Abstract
One explanation given for the high microbial diversity found in soils is that they contain a large inactive biomass that is able to persist in soils for long periods of time. This persistent microbial fraction may help to buffer the functionality of the soil community during times of low nutrients by providing a reservoir of specialized functions that can be reactivated when conditions improve. A study was designed to test the hypothesis: in soils lacking fresh root or detrital inputs, microbial community composition may persist relatively unchanged. Upon addition of new inputs, this community will be stimulated to grow and break down litter similarly to control soils. Soils from two of the Detrital Input and Removal Treatments (DIRT) at the H. J. Andrews Experimental Forest, the no-input and control treatment plots, were used in a microcosm experiment where Douglas-fir needles were added to soils. After 3 and 151 days of incubation, soil microbial DNA and RNA was extracted and characterized using quantitative PCR (qPCR) and 454 pyrosequencing. The abundance of 16S and 28S gene copies and RNA copies did not vary with soil type or amendment; however, treatment differences were observed in the abundance of archaeal ammonia-oxidizing amoA gene abundance. Analysis of ∼110,000 bacterial sequences showed a significant change in the active (RNA-based) community between day 3 and day 151, but microbial composition was similar between soil types. These results show that even after 12 years of plant litter exclusion, the legacy of community composition was well buffered against a dramatic disturbance.
INTRODUCTION
The high diversity of microorganisms in soil has proven challenging to investigations of community dynamics and persistence. Numerous DNA- and lipid-based investigations of soil microbial communities have suggested high spatial and seasonal variability, but these methods are often limited to the dynamics of the most dominant taxa and so leave open questions concerning persistence of rare taxa and the ability of a community to maintain key functions. Emerging evidence suggests that the majority of environmental microorganisms may be dormant (1, 2), which may account for the striking numbers of individual taxa that compose the rare biosphere. Furthermore, at least one deep sequencing effort recently revealed that microbial community membership may be stable for long time frames (3). Caporaso et al. (3) observed seasonal shifts in the abundance of dominant species but showed that all operational taxonomic units (OTUs) were present at each sampling time during a 6-year period.
There is likely a spectrum of dormancy within the soil microbial community, ranging from nonviable cells to cells that were recently active. Clearly distinguishing between active and dormant cells is complex but could be aided by the analysis of rRNA rather than rRNA genes (4). Although the dynamics of rRNA in the environment are not completely known, sporulation in Bacillus sp., one of the best-described mechanisms for dormancy, results in rRNA degradation immediately following sporulation that continues for a few days after sporulation initiates (5). An examination of both rRNA and rRNA genes showed phylogenetically related clusters in rRNA sequences only, suggesting that the active community might be closely related bacteria with similar ecological strategies (6). This also accounts for observations of low-abundance taxa in DNA sequence libraries found to be highly abundant in rRNA libraries (7). Focusing on the composition of rRNA may therefore be more reflective of a community that is engaged in litter decomposition and nutrient cycling.
The Detrital Input and Removal Treatments (DIRT) were established in the H. J. Andrews Experimental Forest, OR, in 1997 to test the effects of above- and below-ground plant inputs on soil organic matter stabilization. Included in the experimental manipulations is a no-input (NI) treatment where both roots and above-ground litter have been excluded from the plots. After 12 years of the field experiment, these sites provided a unique opportunity to assess the persistence of a microbial community able to use new additions of plant litter. Previous research at this location has characterized the microbial communities based on lipid (8) and DNA analyses (9) and showed that NI and control (CO) soils varied in composition after 6 years (Table 1). These investigations were likely limited to dominant OTUs and included both active and dormant microbial members. The present study examined extracted soil rRNA, characterized the community using a more intensive sequencing effort, and included a microcosm experiment where new litter additions were provided to soil microorganisms. We hypothesized that following 12 years of no above- or below-ground inputs from plants, active communities in CO and NI soils would differ, but when provided with new inputs, active communities in CO and NI soils would be more similar after a 5-month incubation.
Table 1.
Summary of previous findings comparing DIRT no-input and control treatments
| Method of characterization | Summary of findings | Reference |
|---|---|---|
| Phospholipid fatty acid (PLFA) | Microbial biomass was the same across treatments. Composition was significantly different between CO and NI soils. Fungal/bacterial ratios were significantly lower in NI soils. | 8 |
| DNA-based length heterogeneity (LH) PCR | Bacterial 16S rDNA and fungal ITS compositions were significantly different between CO and NI soils. | 9 |
| DNA-based pyrosequencing | Bacterial 16S rDNA compositions differed between CO and NI (1,100 sequences). Fungal ITS compositions differed between CO and NI (475 sequences). | D. D. Myrold, unpublished data |
MATERIALS AND METHODS
Sample collection.
Soils were collected from the DIRT plots located in the H. J. Andrews Experimental Forest (44°15′N, 122°10′W, 531-m elevation). The DIRT plots were established in 1997 at an undisturbed, forested old-growth stand containing Douglas-fir (Pseudotsuga menziesii) and western hemlock (Tsuga heterophylla); they have been described previously (8, 10, 11). Soils at the site have been classified as coarse loamy mixed mesic Typic Hapludands (12). Experimental manipulations were imposed on three replicate plots, each measuring 10 m by 15 m. For the purposes of this study, soils were collected from two experimental treatments: NI and CO. The perimeter of the NI plots had been trenched to 1 m to allow the installation of an impenetrable barrier, excluding roots, and a 1-mm mesh was installed across the surface and periodically swept to prevent above-ground litter accumulation.
In August 2009, 30 soil cores (2.2-cm diameter to a depth of 10 cm) were collected from NI and CO plots. In CO plots, surface litter was removed prior to sampling, so all samples consisted only of mineral soil. Soil was sieved to 4 mm, and all cores for a given treatment were bulked, creating two composite samples. Subsamples of composited soils were extracted for DNA to establish a background gene quantity. The remaining soil was preincubated at 25°C for 10 days in accordance with standard protocols for long-term ecological reserve (LTER) sites (13).
Soil microcosms.
Microcosms were established using composited soils. Each microcosm contained 30 g of soil kept at field moisture. Half of the microcosms received 0.78 g of 15N-labeled Douglas-fir needles (1.25 mg C g−1 soil), double the rate of average field inputs (14). Before additions, the needles were leached to remove readily soluble organic compounds and chopped to mimic soil animal processing. The litter was homogeneously mixed into soil. The experiment included control/no litter (CO-NL), control/Douglas-fir (CO-DF), no input/no litter (NI-NL), and no input/Douglas-fir (NI-DF). The microcosms were sealed in glass jars to prevent drying and periodically vented to maintain aerobic conditions. Microcosms were incubated for a total of 151 days. At day 3 and day 151, four microcosms of each treatment were destructively sampled. Soil (2 g) was preserved using LifeGuard soil preservation solution (MO BIO Laboratories, Carlsbad, CA) and stored at −20°C until extraction. Due to low extraction recovery in some samples, three replicates per treatment were analyzed.
Soluble C+N and microbial biomass C+N.
At each harvest time, 15 g (dry weight) soil was extracted with 50 ml of 0.05 M K2SO4 and shaken for 1 h before being filtered through Whatman no. 2 filter paper. Ammonium (NH4+) and nitrate (NO3−) were measured by automated colorimetry (Astoria-Pacific 300 series autoanalyzer; Clackamas, OR). Microbial biomass C and N were determined using a 24-h chloroform fumigation followed by extraction (15, 16). Combustion oxidation was used to measure total soluble C, microbial biomass C, and microbial biomass N (Shimadzu TOC-V CSH with TNM-1 module; Tokyo, Japan). A correction factor of 0.45 was used for biomass C (15), and a correction factor of 0.54 was used for biomass N (16).
Quantitative PCR.
Soil RNA and DNA were extracted from each sample using the RNA PowerSoil total RNA isolation kit and the RNA PowerSoil DNA elution accessory kit (MO BIO Laboratories, Carlsbad, CA), according to the manufacturer's instructions. RNA extracts were treated with DNase I (5Prime Inc., Gaithersburg, MD) and then checked by PCR amplification to ensure complete removal of DNA. RNA extracts were reverse transcribed using SuperScript III first-strand synthesis system with random hexamers (Life Technologies, Grand Island, NY).
Both cDNA and DNA were quantified using a Qubit 1.0 fluorometer (Life Technologies, Grand Island, NY). cDNA and DNA extracts were diluted to 1.25 ng μl−1 in preparation for quantitative PCR (qPCR). Brilliant SYBR green qPCR master mix (Stratagene, La Jolla, CA) was used with an ABI 7500 sequence detection system (Life Technologies, Grand Island, NY). Bacterial 16S gene copy numbers were determined using primers Eub338/Eub518, according to published protocols (17, 18). Fungal 28S gene copy numbers were determined using primers LR3/LROR (19) with the following settings: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 1 min. Data were collected at 72°C. Ammonia-oxidizing bacteria (AOB) were quantified using primers amoA-1F/amoA-2R (20) and ammonia-oxidizing archaea (AOA) were quantified using primers Arch-amoAF and Arch-amoAR (21, 22). Plasmids containing primer-specific inserts were used as standards for all assays, except AOB, which were quantified using dilutions of Nitrosomonas europea genomic DNA. Efficiency for all PCR runs were 95 to 105%, and standard curves had an r2 value of >0.98. Disassociation curves were run to ensure specific amplification.
Barcoded pyrosequencing of the 16S and 28S rRNA genes.
PCR was performed using fusion primers containing Titanium A or B adaptors (Roche, Basel, Switzerland), a sample-specific barcode, and gene-specific primers. Bacterial and archaeal 16S genes were amplified using F515/R806 (23) targeting the V4 hypervariable region, and fungal 28S was amplified using LR3/LROR (19) targeting regions D1 and D2. Approximately 25 ng of cDNA was amplified using Invitrogen Platinum Taq (Life Technologies, Grand Island, NY) with the following reagent concentrations: 1× buffer, 2 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.4 μM each primer, and 0.5 U of Taq. Amplifications were cleaned using Agencourt AMPure XP beads (Beckman Coulter Inc., Danvers, MA), by following the Roche 454 Technical Bulletin 2011-002. Prior to pooling, each sample was quantified using the Qubit fluorometer to ensure that equimolar amounts of each sample would be added into a single sequencing reaction. A single pooled sample was submitted to the Oregon State University Center for Genome Research and Biocomputing and analyzed using an FLX Junior (Roche, Basel, Switzerland).
Sequence and statistical analysis.
Univariate data, including qPCR and OTU abundance, were analyzed by three-way analysis of variance (ANOVA) with pairwise comparisons calculated by Tukey's honestly significant difference (HSD) test (P values of <0.05 were considered significant). Univariate statistics were analyzed using JMP version 10.0 (SAS Institute Inc., Cary, NC). qPCR data were log transformed to achieve normality. All sequence analysis was conducted using the QIIME pipeline (24). The sequence library was first separated into 16S and 28S rRNA sequences and then demultiplexed. After quality filtering, the rRNA sequence library contained 108,315 16S sequences and 2,523 28S sequences. (The low number of 28S sequences was likely a result of preferential emulsion PCR of the shorter 16S target compared to the 28S.) OTUs were identified at 97% sequence similarity using Silva 108 (25). Pairwise distances were calculated using UniFrac (26), and these data were visualized using principal coordinate plots (PCoA). The robustness of the sequencing effort was investigated by jackknife analysis. OTU tables were further analyzed with multiresponse permutation procedures (MRPP) to test for significant differences among treatments (PC-ORD version 6; Gleneden Beach, OR).
RESULTS
Soluble C+N and microbial biomass C+N.
Soluble C was significantly different in CO (34 mg kg−1 soil) and NI (24 mg kg−1 soil) soils at day 3 (P = 0.02) but did not differ at day 151 (Table 2). Soluble mineral N (the sum of NH4+ and NO3−-N) was significantly different among soil types (P = 0.001) and litter addition (P = 0.03), but both parameters had significant statistical interactions with sampling day due to the large observed increased in mineral N over the 151-day incubation. Mineral N concentration was highest in the NI-NL incubations at day 151 (74.8 mg kg−1 soil).
Table 2.
Concentrations of total soluble C, total soluble N, microbial biomass C, microbial biomass N, and extracted DNA for each treatment
| Treatment | Harvest day | Concna |
||||
|---|---|---|---|---|---|---|
| Soluble C (mg C kg−1 soil) | Soluble mineral N (mg N kg−1 soil) | Microbial biomass C (mg C kg−1 soil) | Microbial biomass N (mg N kg−1 soil) | Total DNA extracted (μg g−1 soil) | ||
| CO-NL | 3 | 38.0 A | 3.7 C | 379 AB | 31 A | 7.6 A |
| 151 | 27.4 AB | 40.2 B | 304 BC | 28 B | 4.3 A | |
| CO-DF | 3 | 29.3 A | 3.0 C | 427 A | 33 A | 6.5 A |
| 151 | 26.9 AB | 33.4 B | 293 C | 26 B | 2.9 A | |
| NI-NL | 3 | 26.3 B | 6.4 C | 349 AB | 33 A | 3.0 A |
| 151 | 29.4 AB | 74.8 A | 326 BC | 25 B | 3.0 A | |
| NI-DF | 3 | 22.4 B | 5.5 C | 406 A | 35 A | 2.6 A |
| 151 | 26.0 AB | 47.8 B | 288 C | 21 B | 2.2 A | |
Letters show Tukey's HSD comparisons (P < 0.05).
Microbial biomass C significantly decreased from day 3 to day 151 (Table 2). The most dramatic decrease occurred in soils amended with Douglas-fir needles, where day 3 Douglas-Fir treatments averaged 417 mg C kg−1 soil and at day 151 averaged 290 mg C kg−1 soil. A similar trend was observed in microbial biomass N (P = 0.002), with an average decrease of 8 mg N kg−1 soil from day 3 to day 151. In addition to the chloroform fumigation determination of microbial C and N, total extracted DNA was also compared. There were no statistical differences in total DNA among plot, litter, or time; CO soils at day 3 tended to contain more DNA, but yields were variable among extractions.
Gene abundance for bacteria, fungi, and ammonia oxidizers.
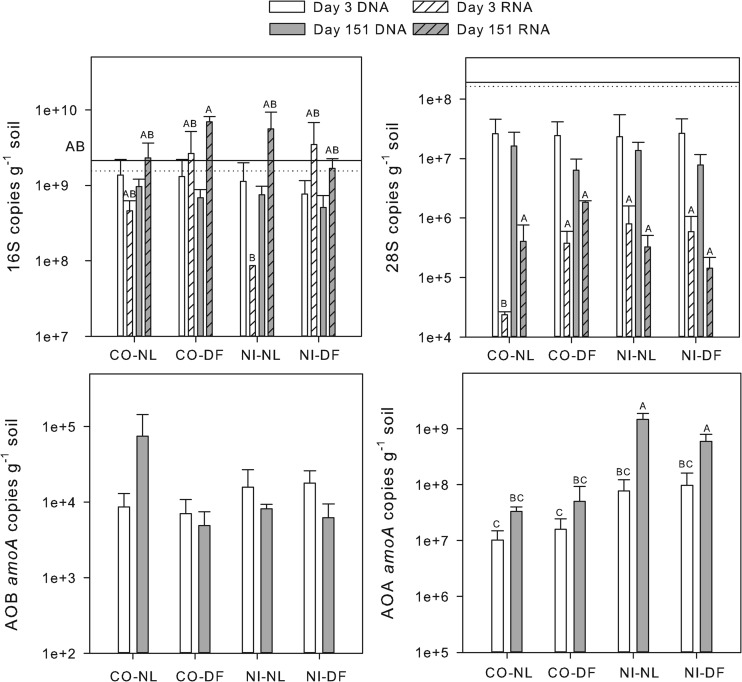
Bacterial 16S ribosomal gene copy numbers were measured in soils immediately after sieving but did not differ between CO (2.12 × 109 copies g−1 soil) and NI (1.55 × 109 copies g−1 soil). Soils from the incubation experiment averaged 1.3 × 109 copies g−1 soil and also did not differ among soil type, litter addition, or sampling time. Bacterial 16S rRNA copy numbers averaged 3.2 × 109 copies g−1 soil, significantly more than DNA. The rRNA copy numbers did not vary by soil type, treatment, or time, but a significant interaction among the three main factors was observed. CO-DF at day 151 did differ from NI-NL at day 3, with all other treatments intermediate (Fig. 1).
Fig 1.
Gene abundance and ribosomal abundance for bacteria (16S), fungi (28S), ammonia-oxidizing bacteria (AOB), and ammonia-oxidizing archaea (AOA). Lines on the 16S and 28S graphs indicate the gene copy numbers in freshly sieved control (solid line) and no-input (dotted line) soils. Dashed lines in the AOB and AOA graphs show the assay detection limits (note that all amoA mRNA values were below detection). Treatments include control/no litter (CO-NL), control/Douglas-fir (CO-DF), no input/no litter (NI-NL), and no input/Douglas-fir (NI-DF). Bars represent treatment means (n = 3) with standard errors. Letters indicate significant treatment effects (P < 0.05).
No differences between soil types or litter additions were observed in 28S gene copy numbers. 28S gene copy numbers tended to be lower at day 151 than at day 3, but this was also not significant (P = 0.1). Gene copy numbers were an order of magnitude lower in incubated soils than in extractions of fresh soil, however (Fig. 1; P < 0.001). Gene copy numbers averaged 1.8 × 107 copies g−1 soil, and RNA copy numbers were significantly lower, with an average of 4.2 × 105 copies g−1 soil (Fig. 1). The 28S RNA copy numbers were significantly lower in CO-NL at day 3 than in all other treatments, but other treatments were similar to each other.
Soluble mineral N was highest in NI-NL treatments at day 151 compared to that in all treatments (Table 2); this was mostly due to high concentrations of NO3− (data not shown). These data suggest the presence of an ammonia-oxidizing population that was confirmed with qPCR. AOA amoA gene copy numbers averaged 3.3 × 108 copies g−1 soil, significantly higher than AOB amoA gene copy numbers (8.0 × 103 copies g−1 soil). AOB amoA gene copy numbers did not differ among incubations or sampling time (P = 0.2). AOB amoA transcripts could be detected in some extracts, but amplification was not consistent in the assay. AOA amoA gene copy numbers were more abundant in NI than in CO soils (2.5 × 108 copies g−1 soil and 1.5 × 107 copies g−1 soil, respectively; P < 0.001). AOA amoA gene copy numbers were also higher at day 151 (P = 0.001) than at day 3. Similar to AOB amoA transcripts, AOA amoA transcripts did not consistently amplify.
Bacterial and fungal community composition.
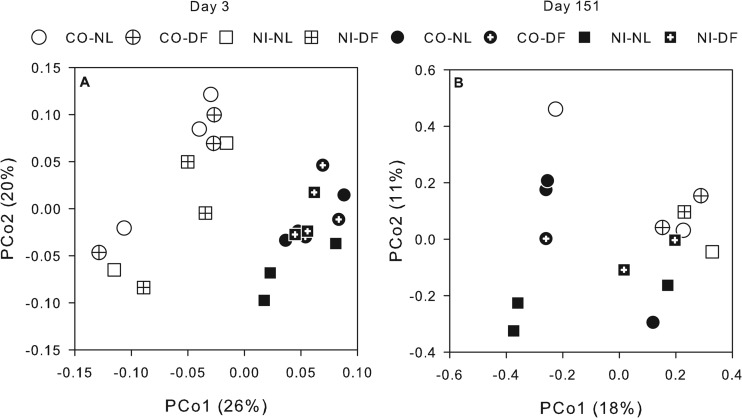
Bacterial community compositions based on 16S rRNA were significantly different between day 3 and day 151 for all treatments (Fig. 2; MRPP, P < 0.001). Each sample was represented by an average of 5,240 sequences or ∼15,000 sequences per treatment. Day 3 samples did not vary based on soil or litter addition, but at day 151, CO and NI bacterial composition tended to be different (MRPP, P = 0.06). Pairwise comparisons revealed that CO-DF significantly differed from NI-NL at day 151 (Fig. 2; MRPP, P = 0.03). Jackknife analysis resulted in little variation within each sample, suggesting that these patterns were robust (data not shown).
Fig 2.
Unifrac-weighted principal coordinate analysis (PCoA) ordinations of the 16S (A) and 28S (B) rRNA genes. Each point represents the average distance for an individual incubation. Percentages are the amount of data explained along each axis.
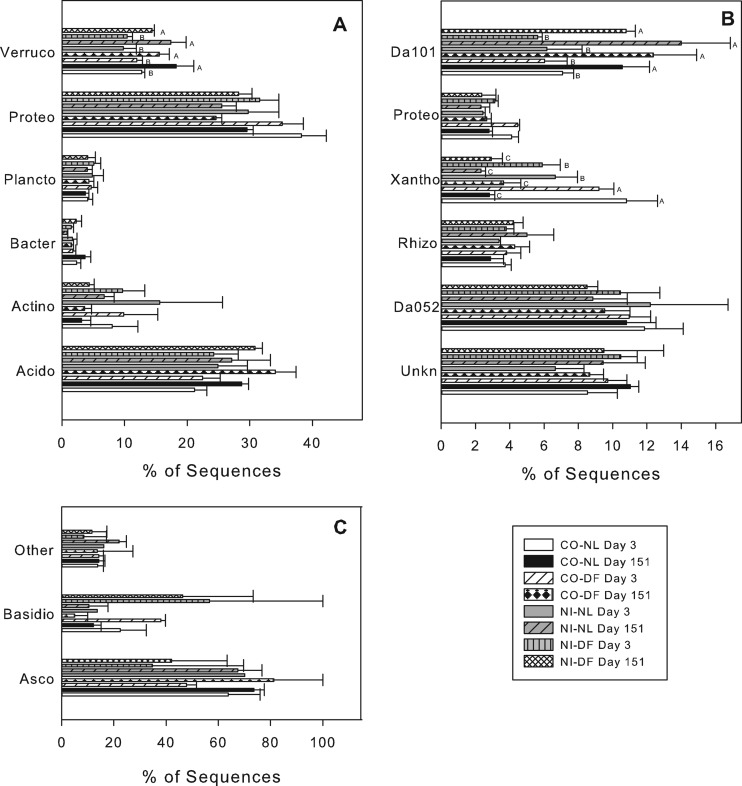
At a ≥99% sequence similarity, 483 OTUs were identified across all treatments, with 85 OTUs found only once and 337 occurring at least twice within replicate samples. When the 337 OTUs were each analyzed for treatment effects, 68 OTUs varied between day 3 and day 151. These OTUs included putatively identified Chthoniobacterales Da101 and an uncultured Xanthomonadales (Fig. 3B). Indicator species analysis identified ∼50 OTUs that differed between CO and NI soils at day 151, most of which were present in both soils but varied in percent abundance and matched those identified by univariate analysis. Indicator species analysis could not be used to identify OTUs varying between plot or litter type, because they were not statistically different overall. Univariate analysis revealed 15 OTUs that varied between plot, and only six varied between litter treatments (data not shown).
Fig 3.
Percent of sequencing matching putatively identified OTUs. (A) The six most abundant bacteria phyla: Acidobacteria (Acido), Actinobacteria (Actino), Bacteroidetes (Bacto), Planctomycetes (Plancto), Proteobacteria (Proteo), Verrucomicrobia (Verruco); (B) the six most abundant bacterial OTUs at 99% sequence similarity: unknown phyla (Unkn), Acidobacteria Da052 (Da052), Rhizobiales unknown family (Rhiz), Xanthomonadales (Xantho), Proteobacteria unknown class (Proteo), Chthoniobacterales Da101 (Da101); (C) the distribution of Ascomycota (Asco), Basidiomycota (Basidio), and other fungi. Treatments include control/no litter (CO-NL), control/Douglas-fir (CO-DF), no input/no litter (NI-NL), and no input/Douglas-fir (NI-DF). Bars represent treatment means (n = 3) with standard errors. Letters indicate significant treatment effects.
Acidobacteria was the most common phylum across all treatments; percent abundance did not vary among treatments (Fig. 3A). There were 35 OTUs putatively identified within the Acidobacteria, representing groups 1, 3, and 4. The most common OTU was Acidobacteria Da052 (Fig. 3B).
The phylum Proteobacteria was the second most abundant across all treatments and at the phylum level did not vary among treatments. No significant treatment effects were present at the class or order level. The most abundant OTUs were matched to Alphaproteobacteria, with a number of OTUs putatively identified as Rhizobiales, including the OTU shown in Fig. 3B. Eight OTUs were putatively identified as the Gammaproteobacteria Xanthomonadales. The most abundant Xanthomonadales OTU was more abundant at day 3 than at day 151 (Fig. 3B; P < 0.001). At day 3, the same OTU was more abundant in CO than in NI soils (P = 0.009).
Of the six most abundant bacterial phyla, only Verrucomicrobia differed among treatments (Fig. 3A). The Verrucomicrobia were more abundant at day 151 (P = 0.006). The most abundant Verrucomicrobia OTU, putatively identified as Chthoniobacterales Da101, was the only Verrucomicrobia OTU that significantly differed, increasing from day 3 to day 151 (P < 0.001).
An average of 132 fungal 28S rRNA gene sequences were analyzed per incubation, with 10 of the samples having too few rRNA sequences for further analysis. Fewer sequences and a higher variability measured by jackknife analysis (data not shown) made it difficult to observe consistent patterns. The fungal composition did, however, significantly differ between day 3 and day 151 (Fig. 2B; MRPP, P = 0.03). All sequences matched fungi, but targeting the D1 and D2 regions of the 28S rRNA limited the putative sequencing identification to the phylum level. Putatively identified Ascomycota were most abundant in all treatments (Fig. 3C), with Basidiomycota OTUs comprising most of the other sequences (Fig. 3C). No significant differences were seen in the phylum abundances among treatments.
DISCUSSION
Our original hypothesis that following 12 years of no above- or below-ground inputs from plants, active communities in CO and NI soils would differ, but when provided with new inputs, active communities in CO and NI soils would be more similar after a 5-month incubation, was not supported. Litter addition had no observed effect on the quantity or composition of the bacterial or fungal community, and in fact CO and NI soils were surprisingly similar regardless of additions. Brant et al. (8) did not observe differences in total microbial biomass between CO and NI based on phospholipid fatty acid (PLFA) analysis (Table 1). Microbial C and N as determined by chloroform fumigation agreed with these earlier findings (Table 2). Even after 12 years, CO and NI soils support similar microbial biomass.
There are several possible explanations for these findings: (i) the high concentration of C stored in the soils, (ii) the low concentration of fresh C input added to incubations, (iii) preleaching of litter to remove the soluble fraction, and (iv) standard soil handling procedures.
This old-growth coniferous soil ecosystem, with andic soils, may have an increased capacity to store C compared to soils of other mineralogy. Control soils contained 56.4 g C kg−1 soil, and after 12 years, NI soils still contained 40.4 g C kg−1 soil (27). The large reserve of soil C in NI soils may continue to sustain an active decomposer community. We are currently following up with similar experiments in soils that differ in C content to understand how C storage may impact microbial persistence.
To mimic double the rate of yearly input, 1.25 mg C kg−1 soil was added in the form of preleached litter. These additions did not substantially change the size of the soluble C pool (Table 2) and presumably by removing the soluble fraction did not stimulate a community that would take advantage of readily available, small molecular weight molecules. It is worth noting, however, that DF additions did result in higher minimal bactericidal concentrations (MBC) at day 3 in both soil types, but in both cases MBC had significantly decreased at day 151 (Table 2).
Soils used in incubations were collected and handled in accordance to standard soil handling procedures at LTER locations (13). These guidelines suggest preincubating soils at constant moisture to stabilize the soil microbial biomass. Peterson and Klug (28) examined the effects of soil handling (sieving, incubation, and temperature) on microbial communities using PLFAs. Sieving and incubation resulted in limited changes in PLFA profiles, but a decrease in the fungal marker 18:2ω6,9 was observed. Similarly, 28S rRNA gene copy numbers significantly decreased in incubated soils at day 3 compared to freshly sieved soils (Fig. 1). The mechanism for decreased 28S rRNA gene copies during preincubation is not known. Forest fungal populations, dominated by ectomycorrhizal (EcM) fungi, are sensitive to changes in plant inputs (29, 30), and so simply removing the CO soils could have resulted in this decrease. Decreases were also observed in NI soils, however, so perhaps sieving destroyed hyphae. Additional soil handling may have resulted in increased competition between bacteria and fungi (31). Therefore, it remains unclear if the preincubation created artifacts that masked the effects of litter additions on CO and NI soils, but even on fresh samples CO and NI soils did not vary in 16S or 28S rRNA gene copy numbers.
Consistently low rRNA copy numbers and difficulty in amplifying 28S rRNA for sequencing suggest that few fungi were active during the incubation. Few studies have targeted soil rRNA of the fungi (32), and fewer still have included sequencing to determine the identity of OTUs (7). Given the important roles of EcM fungi (belonging primarily to the Basidiomycota), we expected to observe higher proportions of these fungi in the CO soils, but the abundance of Basidiomycota sequences did not vary among treatments. The lack of relative abundance differences, within the fungi and most of the bacteria, may be due to the methodological constraints of 454 sequencing (33). Recent studies have observed that 454 sequencing data are not quantitative (33) and that low sampling depth may result in high within-replicate variability (34). Variability in the abundance of fungal sequences was higher than bacterial sequences, and sequencing depth was much lower.
Forty percent of bacterial sequences matched one of six OTU groupings (Fig. 3B). The dominance of so few organisms in the rRNA sequence library was somewhat surprising, given previous estimates of diversity in soils from these forest types using DNA-based clone libraries (17). Baldrian et al. (7) reported decreased diversity and evenness in rRNA compared to those in rRNA genes in a forest soil; this observation could account for the patterns observed in H. J. Andrews soils. These comparisons are also consistent with the notion of a small subpopulation of active microorganisms (1, 6).
Bacterial composition did not vary at day 3; by day 151, active bacteria in NI-NL soils were significantly different from other treatments. No clear patterns in bacterial sequence abundance emerged that were consistent with the total community analysis. Sequences were analyzed both with (Fig. 2) and without (data not shown) taxonomic weighting. No differences were detected in the unweighted analysis, similar to the observations of Lemos et al. (35). These results point to the potential importance of taxonomic relatedness to fulfill similar ecological niches (6).
Verrucomicrobia was the only phylum that significantly differed among treatments, decreasing from day 3 to day 151 (Fig. 3A). This was due primarily to differences in Chthoniobacterales Da101 (Fig. 3B). Chthoniobacterales is one of three described orders in the phylum Verrucomicrobia. Most members of the order appear to be soil-inhabiting bacteria, but some are endosymbionts of nematodes (36). The type strain Chthoniobacter flavus is a chemoheterotroph capable of growing on a variety of polymeric substances, at pH 4 to 7, and has a low growth rate, requiring 3 months between culture transfers (36).
The representative sequences Chthoniobacter Da101 and Acidobacteria Da052 were both recovered as rRNA from Dutch pasture soils (37, 38). Although the ubiquity of members of the Acidobacteria and Verrucomicrobia across soils is well known, the high similarity of sequences in our study to those also recovered during rRNA analysis in Dutch soils may point to a more narrowly defined collection of ubiquitous soil bacteria that are active most of the time. Like Chthoniobacter, most cultured Acidobacteria are slow-growing aerobic chemoheterotrophs (39, 40). A meta-analysis of several DNA-based studies also pointed to higher relative abundance of Verrucomicrobia and Acidobacteria in bulk soils than in rhizosphere soils (41).
Although DNA clone libraries at a nearby site identified several OTUs putatively matched to Bradyrhizobium (member of the Alphaproteobacteria) (17, 42), current sequence libraries contained <10 sequences matching to this genus. These results highlight the need to further investigate microbial communities based on their active or recently active members. The OTU putatively identified as an uncultured Xanthomonadales was more abundant in CO than in NI soils at day 3 and decreased in all treatments at day 151. Although the putatively identified sequences matched an uncultured organism, other members of Xanthomonadales are well known plant pathogens infecting conifers, among other plant types (43).
Although few differences were observed between the general bacterial and fungal communities, AOA data suggest that the ability to persist and fulfill defined ecological niches may be highly variable between taxa. AOA abundance was highest in NI soils (Fig. 1) but was several orders of magnitude higher than AOBs in both soil types. In a previous study, AOAs were not detected at a nearby location (20), but this result could be due to the use in the previous study of an early AOA primer pair that has since been redesigned based on a quickly expanding sequence collection (22). More research is needed to understand the role of AOAs in forest soils. The high abundance of AOA gene copies in NI soils was consistent with studies that have shown increased AOA populations in microcosms containing low NH4+ (≤20 mg NH4+-N kg−1 soil) (44). For comparison, NI and CO soils averaged 0.9 mg NH4+-N kg−1 soil. Although mRNA could not be detected, the increase in gene abundance suggests that AOAs were reproducing under these conditions and resulted in increased mineral N (Table 2). No attempt was made in this study to remove rRNA or use amoA primers during reverse transcription, strategies that may enrich for transcript. Griffiths et al. (45) reported higher AOA abundance in fallow versus cropped soils. These trends suggest a plant-microbe interaction that warrants further investigation.
The legacy of plant inputs that contribute to high soil OM in this ecosystem continued to shape the active microbial community even a decade after alteration. In spite of 12 years of soils not receiving plant inputs, the membership of the soil microbial community had not changed, and similar populations of microorganisms were active in both CO and NI soils. It will be important to monitor these sites in the coming decades to observe if CO and NI soils eventually begin to diverge in microbial composition. This speaks to the importance of long-term study plots such as the DIRT experiment to understand microbial dynamics. Periodic microbial characterization of soils in the DIRT experiment in the future will shed light on the extent of microbial persistence.
ACKNOWLEDGMENTS
We thank the staffs of the H. J. Andrews Experimental Forest, the Willamette National Forest, and the LTER for support of the DIRT plots. We also thank Peter Bottomley for discussions concerning experimental design.
The H. J. Andrews LTER provides the faculty SEED grant that funded this work. Elizabeth Brewer was supported by an NSF IGERT fellowship and NSF DEB-0616629.
Footnotes
Published ahead of print 21 December 2012
REFERENCES
- 1. Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9:119–130 [DOI] [PubMed] [Google Scholar]
- 2. Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. 2012. Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl. Environ. Microbiol. 78:3221–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. 2012. The Western English Channel contains a persistent microbial seed bank. ISME J. 6:1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 107:5881–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Segev E, Smith Y, Ben-Yehuda S. 2012. RNA dynamics in aging bacterial spores. Cell 148:139–149 [DOI] [PubMed] [Google Scholar]
- 6. DeAngelis KM, Firestone MK. 2012. Phylogenetic clustering of soil microbial communities by 16S rRNA but not 16S rRNA genes. Appl. Environ. Microbiol. 78:2459–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baldrian P, Kolarik M, Stursova M, Kopecky J, Valaskova V, Vetrovsky T, Zifcakova L, Snajdr J, Ridl J, Vlcek C, Voriskova J. 2012. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 6:248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brant JB, Myrold DD, Sulzman EW. 2006. Root controls on soil microbial community structure in forest soils. Oecologia 148:650–659 [DOI] [PubMed] [Google Scholar]
- 9. Kageyama SA. 2005. Effects of vegetation and disturbance on fungal communities in the western Cascades of Oregon. Ph.D. dissertation Oregon State University, Corvallis, OR [Google Scholar]
- 10. Nadelhoffer KJ, Boone RD, Bowden RD, Canary J, Kaye J, Lajtha K, McDowell W, Micks P, Ricca A. 2005. The DIRT experiment: litter and root influences on forest soil organic matter stocks and function. In Foster DR, Aber JD. (ed), Forests in time: the environmental consequences of 1,000 years of change in New England. Yale University Press, New Haven, CT [Google Scholar]
- 11. Lajtha K, Crow SE, Yano Y, Kaushal SS, Sulzman EW, Sollins P, Spears JDH. 2005. Detrital controls on soil solution N and dissolved organic matter in soils: a field experiment. Biogeochemistry 76:261–281 [Google Scholar]
- 12. Dixon JJ. 2003. Applying GIS to soil geomorphologic landscape mapping in the Lookout Creek valley, Western Cascades, Oregon. Oregon State University, Corvallis, OR [Google Scholar]
- 13. Paul EA, Harris D, Klug MJ, Ruess RW. 1999. The determination of microbial biomass, p 291–317 In Robertson GP, Coleman DC. (ed), Standard soil methods for long-term ecological research (long-term ecological research network series). Oxford University Press, New York, NY [Google Scholar]
- 14. Holub SM, Lajtha K, Spears JDH, Tóth JA, Crow SE, Caldwell BA, Papp M, Nagy PT. 2005. Organic matter manipulations have little effect on gross and net nitrogen transformations in two temperate forest mineral soils in the U.S.A. and central Europe. Forest Ecol. Manag. 214:320–330 [Google Scholar]
- 15. Joergensen RG. 1996. The fumigation-extraction method to estimate soil microbial biomass: calibration of the Kec value. Soil Biol. Biochem. 28:25–31 [Google Scholar]
- 16. Brookes PC, Landman A, Pruden G, Jenkinson DS. 1985. Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 17:837–842 [Google Scholar]
- 17. Yarwood SA, Bottomley PJ, Myrold DD. 2010. Soil microbial communities associated with Douglas-fir and red alder stands at high- and low-productivity forest sites in Oregon, U.S.A. Microb. Ecol. 60:606–617 [DOI] [PubMed] [Google Scholar]
- 18. Fierer N, Jackson JA, Vilgalys R, Jackson RB. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. 2010. Soil microbial community responses to multiple experimental climate change drivers. Appl. Environ. Microbiol. 76:999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boyle-Yarwood SA, Bottomley PJ, Myrold DD. 2008. Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ. Microbiol. 10:2956–2965 [DOI] [PubMed] [Google Scholar]
- 21. Zeglin LH, Taylor AE, Myrold DD, Bottomley PJ. 2011. Bacterial and archaeal amoA gene distribution covaries with soil nitrification properties across a range of land uses. Environ. Microbiol. Reports. 3:717–726 [DOI] [PubMed] [Google Scholar]
- 22. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S.A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brewer EA. 2010. Response of soil microbial communities and nitrogen cycling processes to changes in vegetation inputs. Oregon State University, Corvallis, OR [Google Scholar]
- 28. Peterson SO, Klug MJ. 1994. Effects of sieving, storage, and incubation temperature on the phospholipid fatty acid profile of a soil microbial community. Appl. Environ. Microbiol. 60:2421–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yarwood SA, Myrold DD, Högberg MN. 2009. Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microbiol. Ecol. 70:151–162 [DOI] [PubMed] [Google Scholar]
- 30. Högberg MN, Högberg P, Myrold DD. 2007. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia 150:590–601 [DOI] [PubMed] [Google Scholar]
- 31. Boyle SA, Yarwood RR, Bottomley PJ, Myrold DD. 2008. Bacterial and fungal contributions to soil nitrogen cycling under Douglas fir and red alder at two sites in Oregon. Soil Biol. Biochem. 40:443–451 [Google Scholar]
- 32. Anderson IC, Parkin PI. 2007. Detection of active soil fungi by RT-PCR amplification of precursor rRNA molecules. J. Microbiol. Meth. 68:248–253 [DOI] [PubMed] [Google Scholar]
- 33. Zhou J, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, Xie J, Van Nostrand JD, He Z, Yang Y. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 5:1303–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pinto AJ, Raskin L. 2012. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7:e43093 doi:10.1371/journal.pone.0043093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lemos LN, Fulthorpe RR, Roesch LF. 2012. Low sequencing efforts bias analyses of shared taxa in microbial communities. Folia Microbiol. (Praha) 57:409–413 [DOI] [PubMed] [Google Scholar]
- 36. Sangwan P, Chen X, Hugenholtz P, Janssen PH. 2004. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl. Environ. Microbiol. 70:5875–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Felske A, Akkermans ADL. 1998. Prominent occurence of ribosomes from an uncultured bacterium of the Verrucomicrobiales cluster in grassland soils. Lett. Appl. Microbiol. 26:219–223 [DOI] [PubMed] [Google Scholar]
- 38. Felske A, de Vos WM, Akkermans ADL. 2000. Spatial distribution of 16S rRNA levels from uncultured acidobacteria in soil. Lett. Appl. Microbiol. 31:118–122 [DOI] [PubMed] [Google Scholar]
- 39. Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eichorst SA, Kuske CR, Schmidt TM. 2011. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 77:586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364 [DOI] [PubMed] [Google Scholar]
- 42. Kluber LA, Smith JE, Myrold DD. 2011. Distinctive fungal and bacterial communities are associated with mats formed by ectomycorrhizal fungi. Soil Biol. Biochem. 43:1042–1050 [Google Scholar]
- 43. Lopez NI, Haedo AS, Mendez BS. 1999. Evaluation of Xanthomonas campestris survival in a soil microcosm system. Int. Microbiol. 2:111–114 [PubMed] [Google Scholar]
- 44. Allison SD, Martiny JB. 2008. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105(Suppl 1):11512–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]