/ER acronym, derived from “Beggiatoa orange Guaymas”). Additional sequence analysis was carried out using a combination of the J. Craig Venter Institute (JCVI)-supplied annotation; the IMG/ER (13) and RAST (14) platforms; and BLASTN, BLASTX, BLASTP, and PSIBLAST searches of the GenBank nr databases. Tests of genome completeness are described elsewhere (MacGregor et al., submitted).
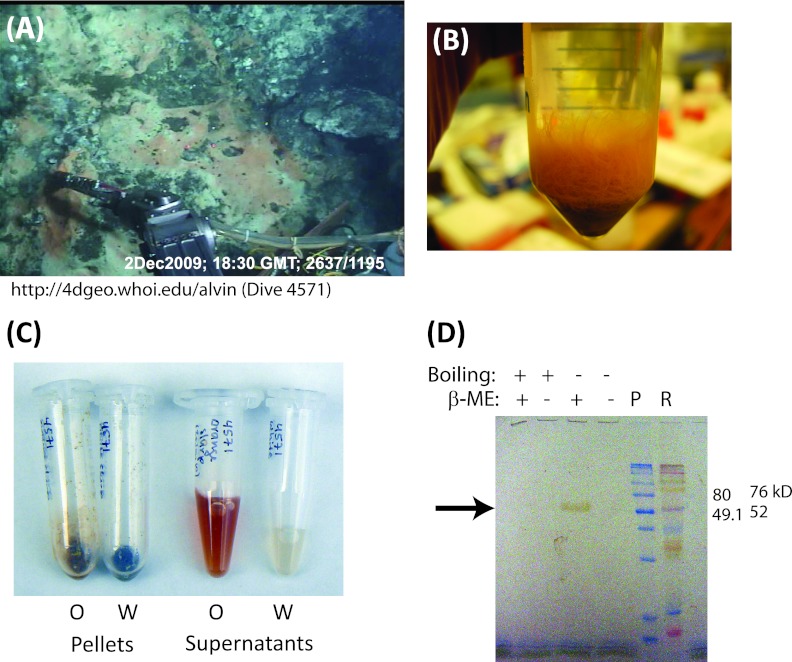
Fig 1.
(A) Slurp gun collection on dive 4571 of a mat sample used for protein characterization, showing orange and white Beggiatoaceae mats, mineral deposits (top right), and the Alvin arm with slurp gun. (B) Orange Beggiatoa (“Candidatus Maribeggiatoa”) tuft. Note that individual filaments are visible. (C) Cell pellets and supernatants from orange (O) and white (W) mats showing the strong coloration of the orange-mat supernatant. (D) Unstained polyacrylamide gel showing a single pigmented ∼50-kDa band (arrow) in the orange-mat supernatant (SDS-14% PAGE, 0.1% SDS; 5 μl orange-mat supernatant per lane). P, prestained molecular mass markers (Amersham); R, Rainbow molecular weight markers (GE Healthcare); β-ME, β-mercaptoethanol.
Protein identification.
Orange and white mat samples were collected with a slurp gun on Alvin dive 4571 (Fig. 1A and B), 2 December 2009, from the Busted Mushroom area (latitude 27°0.647400′N, longitude 11°24.405780′W; depth, 1,990 m). Samples were stored frozen at −80°C for approximately 1 month, and subsamples were scraped off with a sterile spatula. The orange-mat supernatant (∼500 μl) was collected by centrifugation (6,400 × g; 3 min; 4°C) from ∼0.5 g of mat material (Fig. 1C). Ten microliters of supernatant was separated by SDS-PAGE minigel electrophoresis (10% polyacrylamide, 0.1% SDS, no reductant; 80 V through stacking gel and 150 V thereafter for 1.25 h) (Fig. 1D), and the most strongly Coomassie-staining band, which was also visibly pigmented, was excised for identification. Protein identification was performed at the Harvard Microchemistry and Proteomics Analysis Facility by microcapillary reverse-phase high-performance liquid chromatography (HPLC) nano-electrospray tandem mass spectrometry (μLC–MS-MS) on a Thermo LTQ-Orbitrap mass spectrometer. The MS-MS spectra were correlated with the BOGUAY genome sequence using Sequest (15) and programs developed by the Harvard group (16), reviewed for consensus with known proteins, and manually confirmed for fidelity. The signal sequence was predicted by SignalP via the SMART interface (17, 18), and the protein's subcellular location was predicted by PSORTb v3.2 (19), in both cases using the default bacterial settings.
In-gel and in-solution activity measurements.
Protein concentrations were determined by the bicinchoninic acid (BCA) assay (Thermo Fisher). For in-gel assays, samples were incubated in loading dye (50 mM Tris-HCl, pH 6.3, 0.2% bromophenol blue, 10% glycerol) containing 0.2% SDS in the absence of reductant for 10 min at 42°C, loaded on a 10% (most assays) or 15% (hydroxylamine oxidase assay) SDS-PAGE gel, and electrophoresed at 120 V for 90 min or (for hydroxylamine oxidase assays) until the ammonium sulfate in the mat supernatant fractions had run off the gel. For hydroxylamine and hydrazine oxidase activity staining, the assay solution was composed of equal volumes of 0.2 mM phenazine methosulfate (PMS) in 50 mM Tris-HCl, pH 8.0 (made fresh each time), and 0.4 mg ml−1 3[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) in the same buffer (20). The solutions were mixed immediately before use, and hydroxylamine or hydrazine was added to a final concentration of 5 mM. Gels were incubated in the assay solution for 30 min with rocking in the dark before being scanned. For heme staining (39), the gels were washed for 10 min in 3:7 (vol/vol) methanol-sodium acetate (0.25 M, pH 5.0) and incubated for 20 min in the dark in 3:7 (vol/vol) sodium acetate (0.25 M, pH 5.0) containing 6 mM 3,3′-dimethoxybenzidine dihydrochloride in methanol (21). Hydrogen peroxide was then added to make a 60 mM final concentration, and the gel was incubated an additional 60 min. The gel was then washed for 30 min in 8:1:1 (vol/vol/vol) double-distilled water (ddH2O)-methanol-acetic acid (21), scanned, and then equilibrated in water before drying.
In vitro spectrophotometric assays were performed anaerobically in air-tight Schlenk cuvettes using a Hewlett-Packard 8453 spectrophotometer equipped with a multicuvette sample holder. All reactions were performed in triplicate and prepared in an anaerobic Coy Chamber. The hydroxylamine and hydrazine assay solutions consisted of 3.0 ml of 100 mM potassium phosphate buffer (pH 8.0) containing 1.2 mM MTT, 2.0 mM PMS, and 1.5 mM either hydrazine or hydroxylamine (22–24). The reactions were initiated by the addition of 100 μl of enzyme solution (3.1-ml total volume), and the progress of the reactions was monitored spectrophotometrically by collecting spectra every minute for 10 min. Initial rates were calculated from the change in absorbance over time at 578 nm after subtracting the rate of the blank. The nitrite reductase assay solution consisted of 3.0 ml of 100 mM potassium phosphate buffer (pH 7.0) containing 0.6 mM methyl viologen, 10 mM NaNO2, and enzyme. The reaction was initiated by the addition of sodium dithionite to a final concentration of 3.23 mM. Reaction progress was monitored as described above using the change in absorbance at 500 nm (25).
RESULTS
Identification of an abundant orange protein.
Supernatants of cell pellets from orange-mat samples collected in 2009 and stored frozen were a deep orange-red, in contrast to the relatively colorless supernatants from white-mat samples (Fig. 1C). Denaturing gel electrophoresis of β-mercaptoethanol-reduced orange-mat supernatant revealed a single pigmented band (Fig. 1D), which was identified by tandem mass spectroscopy and partially purified for in vitro characterization, as described below.
The pigmented protein is encoded by ORF 00024_0691.
The pigmented band was excised from a Coomassie-stained SDS-PAGE gel for identification. Peptide fragment sequences were identified by μLC–MS-MS and compared to the protein sequences predicted from the BOGUAY genome obtained from an orange filament collected from a different mat (MacGregor et al., submitted). Of 1,272 spectra obtained, a total of 431 (89 unique) could be assigned to 00024_0691 (versus a total of 78 [10 unique] for the conserved hypothetical protein 00871_2242, the second most represented), covering 61.3% of the protein. These spectra accounted for 87% of the total ion current for all fragments (versus 2.4% for hypothetical protein 01444_5033) and 22% of the average ion currents per contig (versus 9.2% for 01444_5033) (see Fig. S1 in the supplemental material). From the single-filament genome, open reading frame 00024_0691 is by far the best match to the pigmented protein from the bulk orange mat and was originally annotated as “hypothetical protein.” The apoprotein encoded, without the predicted signal peptide (see below), has a predicted molecular mass of 54.9 kDa (26), to which eight hemes would add approximately 5.5 kDa (27). This is consistent with its apparent molecular mass by SDS-PAGE (Fig. 1D).
Open reading frame 00024_0691 is predicted to encode a secreted octaheme cytochrome related to hydroxylamine and hydrazine oxidases.
The first 27 amino acids (aa) of the predicted 00024_0691 amino acid sequence represent a possible signal sequence (17, 18), and the protein is predicted from its sequence to be periplasmic (19); specific signal sequences for secretion to the vacuole, if any, are not known. Its inferred phylogeny is shown in Fig. S2 in the supplemental material; close relatives include a second BOGUAY sequence (00935_1708), which in turn is related to a sequence from the genome of the coastal Beggiatoa (“Candidatus Isobeggiatoa”) strain PS. The 00024_0691 sequence is affiliated with a metagenomic sequence assigned to the SUP05 cluster found in marine oxygen minimum zones (28), as well as with two sequences from gammaproteobacterial endosymbionts of hydrothermal vent organisms. Of the limited number of BOGUAY protein phylogenies examined to date, the general pattern is most similar to that found for ATP citrate lyase (MacGregor et al., submitted), a key enzyme in the reductive tricarboxylic acid (TCA) cycle.
Heme-binding sites, active-site residues, and a possible subunit interaction residue were identified by comparison with the hydroxylamine and hydrazine oxidase protein alignment of Klotz et al. (29). The predicted amino acid sequence falls into group II.2, along with putative octaheme proteins identified in other genome-sequencing projects. A representative selection of these is shown in Fig. 2, excluding for simplicity two additional Shewanella sp. sequences. The 00024_0691 amino acid sequence has eight predicted heme-binding sites (CXXCH). Six possible heme axial ligands (all histidines) can be identified by comparison with the characterized proteins. The axial ligand for heme 7 cannot, but a histidine conserved among the group II.2 sequences several amino acids downstream from the ligand in the characterized proteins is a candidate for the role. Similarly, a tyrosine implicated in subunit interaction in the characterized proteins is missing in group II.2, but there is a conserved tyrosine several amino acids downstream.
Fig 2.
Amino acid sequence alignment of putative BOGUAY genome HAO-like octaheme cytochromes 00024_0691 and 00935_1708 with related proteins and predicted proteins. Homologs of the two closely related HAO-like BOGUAY sequences (boldface) were identified by BLASTP searches of the NCBI and IMG/ER databases, and the central regions of each were aligned in accordance with the alignment of Klotz et al. (29). Nitrosomonas europaea HAO and Kuehenia stuttgartiensis HZO are included for comparison. Possible heme-binding sites and axial iron ligands are underlined, and active-site residues are underlined and italicized. A conserved arginine is proposed to hydrogen bond with the propionate group of heme 3. A tyrosine acts as an intersubunit cross-link in the N. europaea HAO and putatively in the K. stuttgartiensis HZO, a conserved tyrosine in the other sequences is a possible homolog, and a nearby conserved histidine might serve as the heme 7 axial ligand (question marks). Accession numbers for GenBank sequences: SUP05_FGYC13F180042, ACX30478; Shal_1091, YP_001673319; BgP_3120, ZP_01999520.1; Rfer_0059, YP_521352.1; Mmc1_3025, YP_866922.1; HAO, CAD84873; HZO, CAJ71439. The remaining sequences can be found in IMG/ER (http://www.jgi.doe.gov) using the locus tags shown (e.g., BOGUAY_0691).
In addition to 00024_0691 and 00935_1708, the BOGUAY genome includes proposed genes for three other multiheme cytochromes. One octaheme cytochrome (01341_2386) may be a periplasmic nitrite reductase (MacGregor et al., submitted). ORF 00614_2851 is predicted to encode a soluble periplasmic octaheme cytochrome and 00935_1710 a membrane-anchored one, as characterized by IMG/ER (13). Their predicted molecular masses (26) (plus 8 heme groups [27]) are as follows: 00935_1710, 41.2 kDa (46.7 kDa); 01341_2386, 61.2 kDa (66.7 kDa); 00614_2851, 47.1 kDa (52.6 kDa); and 00935_1708, 64.9 kDa (70.4 kDa). The putative pentaheme cytochrome (01100_4349) is predicted to be membrane anchored.
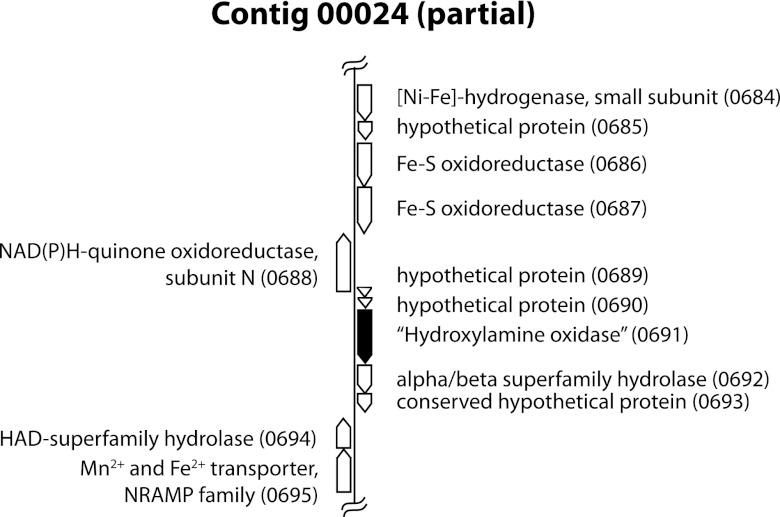
The gene neighborhood of 00024_0691.
The genomic neighborhood of 00024_0691 was examined for possibly related genes (Fig. 3). As annotated by IMG/ER and RAST, it is in a group of five ORFs in the same orientation (0689 to 0693). 00024_0689 and 00024_0690 both encode short hypothetical proteins with no close database matches (as of March 2012). 00024_0692 appears to encode an alpha/beta superfamily hydrolase. This is specified as an acetyl xylan esterase by IMG/ER, but given the diverse activities of this enzyme family, the more general designation seems advisable. None of its close homologs are adjacent to annotated cytochrome genes. 00024_0694, downstream and transcribed in the opposite direction, encodes a putative haloacid dehalogenase superfamily hydrolase (COG0637 by IMG/ER and BLASTP), another hydrolase group with a broad range of functions. 00024_0695 is predicted to encode an Mn2+/Fe2+ transporter of the NRAMP (natural resistance-associated macrophage protein) family, divalent cation transporters widespread in both prokaryotes and eukaryotes. These two genes do not seem to be generally associated with each other, or with multiheme cytochromes, in other sequenced microbial genomes. ORF 00024_0693, downstream of the putative alpha/beta family hydrolase gene, may be derived from a mobile element (B. J. MacGregor, J. F. Biddle, A. G. Matthysse, and A. Teske, submitted for publication).
Fig 3.
Gene neighborhood of ORF 00024_0691. Annotation is based on IMG/ER and JCVI results, which were similar, except that IMG/ER (here and in general) annotates more short hypothetical proteins.
Directly downstream of 00024_0691 on the opposite strand is a putative NuoN gene (Fig. 3), encoding a subunit of the membrane complex of the respiratory NADH-quinone oxidoreductase (complex I). In bacteria and mitochondria, this complex transfers electrons from NADH via the quinone pool to respiratory electron transport chains. In those bacteria in which it has been studied, this consists of a quinone-reducing membrane complex (NuoA, -H, -J, -K, -L, -M, and -N), a connecting FeS protein (Nuo B, -C, -D, and -I), and a cytoplasmic NADH dehydrogenase (NuoE, -F, and -G). The genes encoding these are sometimes found together as an operon (30, 31), but in the BOGUAY genome they are split among several contigs (see Table S1 in the supplemental material) in such a way that they cannot be strung together. There are two nonidentical copies of several of them (NuoB, -C, -D, -F, and -H). The contig 00322 ORFs, which are all on the same strand, are preceded by a putative antitoxin/toxin gene pair (00322_3114 and 00322_3115), while the series of Nuo genes on contig 00285 is interrupted by a putative transposase gene (00285_1232), suggesting that an ancestral operon might have been disrupted or that the genes may have been introduced by lateral transfer. Contig 00322 also includes a possible hybrid cluster protein (Hcp) gene, discussed elsewhere (MacGregor et al., submitted).
Downstream of the NuoN gene, on the opposite strand, are four ORFs putatively encoding a (NiFe) hydrogenase small subunit (00024_0684); a protein related to the membrane-anchor subunit of nitrate reductase, NarI (00024_0685); and two similar, but not identical, FeS oxidases (00024_0686 and 0687). In numerous related sulfur-oxidizing species, including Thiothrix nivea JP2, Beggiatoa alba B18LD, and Thiorhodovibrio sp. strain 970, homologs of the first three genes are followed by a gene putatively encoding the large subunit of a (NiFe) hydrogenase. The cognate gene in the BOGUAY genome (00336_4411) is found separately at the beginning of a small contig with only two other ORFs not obviously related to the hydrogenase function (one putative ApaH gene, part of purine metabolism, and one 26-aa fragment with no good database matches). This may be further evidence of genome rearrangement in the vicinity of 00024_0691.
The partially purified pigmented protein has peroxidase, hydroxylamine oxidase, hydrazine oxidase, and nitrite reductase activities.
The pigmented protein was enriched from orange-mat supernatant by ammonium sulfate precipitation and tested for peroxidase activity (characteristic of cytochromes), hydroxylamine and hydrazine oxidation, and nitrite reduction. An ∼50-kDa protein in the 80% ammonium sulfate precipitate comigrated on SDS-PAGE gels with bands having peroxidase, hydrazine oxidase, and hydroxylamine oxidase activities (Fig. 4). The hydroxylamine oxidase assay (Fig. 4D) was carried out on a lower-percentage acrylamide gel in order to run off a second positively reacting protein or reagent, which has not been further identified. Bovine serum albumin (BSA) was included as a control for nonspecific activity.
Fig 4.
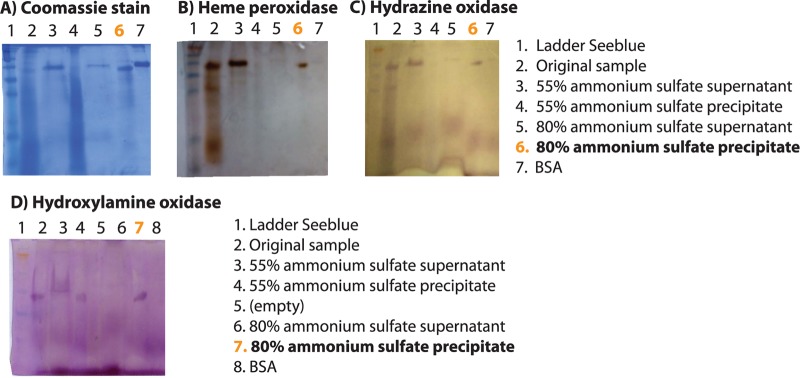
Partial purification and activity assays of the abundant orange protein.
The hydroxylamine oxidase, hydrazine oxidase, and nitrite reductase activities of the original mat supernatant and the partially purified protein were then assayed spectrophotometrically (Table 1) (no in-gel assay for nitrite reductase activity is available). All three activities were detected and were purified to similar extents, suggesting they are carried out by the same protein.
Table 1.
Spectrophotometric enzyme activity assays
| Assay | Activitya |
Fold purification | |
|---|---|---|---|
| Original sampleb | Partially purifiedc protein | ||
| Hydroxylamine oxidased | 0.02 ± 0.006 | 0.16 ± 0.1 | 7.2 |
| Hydrazine oxidased | 0.09 ± 0.04 | 0.46 ± 0.1 | 5.1 |
| Nitrite reductasee | 0.052 ± 0.025 | 0.31 ± 0.1 | 6.0 |
For all assays, activities are shown as units ± standard deviation (n = 3).
Protein concentration, 2.15 mg ml−1.
Protein concentration, 0.15 mg ml−1.
One unit is defined as (1 μmol MTT/min)/(mg protein).
One unit is defined as (1 μmol methyl viologen/min)/(mg protein).
DISCUSSION
Function of the abundant multiheme cytochrome.
By sequence database comparisons, the abundant orange protein falls within a large group of octaheme cytochrome hydrazine and hydroxylamine oxidases, most of which are known only from genomic sequences. Neither function seems likely, from what is known of Beggiatoaceae physiology or from the pathways putatively encoded by the genome (MacGregor et al., submitted). Hydrazine oxidation is known as a step in anaerobic ammonia oxidation (anammox) (32), found in only a limited group of bacteria. Anammox also requires a hydrazine synthase, not identified in the BOGUAY genome. Hydroxylamine oxidases are involved in aerobic ammonia oxidation, which likewise is known to be carried out by only a few microbial groups (33). No BOGUAY homolog for the ammonia monooxygenase (AmoA) required by all identified ammonia oxidizers could be found in the genome.
The genomic neighborhood of ORF 00024_0691 provides few obvious clues to the function of the abundant orange protein (Fig. 3). There are two short ORFs encoding hypothetical proteins upstream on the same strand and a possible hydrolase (00024_0692) and mobile element or remnant thereof (00024_0693) (MacGregor et al., submitted) downstream. The latter might suggest that the cytochrome gene was once located elsewhere in the genome or acquired by gene transfer. There is no evidence in IMG/ER that close relatives of the putative hydrolase gene are associated with cytochromes; they are most often found adjacent to other possible hydrolase genes. The BgP homolog of BOGUAY 00024_0691 (BgP_3120) is on a short contig with only three other annotated ORFs, which bear no apparent sequence or functional similarity to the 00024_0691 gene neighborhood; they are annotated as a sigma-54-dependent response regulator (BgP_3117); a potassium uptake protein, TrkA (BgP_3118); and a short hypothetical protein (BgP_3119). The first two are found together in several related species (including B. alba B18LD), but not in BOGUAY, and not in association with multiheme cytochrome genes.
Activities and abundance of multiheme cytochromes.
In vitro characterization of the partially purified orange Guaymas Beggiatoa protein identified no definitive function. Specific activity in all three assays tested—hydroxylamine oxidation, hydrazine oxidation, and nitrite reduction—increased by approximately the same factor with purification, and the two activities that could be assayed on gels (hydroxylamine and hydrazine oxidation) both comigrated with the strongly Coomassie- and heme-staining band. The in vivo activity (or activities) of the protein may, of course, be different, depending on its subcellular location, local environment, and interaction partners. Alternatively, it is theoretically possible that some minor contaminant may be responsible for the activity. However, the fact that a single ORF could be identified in the genome from a band cut from a similar SDS-PAGE gel supports the attribution of these activities to a single protein. Multiple in vitro activities have been demonstrated for other multiheme cytochromes, including the Thioalkavibrio nitratireducens (34) and Shewanella oneidensis (35, 36) cytochromes discussed above. The T. nitratireducens enzyme also has sulfite reductase activity, which was not tested for in this study (the BOGUAY genome does encode a possible SorAB sulfite oxidase [MacGregor et al, submitted]).
Abundant multiheme cytochromes with hydroxylamine and/or hydrazine oxidase activity have been noted in enrichment cultures of anammox bacteria. A protein classified as an HAO/HZO (23, 24) accounted for as much as 9% of the total soluble protein in a “Candidatus Brocadia anammoxidans” enrichment, and an HAO accounting for 7% of the total protein was purified from a cell extract of an anaerobic ammonia-oxidizing sludge dominated by strain KSU-1 (24). The functions of these proteins are not known, although they are considered to play a role in anammox. The apparent abundance of the 00024_0691 gene product in Coomassie-stained mat supernatants (Fig. 4A) and its straightforward MS-MS identification are consistent with its being a major orange Guaymas Beggiatoa protein, although other explanations are possible; for example, it might have unusually high extracellular stability as mat samples are frozen and thawed. If it functions primarily as a hydroxylamine (or hydrazine) oxidase, it is difficult to find a role for it in Beggiatoaceae nitrogen metabolism as currently understood. Its most likely role at present seems to be as a respiratory or perhaps detoxifying nitrite reductase in the vacuole and/or periplasm. The large volume of these compartments compared to the periplasmic space of nonvacuolar bacteria could require production of large amounts of soluble proteins to achieve sufficient local concentrations. Alternatively, or in addition, it could serve to detoxify other reactive nitrogenous compounds that might accumulate transiently in the shifting hydrothermal environment.
Perspectives.
Marine vacuolate Beggiatoaceae have so far proven difficult to obtain in pure culture, which limits the possible future experimental approaches. Comparison with the genomes of other single filaments, particularly white ones, should reveal differences in the potential cytochrome repertoire and possible nitrogen respiration pathways. Immunolocalization using antipeptide antibodies and freshly collected mat material could clarify whether the 00024_0691 protein is found in the periplasm, vacuole, or both. Hyperspectral microscopy (37), which allows measurement of pigment spectra at very high spatial resolution, is also a possibility if periplasm and vacuole can be distinguished. While technically challenging, in situ measurement of nitrogen compound concentrations in Guaymas Basin microbial mats over time could reveal whether detoxification is likely to be required; perhaps the OsmoSampler technique (38) could be adapted to this purpose.
In conclusion, genomics-based methods allowed identification of the gene encoding a protein whose abundance in microbial mats suggests an important role, and this abundance allowed relatively straightforward partial protein purification and initial characterization. However, proof of its in vivo function will likely still require culture- or enrichment-based experiments.
Supplementary Material
ACKNOWLEDGMENTS
We thank the captains and crews of the RV Atlantis and HOV Alvin for two very enjoyable and productive (if muddy) cruises and the shipboard parties of legs AT 15-40 and AT 15-56. Genome sequencing was performed by the J. Craig Venter Institute, with funding from the Gordon and Betty Moore Foundation Marine Microbial Genome Sequencing Project. Protein identification was carried out by William S. Lane of the Harvard Microchemistry and Proteomics Analysis Facility, Cambridge, MA. The students and auditors of Ann Matthysse's 2010 and 2011 Bacterial Genetics (Biology 522) classes, Sarah Allen, Anke Dopychai, Paul Richard Dunbar, Christopher Harbort, Stuart Hoyle, Stephanie Lambeth, Alex Lawler, Elizabeth Littauer, Nicholas Panchy, Nikolas Stasulli, Lisa Nigro, Lindsay D'Ambrosio, Luke McKay, and TingTing Yang, helped with genome annotation.
The use of RAST was supported in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (NIAD) under contract HHSN266200400042C. The Guaymas Basin project was funded by NSF OCE 0647633.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02538-12.
REFERENCES
- 1. Jannasch HW, Nelson DC, Wirsen CO. 1989. Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site.Nature 342:834–836 [Google Scholar]
- 2. Albertin ML. 1989. Interpretations and analysis of Guaymas Basin multi-channel seismic reflection profiles: implications for tectonic history. M.A. thesis University of Texas at Austin, Austin, TX [Google Scholar]
- 3. Aragón-Arreola M, Morandi M, Martín-Barajas A, Delgado-Argote L, González-Fernández A. 2005. Structure of the rift basins in the central Gulf of California: kinematic implications for oblique rifting. Tectonophysics 409:19–38 [Google Scholar]
- 4. McKay LJ, MacGregor BJ, Biddle JF, Albert DB, Mendlovitz HP, Hoer DR, Lipp JS, Lloyd KG, Teske AP. 2012. Spatial heterogeneity and underlying geochemistry of phylogenetically diverse orange and white Beggiatoa mats in Guaymas Basin hydrothermal sediments. Deep Sea Res. 67:21–31 [Google Scholar]
- 5. Hinck S, Mußmann M, Salman V, Neu TR, Lenk S, de Beer D, Jonkers HM. 2011. Vacuolated Beggiatoa-like filaments from different hypersaline environments form a novel genus. Environ. Microbiol. 13:3194–3205 [DOI] [PubMed] [Google Scholar]
- 6. Otte S, Kuenen JG, Nielsen LP, Paerl HW, Zopfi J, Schulz HN, Teske A, Strotmann B, Gallardo VA, Jørgensen BB. 1999. Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl. Environ. Microbiol. 65:3148–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McHatton SC, Barry JP, Jannasch HW, Nelson DC. 1996. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62:954–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Albuquerque JP, Keim CN, Lins U. 2010. Comparative analysis of Beggiatoa from hypersaline and marine environments. Micron 41:507–517 [DOI] [PubMed] [Google Scholar]
- 9. Hinck S, Neu TR, Lavik G, Mussmann M, De Beer D, Jonkers HM. 2007. Physiological adaptation of a nitrate-storing Beggiatoa sp. to diel cycling in a phototrophic hypersaline mat. Appl. Environ. Microbiol. 73:7013–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nelson DC, Wirsen CO, Jannasch HW. 1989. Characterization of large, autotrophic Beggiatoa spp. abundant at hydrothermal vents of the Guaymas Basin. Appl. Environ. Microbiol. 55:2909–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prince RC, Stokley KE, Haith CE, Jannasch HW. 1988. The cytochromes of a marine Beggiatoa. Arch. Microbiol. 150:193–196 [Google Scholar]
- 12. Nikolaus R, Ammerman JW, MacDonald IR. 2003. Distinct pigmentation and trophic modes in Beggiatoa from hydrocarbon seeps in the Gulf of Mexico. Aquatic Microb. Ecol. 32:85–93 [Google Scholar]
- 13. Markowitz VM, Mavromatis K, Ivanova NN, Chen I-MA, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278 [DOI] [PubMed] [Google Scholar]
- 14. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 [DOI] [PubMed] [Google Scholar]
- 16. Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. 1998. Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37:10866–10870 [DOI] [PubMed] [Google Scholar]
- 17. Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nejidat A, Shmuely H, Abeliovich A. 1997. Effect of ammonia starvation on hydroxylamine oxidoreductase activity of Nitrosomonas europaea. J. Biochem. 121:957–960 [DOI] [PubMed] [Google Scholar]
- 21. Tong Y, Guo ML. 2007. Cloning and characterization of a novel periplasmic heme-transport protein from the human pathogen Pseudomonas aeruginosa. J. Biol. Inorg. Chem. 12:735–750 [DOI] [PubMed] [Google Scholar]
- 22. Schalk J, de Vries S, Kuenen JG, Jetten MSM. 2000. Involvement of a novel hydroxylamine oxidoreductase in anaerobic ammonium oxidation. Biochemistry 39:5405–5412 [DOI] [PubMed] [Google Scholar]
- 23. Shimamura M, Nishiyama T, Shigetomo H, Toyomoto T, Kawahara Y, Furukawa K, Fujii T. 2007. Isolation of a multiheme protein with features of a hydrazine-oxidizing enzyme from an anaerobic ammonium-oxidizing enrichment culture. Appl. Environ. Microbiol. 73:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shimamura M, Nishiyama T, Shinya K, Kawahara Y, Furukawa K, Fujii T. 2008. Another multiheme protein, hydroxylamine oxidoreductase, abundantly produced in an anammox bacterium besides the hydrazine-oxidizing enzyme. J. Biosci. Bioeng. 105:243–248 [DOI] [PubMed] [Google Scholar]
- 25. Kostera J, McGarry J, Pacheco AA. 2010. Enzymatic interconversion of ammonia and nitrite: the right tool for the job. Biochemistry 49:8546–8553 [DOI] [PubMed] [Google Scholar]
- 26. Stothard P. 2000. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104 [DOI] [PubMed] [Google Scholar]
- 27. National Center for Biotechnology Information 30 March 2012. posting date PubChem compound database. http://www.ncbi.nlm.nih.gov/pccompound
- 28. Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, Tortell PD, Hallam SJ. 2009. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326:578–582 [DOI] [PubMed] [Google Scholar]
- 29. Klotz MG, Schmid MC, Strous M, den Camp HJMO, Jetten MSM, Hooper AB. 2008. Evolution of an octahaem cytochrome c protein family that is key to aerobic and anaerobic ammonia oxidation by bacteria. Environ. Microbiol. 10:3150–3163 [DOI] [PubMed] [Google Scholar]
- 30. Friedrich T, Weiss H. 1997. Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J. Theor. Biol. 187:529–540 [DOI] [PubMed] [Google Scholar]
- 31. Moparthi VK, Hagerhall C. 2011. The evolution of respiratory chain complex I from a smaller last common ancestor consisting of 11 protein subunits. J. Mol. Evol. 72:484–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Almeida NM, Maalcke WJ, Keltjens JT, Jetten MSM, Kartal B. 2011. Proteins and protein complexes involved in the biochemical reactions of anaerobic ammonium-oxidizing bacteria. Biochem. Soc. Trans. 39:303–308 [DOI] [PubMed] [Google Scholar]
- 33. Junier P, Molina V, Dorador C, Hadas O, Kim OS, Junier T, Witzel K-P, Imhoff JF. 2010. Phylogenetic and functional marker genes to study ammonia-oxidizing microorganisms (AOM) in the environment. Appl. Microbiol. Biotechnol. 85:425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tikhonova TV, Slutsky A, Antipov AN, Boyko KM, Polyakov KM, Sorokin DY, Zvyagilskaya RA, Popov AN. 2006. Molecular and catalytic properties of a novel cytochrome c nitrite reductase from nitrate-reducing haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio nitratireducens. Biochim. Biophys. Acta 1764:715–723 [DOI] [PubMed] [Google Scholar]
- 35. Atkinson SJ, Mowat CG, Reid GA, Chapman SK. 2007. An octaheme c-type cytochrome from Shewanella oneidensis can reduce nitrite and hydroxylamine. FEBS Lett. 581:3805–3808 [DOI] [PubMed] [Google Scholar]
- 36. Mowat CG, Rothery E, Miles CS, McIver L, Doherty MK, Drewette K, Taylor P, Walkinshaw MD, Chapman SK, Reid GA. 2004. Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat. Struct. Mol. Biol. 11:1023–1024 [DOI] [PubMed] [Google Scholar]
- 37. Polerecky L, Bissett A, Al-Najjar M, Faerber P, Osmers H, Suci PA, Stoodley P, de Beer D. 2009. Modular spectral imaging system for discrimination of pigments in cells and microbial communities. Appl. Environ. Microbiol. 75:758–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jannasch HW, Wheat CG, Plant JN, Kastner M, Stakes DS. 2004. Continuous chemical monitoring with osmotically pumped water samplers: OsmoSampler design and applications. Limnol. Oceanogr. Methods 2:102–113 [Google Scholar]
- 39. Schobert M, Dieter J. 2002. Regulation of heme biosynthesis in non-phototrophic bacteria. J. Mol. Microbiol. Biotechnol. 4:287–294 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.