Abstract
Microorganisms are abundant in the upper atmosphere, particularly downwind of arid regions, where winds can mobilize large amounts of topsoil and dust. However, the challenge of collecting samples from the upper atmosphere and reliance upon culture-based characterization methods have prevented a comprehensive understanding of globally dispersed airborne microbes. In spring 2011 at the Mt. Bachelor Observatory in North America (2.8 km above sea level), we captured enough microbial biomass in two transpacific air plumes to permit a microarray analysis using 16S rRNA genes. Thousands of distinct bacterial taxa spanning a wide range of phyla and surface environments were detected before, during, and after each Asian long-range transport event. Interestingly, the transpacific plumes delivered higher concentrations of taxa already in the background air (particularly Proteobacteria, Actinobacteria, and Firmicutes). While some bacterial families and a few marine archaea appeared for the first and only time during the plumes, the microbial community compositions were similar, despite the unique transport histories of the air masses. It seems plausible, when coupled with atmospheric modeling and chemical analysis, that microbial biogeography can be used to pinpoint the source of intercontinental dust plumes. Given the degree of richness measured in our study, the overall contribution of Asian aerosols to microbial species in North American air warrants additional investigation.
INTRODUCTION
Air samples from the lower troposphere contain a substantial microbial component that originates from a variety of marine/terrestrial sources (1–3). Airborne cells can spread genes to distant environments and even influence weather as cloud/ice condensation nuclei (1), but very little is known about microbial diversity and abundance at higher altitudes, where long-range atmospheric transport (i.e., global dispersal) is more efficient (4). Mountaintop observatories can provide access to the upper troposphere and lower stratosphere, making it feasible to capture enough biomass to employ modern molecular assays. Seasonal measurements at such platforms may help clarify the influence of microorganisms on patterns of climate (5), epidemiology (6–8), and biogeography (9).
We collected samples from the Mt. Bachelor Observatory (MBO), a research station 2.8 km above sea level on the summit of an extinct volcano in central Oregon (43.98°N, 121.7°W). In the springtime, windblown plumes of pollution, smoke, and dust from Asia routinely reach the field site after crossing the Pacific Ocean in 7 to 10 days (4, 10, 11). Annually, as much as 64 Tg of Asian aerosols is transported to North America (12). Recently (13), we described two major Asian long-range transport (ALRT) plumes with high concentrations of particulate matter (mostly dust, but also anthropogenic pollution) arriving at MBO. The first event began at 2:00 coordinated universal time (UTC) on 22 April 2011 and lasted 51 h; the second started at 12:00 UTC on 12 May 2011 and ended 80 h later (see Tables 1 and 2). Airborne bacterial concentrations were measured by quantitative PCR, and rRNA sequencing was used to identify cultured species. Average bacterial genomes ranged from 1 to 4 m−3 across the April episode and 2 to 7 m−3 across the May episode, assuming that intact cells (containing 2 to 8 fg DNA) were captured. Several Gram-positive bacterial isolates were identified using culture-based recovery methods, but since so few species can actually be cultivated (9, 13), our goal was to reexamine the air samples with a more comprehensive molecular tool testing the hypothesis that transpacific plumes deliver rich microbial populations to North America.
Table 1.
April plume sample manifest
| Sample | Date (mo/day/yr) and time (UTC) | Category | Avg aerosol level (μg m−3) | DNA concn (ng μl−1) | PCR yield (ng) | No. of OTUs detected |
|---|---|---|---|---|---|---|
| Abk142 | 4/21/2011 20:00 to 4/22/2011 08:00 | Background | 1.56 | 2.2 | 1,394 | 2,325 |
| Adu143 | 4/22/2011 08:00 to 4/22/2011 19:00 | Plume | 23.3 | 15.3 | 1,442 | 2,197 |
| Adu144 | 4/22/2011 19:00 to 4/23/2011 07:00 | Plume | 10.3 | 1.5 | 1,328 | 2,808 |
| Adu145 | 4/23/2011 07:00 to 4/23/2011 19:00 | Plume | 10.7 | 1.7 | 1,347 | 2,620 |
| Adu146 | 4/23/2011 19:00 to 4/24/2011 07:00 | Plume | 10.2 | 1.8 | 1,330 | 2,742 |
| Abk147 | 4/24/2011 07:00 to 4/24/2011 19:00 | Background | 0 | 2.0 | 991 | 2,114 |
Table 2.
May plume sample manifest
| Sample | Date (mo/day/yr) and time (UTC) | Category | Avg aerosol level (μg m−3) | DNA concn (ng μl−1) | PCR yield (ng) | No. of OTUs detected |
|---|---|---|---|---|---|---|
| Tbk175 | 5/12/2011 06:00 to 5/12/2011 19:00 | Background | 0.408 | 1.6 | 1,339 | 2,311 |
| Tdu176 | 5/12/2011 19:00 to 5/13/2011 08:00 | Plume | 9.52 | 0.5 | 1,331 | 2,155 |
| Tdu177 | 5/13/2011 08:00 to 5/13/2011 20:00 | Plume | 8.70 | 2.6 | 1,314 | 2,682 |
| Tdu178 | 5/13/2011 20:00 to 5/14/2011 08:00 | Plume | 7.82 | 1.4 | 1,115 | 2,864 |
| Tdu179 | 5/14/2011 08:00 to 5/14/2011 20:00 | Plume | 5.05 | 1.1 | 1,361 | 2,841 |
| Tdu180 | 5/14/2011 20:00 to 5/15/2011 08:00 | Plume | 5.56 | 1.2 | 1,309 | 2,656 |
| Tbk181 | 5/15/2011 08:00 to 5/15/2011 20:00 | Background | 1.99 | 1.3 | 1,165 | 2,516 |
| Tbk182 | 5/15/2011 20:00 to 5/16/2011 08:00 | Background | 0.161 | 6.7 | 1,259 | 2,235 |
MATERIALS AND METHODS
Sample collection.
Microbes were collected on sterile polyethersulfone filters (pore size, 0.8 μm) connected to a previously described air-sampling device at MBO (13). Briefly, a high-volume pump pulled ∼0.5 m3 min−1 of air through individual filters over 12-h intervals, and then samples were removed from the device and stored at −80°C. During sampling periods, meteorological and atmospheric chemistry data were collected for aerosol elemental composition (e.g., ammonium sulfate [NH4SO4], soil, and trace metals), carbon monoxide (CO), ozone (O3), water (H2O) vapor, total gaseous mercury (THg), temperature, atmospheric pressure, wind speed, and direction. Details of MBO instruments, calibrations, and element concentration calculations have been published elsewhere (4, 10, 13).
Atmospheric modeling.
We calculated 240-h backward trajectories initialized from MBO during peak aerosol periods to establish the long-range transport history of arriving air masses. Trajectories were calculated with the Hybrid Single-Particle Lagrangian Integrated Trajectory (HYSPLIT) model (14), version 4, which uses global meteorological data from the Global Data Assimilation System archive. The data set has a time resolution of 3 h, a spatial resolution of 1° latitude by 1° longitude, and a vertical resolution of 23 pressure surfaces between 1,000 and 20 hPa. Trajectories were run at multiple heights (1,200, 1,500, and 1,800 m above ground level) surrounding the summit of MBO (4).
Transoceanic aerosol plumes were also modeled with the Navy Research Laboratory Aerosol Analysis and Prediction System (NAAPS; http://www.nrlmry.navy.mil/aerosol/) to compare them with HYSPLIT, version 4, long-range transport patterns. The NAAPS model produces a total aerosol forecast at an optical depth of 550 nm that includes sulfate, smoke, dust, and sea salt mass concentrations. We examined air samples from the periods from 15 to 25 April 2011 and from 7 to 17 May 2011.
DNA extraction and PCR amplification.
To extract DNA, samples were processed with MO BIO PowerWater (PW) kits (product 14900-100-NF; MO BIO Laboratories, Inc., Carlsbad, CA). Each quartered filter was placed inside a 5-ml PowerWater bead tube containing 3 ml of solution PW1, and the tube was incubated for 30 min at 65°C, followed by a 15-min vortex (product 12-812; Fisher Vortex Genie 2; Fisher Scientific). All subsequent extraction steps followed the PowerWater manufacturer's guidelines, with solution volumes being proportionally adjusted to match the boosted volume of solution PW1. A centrifuge (product 5804; Eppendorf, Hauppauge, NY) and PowerVac manifold (product 11991; MO BIO Laboratories, Inc.) were used, along with a wash of 800 μl of 100% ethanol, prior to the addition of solution PW4. The final sample elution volume was 40 μl in buffer EB (Qiagen). DNA samples were quantified using NanoDrop spectrophotometry (ND-1000; Thermo Scientific, Wilmington, DE) and PicoGreen (Life Technologies, Grand Island, NY) methods. Next, 16S rRNA gene PCR amplification was performed: bacterial 16S rRNA genes were amplified using forward primer 27F.1 (5′-AGRGTTTGATCMTGGCTCAG-3′) and reverse primer 1492R.jgi (5′-GGTTACCTTGTTACGACTT-3′); archaeal 16S rRNA genes were amplified using forward primer 4Fa (5′-TCCGGTTGATCCTGCCRG-3′) and reverse primer 1492R.jgi (15). Thermocycling program conditions were 95°C for 3 min, followed by 35 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 2 min and an extension at 72°C for 10 min, before holding at 4°C. Amplified products were concentrated and quantified by electrophoresis using an Agilent 2100 bioanalyzer.
Microarray design, experiment, and scoring.
PhyloChip microarrays are commercially available through Second Genome, Inc. (San Bruno, CA), and have been extensively described elsewhere (15–18). Amplified and purified DNA products were fragmented and biotin labeled. Each reaction mixture was injected into the hybridization chamber of version G3 of a PhyloChip array (15). A PhyloChip control mix was included for scaling and normalization. Oligonucleotide targets and probes representing publicly available bacterial and archaeal 16S rRNA genes were synthesized by photolithography on a PhyloChip version G3 array. Other relevant technical details, quality/processing controls, and reproducibility tests for the PhyloChip version G3 array have been published (15). Operational taxonomic units (OTUs) were defined as a group of highly similar 16S rRNA gene sequences (<0.5% divergence). In total, there were 59,959 potential clusters spanning 2 domains (archaea and bacteria), 147 phyla, 1,123 classes, and 1,219 orders. Each OTU was assigned to 1 of 1,464 families (19). Experimentally, hybridization took place for 16 h in an oven at 48°C and 60 rpm before the PhyloChip array was washed, stained, and scanned using a GeneChip 3000 7G scanner (Affymetrix, Santa Clara, CA). Fluorescence intensity was captured using Affymetrix software (GeneChip Microarray Analysis Suite) and calculated using preestablished formulas (17).
Statistical analysis.
Hybridization scores (HybScores) were calculated for each OTU using probe fluorescent intensities. The relative abundance of taxa was measured by comparing intensities against the intensity of the PhyloChip control mix after subtracting from background values. Multivariate statistical analyses were performed with PhyCA-Stats software (Second Genome, Inc.), and algorithms compared the relationship between probes and taxa. Maximum and minimum HybScores were discarded before averaging and scaling of the values so that a HybScore change of 1,000 represented a doubling in probe fluorescent intensity. Data were reduced to a series of predefined filters (15) based on significant taxon patterns, established through either parametric Welch tests or Adonis tests to generate P values. Presence and absence (i.e., incidence) data were transformed into binary metrics, which used the UniFrac distance method for examining changes in communities (20, 21). Abundance metrics used HybScore data for OTUs and a weighted UniFrac method for measuring sample-to-sample distance.
Microarray data accession number.
The entire microarray data set with OTU annotations can be accessed at the Greengenes database (http://greengenes.secondgenome.com/downloads/phylochip_datasets).
RESULTS AND DISCUSSION
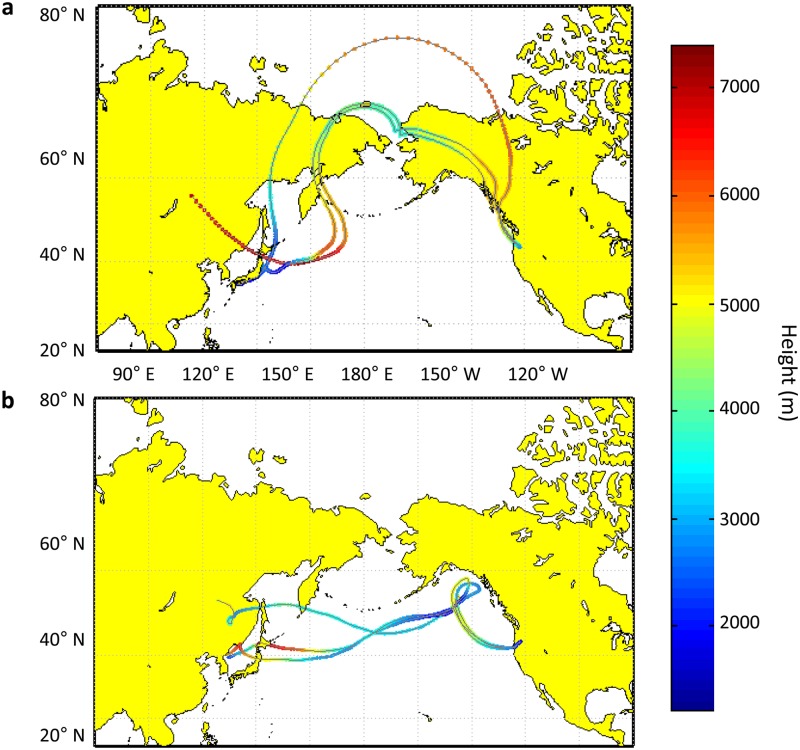
First, it was essential to establish possible source regions and transport histories for the events of interest using meteorological and chemical data. Kinematic back trajectories were modeled for each episode with HYSPLIT, revealing key differences in origin, mixing, and vertical transport. Ten-day back trajectories for the April event began near China, the Korean Peninsula, or Japan and showed the air mass rapidly lifting to an altitude of ∼8 km (Fig. 1a). Low humidity and high O3 indicate that the pollution plume may have mixed with air from the upper troposphere/lower stratosphere during transport (13). Airborne THg and CO can be used as tracers for ALRT (4), and enhancement ratios between the species (see Fig. S1 in the supplemental material) in the April plume were consistent with previously identified Asian pollution events at the collection site (10). Soil and NH4SO4 concentrations were also correlated (see Fig. S2 in the supplemental material), indicating a similar origin and an airborne time sufficient for homogenization (22). The NAAPS model provided another check for ALRT by depicting the transpacific migration of airborne sulfate, dust, smoke, and sea salt at a total aerosol optical depth of 0.1 to 0.2 (see Movie S1 in the supplemental material). For the May episode, 10-day HYSPLIT back trajectories show an air mass originating over the Pacific Ocean and mixing into the marine boundary layer (Fig. 1b). According to the model, the air was swept through a storm loop off the coast of Alaska before ascending into the free troposphere and MBO. Depleted levels of THg/CO (see Fig. S1 in the supplemental material) and NH4SO4/soil (see Fig. S3 in the supplemental material) align with the possibility of a boundary layer excursion. Compared to what was observed in the April episode, the May NAAPS data (see Movie S2 in the supplemental material) show denser emissions in Asia and a larger transpacific plume.
Fig 1.
Ten-day kinematic back trajectories for air arriving at MBO (43.98°N, 121.7°W), depicting transpacific transport patterns from Asia (left) to North America (right). Back trajectories for both episodes were calculated using the HYSPLIT model at the peak plume hour (highest aerosol, CO, and O3 levels). The colored scale bar represents trajectory height (m above sea level). (a) Trajectories ending 22 April 2011 at 12:00 UTC, showing a rapid ascent to the upper troposphere/lower stratosphere prior to sampling; (b) trajectories ending 13 May 2011 at 00:00 UTC, showing possible marine boundary layer mixing and a storm loop off the Alaskan coast prior to sampling.
Air samples (spanning 12-h intervals) from before, during, and after ALRT events were analyzed for microbial richness and abundance using a PhyloChip 16S rRNA microarray (3, 15–18). Tables 1 and 2 provide the aerosol level, microbial concentration (ng DNA), and community richness (number of OTUs) for each sampling interval and category (plume or background). Overall, 2,808 bacterial OTUs were detected at the peak of the April episode (694 above background levels); 2,864 bacterial OTUs were detected at the peak of the May episode (629 above background levels). On the basis of incidence data alone, bacterial richness was highest during plumes and lowest before and after plumes (i.e., from background air). Bacterial taxa spanned a broad range of phyla, including Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes. Six species of archaea were also measured in 8 samples (see Table S1 in the supplemental material).
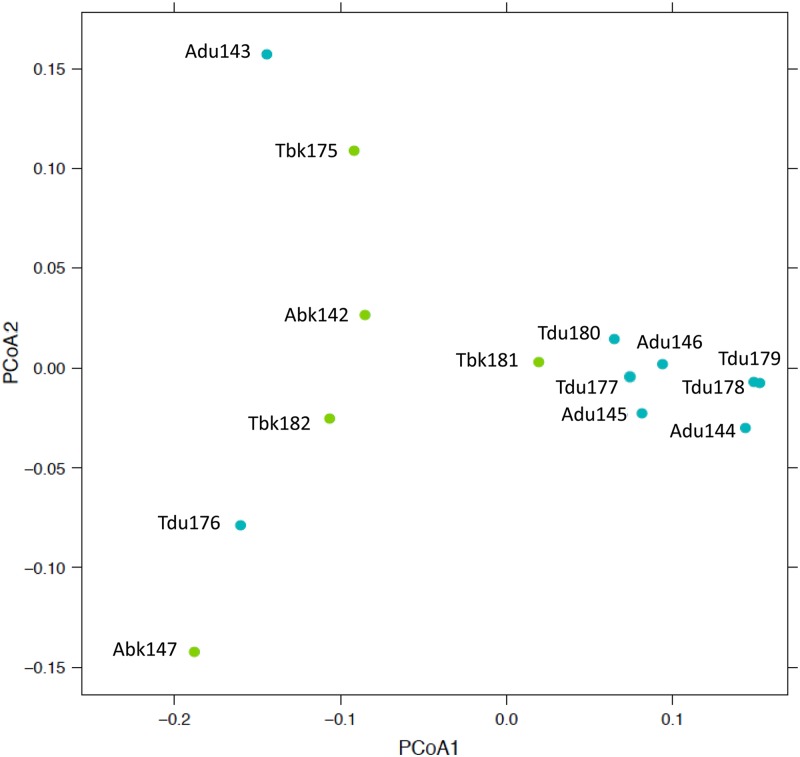
Principle coordinate analysis (PCoA) was used to measure dissimilarity in ALRT plumes. Each point on the ordination plots represents an entire microbial community sample. Figure 2 assembled incidence data from 38,546 possible taxa and partitioned the samples into two distinct categories: plume and background. Significant clusters (P = 0.032) emerged with (i) April and May background samples and (ii) April and May plume samples, supporting the idea that ALRT events were more similar than different. Comparable clustering was observed (P = 0.044) in a follow-up analysis using abundance data from 2,514 possible taxa (see Fig. S4 in the supplemental material). Incidence data for only 86 selected taxa at peak plume sampling intervals (Adu143 to Adu145, Tdu177 to Tdu179) revealed a microbiome contrast between April and May plumes (see Fig. S5 in the supplemental material).
Fig 2.
Principle coordinate analysis of background samples (green) and plume samples (blue). Analysis was based on the unweighted UniFrac distance between samples from 38,546 possible taxa with incidence differences. For axis 1, 29% of the variation was explained; for axis 2, 9% of the variation was explained. The partitioning shows the similarity in community composition between plume samples (abbreviated du) and the similarity in community composition background samples (abbreviated bk), regardless of plume timing (April or May). Note that Adu143 and Tdu176 were transitional samples at the onset of a plume (i.e., a mixture of background and plume).
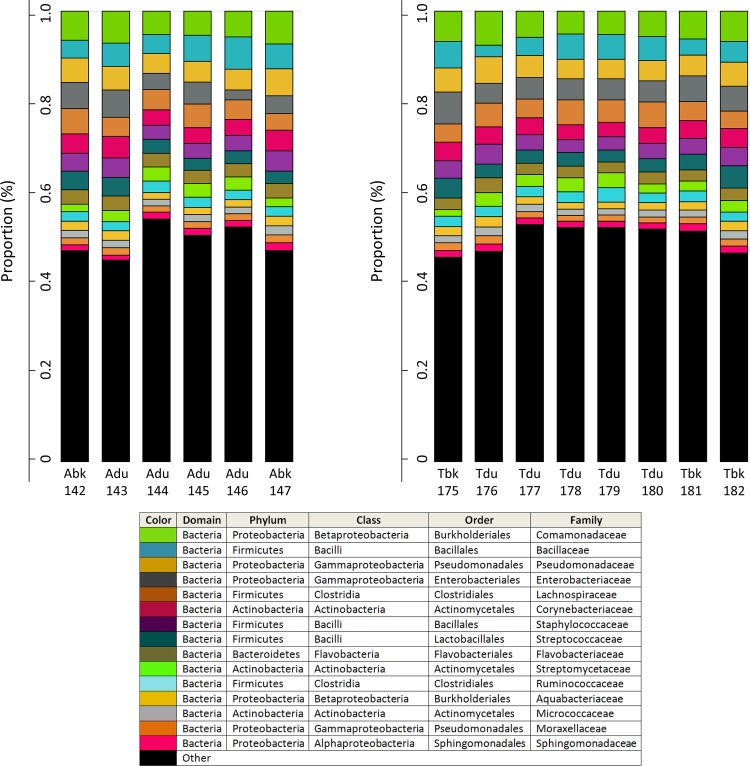
Figure 3 supports the idea that ALRT plumes delivered similar microbiota, despite unique transport histories, in April and May: although species richness levels varied between the events (Tables 1 and 2), the proportion of 15 common bacterial families was essentially parallel. An important point, however, is that while the relative abundance remained steady across ALRT plumes, the absolute abundance of 312 OTUs changed significantly (P < 0.001). To illustrate this observation, a circular tree (see Fig. S6 in the supplemental material) was constructed to display differentially abundant OTUs and their taxonomic relationship on the basis of 16S rRNA gene alignment. Welch test P values were used to reduce the number of significantly different families to 83, and the one OTU with the greatest abundance difference from each family was included in the tree. Heat-map values revealed increases within the class Clostridia and families Sphingomonadaceae, Rhodobacteraceae, and Isosphaeraceae relative to the combined means of baseline samples. Altogether, Proteobacteria (n = 29), Actinobacteria (n = 19), and Firmicutes (n = 19) totaled 80% of the phyla whose abundance increased during ALRT plumes. A higher level of Actinobacteria and Firmicutes is noteworthy because the families include many spore-forming and Gram-positive species capable of surviving extreme conditions associated with long-range upper atmospheric transport. Curiously, 4 out of 5 of the families that decreased in abundance during plumes were Proteobacteria from marine environments (including Alteromonadaceae, Vibrionaceae, and the OM60 family within the Oceanospirillales).
Fig 3.
Relative abundance of 15 common bacterial families across the April plume (left) and May plume (right). The size of each color block (assigned to families in the table below) represents the number of OTUs detected in the family relative to the total number of OTUs detected in that sample. For example, Bacillaceae OTUs accounted for 6.5% of the total OTUs detected in the first April sample (Abk142). Generally, family proportions remained constant across both episodes.
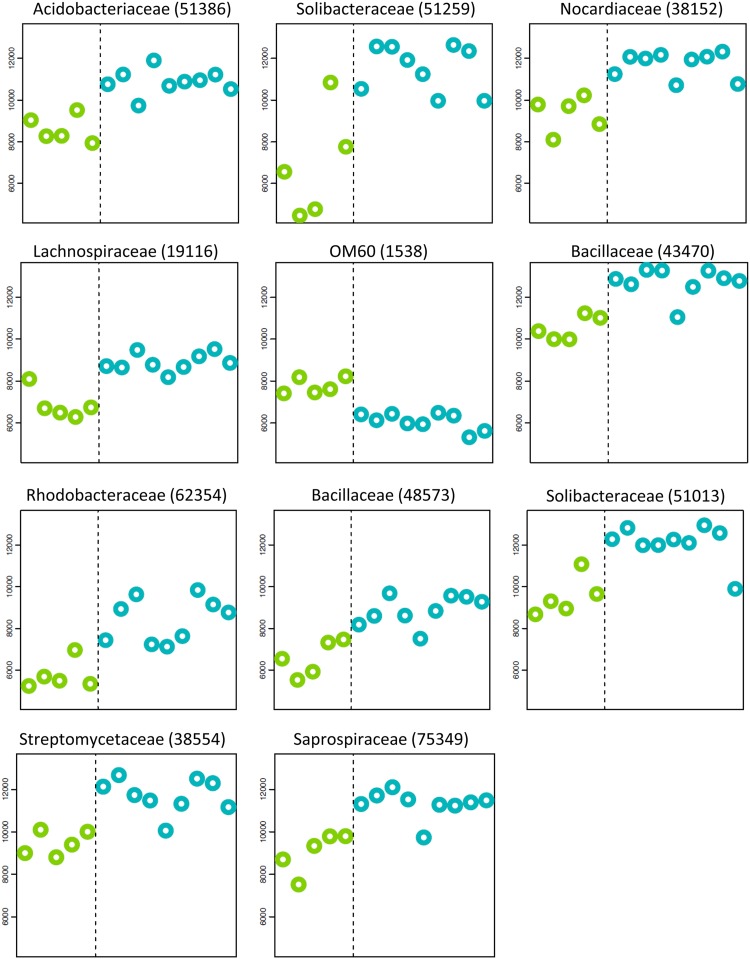
After establishing that ALRT plumes delivered higher concentrations of microbes already present in the North American background air, we focused on variations within specific taxa using prediction analysis for microarrays (PAM) (23). Figure 4 highlights taxa with possible Asian or oceanic origins, including isolates from a Chinese forest (OTU 51259), Dongping Lake sediments (OTU 51013), and marine microbial mats (OTU 75349). The alignment between taxon source regions and probable emission sites identified by atmospheric data was striking; however, annotations from the 16S rRNA sequence database can be inaccurate. Source verification of probe-detected taxa would require sample sequencing (which was outside the scope of our current study). Another possible agreement between atmospheric transport models and microbial biogeography was the detection of Aeropyrum spp. in the May episode. Finding marine archaea (24) in the free troposphere above central Oregon also supports microbial ALRT, but biogeography alone cannot be the only means of inferring distant provenance. Only after considering the location of our field site, prevailing wind direction, and long-range transport validation through a number of independent atmospheric data sets [including (i) HYSPLIT, version 4, kinematic back trajectories, (ii) aged aerosol data (i.e., NH4SO4 and soil), (iii) plume chemical composition (i.e., CO and THg), and (iv) the NAAPS model] could we be confident about the transoceanic origin of air samples.
Fig 4.
Significant abundance variations in specific taxa between background (green) and plume (blue) periods. Combined data for April and May events are shown. HybScores are on the y axis, and the sample order (from left to right) follows the order used in Tables 1 and 2 (e.g., Abk142 is the leftmost data point). Numbers in parentheses are OTU identification numbers.
Soon, it may be useful to think about microorganisms as air pollution (e.g., how aerosols were depicted in Movies S1 and S2 in the supplemental material, moving in plumes through a global background layer). Our main finding—that transpacific dust plumes deliver elevated levels of species already in the background air—suggests that microbes pool like other types of pollution over the Pacific Ocean. However, a transpacific monitoring network with sampling sites in eastern Asia and western North America is needed to establish an aerobiology data set comparable to that for the NAAPS model. Such an undertaking would require seasonal measurements from a variety of natural (desert dust, marine sea spray, etc.) and artificial (livestock feedlots, wastewater-treatment facilities, etc.) upwind emission sources (25), monitored, ideally, through a combination of ground- and aircraft-based platforms. Global sampling efforts using rRNA microarrays might consider employing the same commercial products to reduce false hybridizations and other sources of variation (17). Standardizing air collection techniques, DNA extraction, PCR amplification, and microarray protocols would be useful for comparisons between field sites. Even though microarrays offer improved sensitivity to microbial taxa (our first investigation of the same ALRT samples detected only 18 species of bacteria [13]), culture-based aerobiology data still have value: understanding what species remain viable after intercontinental atmospheric transport informs questions related to disease propagation.
Airborne microorganisms originate from the surface and must eventually return to it. Consequently, the atmosphere has generally been considered a conduit for life rather than a true ecosystem. However, our study revealed a microbial richness that rivals that of surface ecosystems and the presence of many phyla with adaptations for extended viability during atmospheric transport (e.g., spore-forming and Gram-positive bacteria). In addition, the potential for dynamic microbial interactions with the environment, such as in situ metabolism (26), the stimulation of cloud formation and precipitation (5), and selection pressures from UV radiation (27) all support the idea that the atmosphere might be considered an ecosystem in its own right. No matter how it is classified, as desertification injects more dust into the atmosphere (6, 12) and humans grow increasingly vulnerable to changing patterns of weather and disease, it will be important to monitor microbial populations on intercontinental winds.
Supplementary Material
ACKNOWLEDGMENTS
Research funding was provided by National Science Foundation (NSF) Integrative Graduate Education and Research Traineeship (IGERT) at the University of Washington (UW) Astrobiology Program, the National Geographic Society Waitt Grants Program (W177-11), the NASA Astrobiology Institute Director's Discretionary Fund, and the Virtual Planetary Lab at UW. NSF grant ATM-0724327 funded the MBO atmospheric chemistry measurements.
We thank J. Hee, B. Hicks, P. Ball, C. Higginbotham, T. Lomax, J. Baross, T. DeSantis, B. Lidz, C. Kellogg, A. Schuerger, T. Bradshaw, R. Wheeler, and the staff at Mt. Bachelor Ski Resort for support. We are also grateful to three anonymous reviewers for their time and constructive feedback.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03029-12.
REFERENCES
- 1. Burrows SM, Elbert W, Lawrence MG, Pöschl U. 2009. Bacteria in the global atmosphere. Part 1. Review and synthesis of literature data for different ecosystems. Atmos. Chem. Phys. 9:9263–9280 [Google Scholar]
- 2. Fahlgren C, Hagström A, Nilsson D, Zweifel UL. 2010. Annual variations in the diversity, viability, and origin of airborne bacteria. Appl. Environ. Microbiol. 76:3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brodie EL, DeSantis TZ, Moberg Parker JP, Zubietta IX, Piceno YM, Anderson GL. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weiss-Penzias P, Jaffe DA, Swartzendruber P, Dennison JB, Chand D, Hafner W, Prestbo E. 2006. Observations of Asian air pollution in the free troposphere at Mount Bachelor Observatory during the spring of 2004. J. Geophys. Res. 111:D10304 doi:10.1029/2005JD006522 [Google Scholar]
- 5. Christner BC, Morris CE, Foreman CM, Cai R, Sands DC. 2008. Ubiquity of biological nice nucleators in snowfall. Science 319:1214. [DOI] [PubMed] [Google Scholar]
- 6. Kellogg CA, Griffin DW. 2006. Aerobiology and the global transport of desert dust. Trends Ecol. Evol. 21:638–644 [DOI] [PubMed] [Google Scholar]
- 7. Brown JKM, Hovmøller MS. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537–541 [DOI] [PubMed] [Google Scholar]
- 8. Griffin DW. 2007. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 20:459–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Womack AM, Bohannan BJM, Green JL. 2010. Biodiversity and biogeography of the atmosphere. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:3645–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jaffe DA, Prestbo E, Swartzendruber P, Weiss-Penzias P, Kato S, Takami A, Hatakeyama S, Kajii Y. 2005. Export of atmospheric mercury from Asia. Atmos. Environ. 39:3029–3038 [Google Scholar]
- 11. Fischer EV, Hsu NC, Jaffe DA, Jeong MJ, Gong SL. 2009. A decade of dust: Asian dust and springtime aerosol load in the U.S. Pacific Northwest. Geophys. Res. Lett. 36:L03821 doi:10.1029/2008GL036467 [Google Scholar]
- 12. Yu H, Remer LA, Chin M, Bian H, Tan Q, Yuan T, Zhang Y. 2012. Aerosols from overseas rival domestic emissions over North America. Science 337:566–569 [DOI] [PubMed] [Google Scholar]
- 13. Smith DJ, Jaffe DA, Birmele MN, Griffin DW, Schuerger AC, Hee J, Roberts MS. 2012. Free tropospheric transport of microorganisms from Asia to North America. Microb. Ecol. doi:10.1007/s00248-012-0088-9 [DOI] [PubMed] [Google Scholar]
- 14. Draxler RR, Rolph GD. 2003. HYSPLIT (HYbrid Single-Particle Lagrangian Integrated Trajectory). NOAA Air Resources Laboratory READY website. NOAA Air Resources Laboratory, College Park, MD: http://www.arl.noaa.gov/ready/hysplit4.html [Google Scholar]
- 15. Hazen TC, Dubinsky EA, DeSantis TZ, Anderson GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zelma ML, Chakraborty R, Sonnethal EL, D'haeseleer P, Holman H-Y, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208 [DOI] [PubMed] [Google Scholar]
- 16. Wilson KH, Wilson WJ, Radosevich JL, DeSantis TZ, Viswanathan VS, Kuczmarski TA, Anderson GL. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeSantis TZ, Stone CE, Murray SR, Moberg JP, Anderson GL. 2005. Rapid quantification and taxonomic classification of environmental DNA from both prokaryotic and eukaryotic origins using a microarray. FEMS Microbiol. Lett. 245:271–278 [DOI] [PubMed] [Google Scholar]
- 18. DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Anderson GL. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371–383 [DOI] [PubMed] [Google Scholar]
- 19. Hugenholtz P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinf. 7:371 doi:10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clarke KR, Ainsworth M. 1993. A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92:205–219 [Google Scholar]
- 22. Jaffe D, McKendry I, Anderson T, Price H. 2003. Six “new” episodes of trans-Pacific transport of air pollutants. Atmos. Environ. 37:391–404 [Google Scholar]
- 23. Tibshirani R, Hastie T, Narasimhan B, Chu G. 2002. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U. S. A. 99:6567–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sako Y, Nomura N, Uchia A, Ishida Y, Morri H, Koga Y, Hoaki T, Maruyama T. 1996. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100°C. Int. J. Syst. Bacteriol. 46:1070–1077 [DOI] [PubMed] [Google Scholar]
- 25. Burrows SM, Butler T, Jöckel Tost PH, Kerkweg A, Pöschl U, Lawrence MG. 2009. Bacteria in the global atmosphere. Part 2. Modeling of emissions and transport between different ecosystems. Atmos. Chem. Phys. 9:9281–9297 [Google Scholar]
- 26. Sattler B, Puxbaum H, Psenner R. 2001. Bacterial growth in supercooled cloud droplets. Geophys. Res. Lett. 28:239–242 [Google Scholar]
- 27. Smith DJ, Griffin DW, McPeters RD, Ward PD, Schuerger AC. 2011. Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia 27:319–332 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.