Abstract
The diversity and phylogenetic significance of bacterial genes in the environment has been well studied, but comparatively little attention has been devoted to understanding the functional significance of different variations of the same metabolic gene that occur in the same environment. We analyzed the geographic distribution of 16S rRNA pyrosequences and soxB genes along a geochemical gradient in a terrestrial sulfidic spring to identify how different taxonomic variations of the soxB gene were naturally distributed within the spring outflow channel and to identify possible evidence for altered SoxB enzyme function in nature. Distinct compositional differences between bacteria that utilize their SoxB enzyme in the Paracoccus sulfide oxidation pathway (e.g., Bradyrhizobium, Paracoccus, and Rhodovulum) and bacteria that utilize their SoxB enzyme in the branched pathway (e.g., Chlorobium, Thiothrix, Thiobacillus, Halothiobacillus, and Thiomonas) were identified. Different variations of the soxB genes were present at different locations within the spring outflow channel in a manner that significantly corresponded to geochemical conditions. The distribution of the different soxB gene sequence variations suggests that the enzymes encoded by these genes are functionally different and could be optimized to specific geochemical conditions that define niche space for bacteria capable of oxidizing reduced sulfur compounds.
INTRODUCTION
Nearly all transformations within the sulfur cycle are controlled by biosphere processes. In environments with oxic-anoxic interfaces, sulfide is a ubiquitous electron donor that is generated primarily by sulfate-reducing microorganisms. The removal of sulfide through microbial oxidation is an important geochemical and ecosystem process (1–6); however, resolving the spatial, functional, and temporal relationships among bacteria that control the oxidative reactions of the sulfur cycle has been hampered by an overreliance on the 16S rRNA gene to serve as a proxy for microbial metabolism. Combining functional gene analysis with 16S rRNA phylogenies (7–13) can improve the resolution of potential microbially mediated geochemical reactions occurring in the environment. There are several enzymatic systems that have been associated with the oxidation of reduced sulfur compounds (14–16), with the sulfur oxidation (Sox) system being widespread among photosynthetic and nonphotosynthetic bacteria (14, 15, 17).
The Sox system is comprised of four enzyme complexes (SoxXA, SoxYZ, SoxB, and SoxCD) that are responsible for the oxidation of hydrogen sulfide, thiosulfate, elemental sulfur, and sulfite to sulfur intermediates or sulfate (14) in at least two pathways. The Paracoccus sulfur oxidation (PSO) pathway catalyzes the oxidation of thiosulfate to sulfate without the formation of sulfur intermediates using the SoxCD enzyme complex (18–21). The branched pathway, which lacks the SoxCD complex, catalyzes the oxidation of reduced sulfur compounds to intermediate sulfur compounds that are deposited as intra- or extracellular sulfur globules, most commonly as polysulfides (14–16, 22, 23).
The presence/absence of the soxB gene has been used as a marker for the Sox system in environmental bacteria and for phylogenetic studies of sulfur-oxidizing bacteria (10–12, 15, 24–26). Biochemical modeling and analysis of Sox enzymes has focused on a few laboratory bacterial strains (19, 27–30), and a mechanism by which the enzymes of the Sox system oxidize reduced sulfur substrates has been proposed based on spectrometry (31), modeling (32), and crystallization (29) of the Paracoccus pantotrophus SoxB and SoxYZ enzymes and crystallization of the SoxB enzyme from Thermus thermophilus (30). The mechanism is based on the substrate being thiosulfate, which binds to the SoxYZ enzyme (with the aid of the SoxXA enzyme) and remains bound to the SoxYZ enzyme when the sulfone sulfur is hydrolytically cleaved by the SoxB enzyme (30). Whether or not other reduced sulfur compounds can be oxidized in this way is unclear from the literature. However, reconstituted Sox systems of P. pantotrophus have been shown to oxidize different reduced sulfur substrates with different affinities (particularly hydrogen sulfide and sulfite) (19–12). Moreover, Ogawa et al. report that the Sox component of Chlorobium tepidum, which does not possess a SoxCD complex, could oxidize sulfide and sulfite (33). Mutant strains of C. tepidum devoid of a soxB gene showed partial growth on media containing only sulfide relative to the wild type (34), suggesting that the soxB gene is important for function of the Sox enzyme complex in oxidizing reduced sulfur compounds other than thiosulfate.
There is a wide range of amino acid diversity among Sox genes from both cultured and environmental samples. For instance, Petri et al. (24) compared partial-length amino acid sequences of 14 bacterial soxB genes and found the sequences were 41 to 98% similar to one another. This variability among soxB genes calls into question whether different Sox gene variations encode functionally equivalent Sox enzymes or if the sequence diversity yields Sox enzymes with different substrate preferences and reaction rates and/or that are optimized to different geochemical conditions. Further support for this comes from experiments done with the SoxB enzyme of Allochromatium vinosum (A. vinosum does not possess the SoxCD complex) that would not function with SoxXA, SoxYZ, and SoxCD enzyme complexes of P. pantotrophus, despite the SoxXA and SoxYZ enzyme complexes of A. vinosum functioning with the SoxB enzyme of P. pantotrophus (35). This work suggests that there is something different about the SoxB enzyme for bacteria that utilize the branched pathway versus those utilizing the PSO pathway. However, the extent to which the Sox system has been investigated to determine how these differences are manifested in environmental settings is incomplete.
As such, it is possible that in environmental systems where several sulfur-oxidizing bacterial taxonomic groups occupy the same habitat (10, 12, 15, 24–26), Sox gene differentiation could support niche partitioning hypotheses (36). Specifically, we hypothesize that niche space for sulfur-oxidizing bacteria is readily defined by genetic variations in functional genes that alter enzymatic function at specific geochemical conditions. Therefore, the purpose of this study was to examine soxB gene diversity along a sulfidic spring geochemical gradient comprised of phylogenetically different, putative sulfur-oxidizing bacteria to identify how variations of soxB genes were naturally distributed. We also set out to identify possible evidence for altered SoxB enzyme function in nature. Although focused on one sulfur oxidation pathway, the results from this study expand our current knowledge of soxB gene diversity in natural settings and demonstrate that soxB gene sequences vary with geochemical conditions.
MATERIALS AND METHODS
Site description, geochemical characterization, and sample collection.
The main study spring, Rattlesnake Spring, emanates from fractured Ordovician limestone bedrock just north of the Bromide fault near Bromide, Oklahoma, where there is a complex of six private, sulfidic springs (34°25′4″N, 96°29′40″W). The source of the springs is not known, but other sulfidic springs in the Arbuckle Mountains have been traced to a mixture of meteoritic water and brine within the underlying Arbuckle-Simpson aquifer (37–39). All of the springs in the study area have outflow channels at least 5 m long, well-defined geochemical gradients within the outflow channels, and microbial mats. Approximately 2 ml of wet biological material was collected aseptically at each sampling location (described below) and placed on ice for transport.
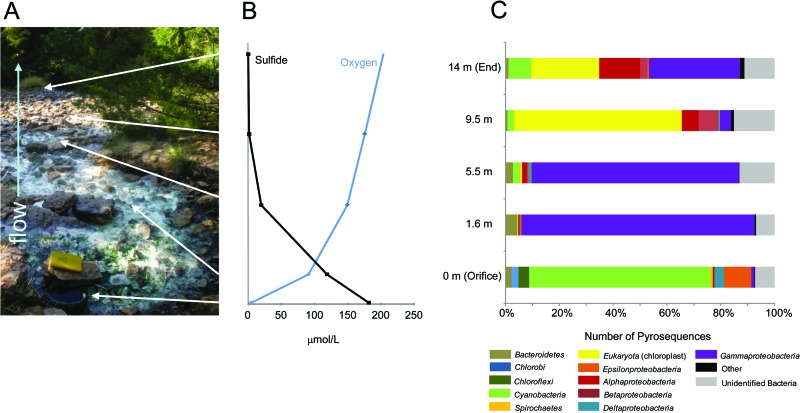
The flow rate at the orifice was 15 cm/s, and the water depth was ∼5 cm. Water depth was greatest in the upper reach of the outflow channel for the first ∼5.5 m, where microbial mats filled the water column and coated rock surfaces, sediment, and vegetation (e.g., leaves and branches that fell into the channel) (Fig. 1A). At 1.6 m, a thick white microbial mat proliferated at a carbonate ledge. At ∼5.5 m from the orifice, the white microbial mat began to dissipate as the outflow channel increased in width, from ∼3.7 m to as much as 7.5 m. This widening caused the water depth to decrease to <1 cm and flow rate to become unperceivably slow. At 9.5 m, there was no visual evidence of white microbial mats; instead, patchy, thin, brown-to-olive-green biofilms were associated with muddy sediment alongside algal mats and vegetation until the last practical sampling point 14 m downstream (Fig. 1A). It is important to note that our main efforts focused on sampling mats horizontally along the flowpath, and we did not attempt to account for any vertical stratification within the mats, biofilms, and sediments.
Fig 1.
(A) Geochemical and microbiological characterization of Rattlesnake Spring, Oklahoma. White arrows designate geochemical and biological sampling locations. (B) Sulfide (black) and dissolved-oxygen (blue) concentrations along the sampling transect. (C) Taxonomic classifications of the 16S rRNA pyrosequences collected from the sampling locations in Rattlesnake Spring.
The spring was sampled in July and October 2010, and water and microbial mat samples were collected at the five transition points along the outflow channel. pH, temperature, and specific conductance were measured in the field using standard electrode methods (40). Total dissolved sulfide and dissolved oxygen were measured in the field using the methylene blue and rhodazine D colorimetric methods, respectively, using CHEMetrics chemistries (Calverton, VA) (40). Water was filtered through 0.22-μm polyvinylidene fluoride filters into separate bottles for anions and acid-preserved metals for major ion determination. In the laboratory, ion concentrations were determined using a dual-column Dionex ICS-3000 reagent-free ion chromatograph. Standard checks were accurate within ±2 standard deviations, except for fluoride, sodium, and calcium (±3 standard deviations). Alkalinity as total titratable bases, and representing bicarbonate (HCO3−) concentration based on the pH and concentration of titratable bases in the fluids, was determined in the field from water passed through 0.2-μm-pore-size filters by endpoint titration using 0.1 N sulfuric acid to pH 4.3 (40).
DNA extraction from the microbiological materials was done in duplicate for each sample, utilizing the PowerSoil DNA isolation kit (MO BIO Laboratories, Carlsbad, CA) according to the manufacturer's instructions. Extractions were stored in Tris-EDTA (TE) buffer at −20°C until use.
454 pyrosequencing and analysis of 16S rRNA genes.
DNA extractions were purified and 16S rRNA genes sequenced by Research and Testing Laboratories (RTL) (Lubbock, TX) using massively parallel bacterial tag-encoded FLX amplicon pyrosequences (bTEFAP), as described previously (41, 42), with the forward primer 28F (5′-TTTGATCNTGGCTCAG-3′) and reverse primer 519r (5′-GTNTTACNGCGGCKGCTG-3′). Following sequencing, all failed sequence reads, low-quality score sequences (<20), and nonbacterial ribosomal sequences were removed. Pyrosequences from July and October were combined into a single data set to provide an assessment of the alpha diversity for each sampling location. Barcodes and adaptors were trimmed, and the resulting sequences were aligned in the Ribosomal Database Project (RDP) pipeline (http://pyro.cme.msu.edu) (43). Pyrosequences from each sample were checked for chimeras using Bellerophan (44) and UCHIME (45), and potential chimeric sequences were removed from the data set. Taxonomy was assigned using the RDP classifier (46). Sequences with <80% similarity were classified as unidentified bacteria, and proteobacterial sequences with <85% similarity were classified as unidentified Proteobacteria. Only sequences with at least 94% similarity were identified to the genus level. The RDP classifier does not classify sequences to the species level. 16S rRNA pyrosequences were clustered using the RDP complete linkage clustering program (43) at a 96% cutoff to define operational taxonomic units (OTUs) and to calculate rarefaction curves (see Fig. S1 in the supplemental material). Shannon diversity scores (H′) were calculated using the RDP Shannon index and Chao1 estimator, including evenness (E).
soxB gene amplifications, cloning, and sequencing.
soxB genes were PCR amplified from DNA extractions for each sample using the soxB693F (5′-ATCGGNCARGCNTTYCCNTA-3′) and soxB1446B (5′-CATGTCNCCNCCRTGYTG-3′) primers (24). After optimization of reagent concentrations, annealing temperature, and product fragment length, amplifications were done with 50 ng of DNA and 5 U/μl Taq DNA polymerase (5′; ThermoFisher Scientific) and executed using an MJ Research Dyad Disciple thermal cycler for 25 cycles under the following conditions after a hot start for 5 min at 94°C: denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and elongation for 1 min at 72°C, followed by a final extension at 72°C for 10 min. Target-size (∼750 bp) products were excised from 1.5% Tris-acetate-EDTA (TAE) low-melt agarose electrophoresis gels and purified with a Promega Wizard miniprep DNA purification kit (Madison, WI) according to the manufacturer's instructions. Sequences were cloned using the Invitrogen TOPO Top 10F cloning kit according to the manufacturer's instructions (Carlsbad, CA). Clones with plasmids containing correct-sized inserts were lysed in TE buffer at 96°C for 10 min, and insert screening was done by Tris-borate-EDTA (TBE) agarose gel electrophoresis. Fragments of the correct size were sequenced with the manufacturer primers M13(−20) (5′-GTAAAACGACGGCCAGT-3′) and M13(−24) (5′-AACAGCTATGACCATG-3′) by the High-Throughput Genomics Unit at the University of Washington in Seattle, WA.
soxB sequence phylogeny and analysis.
Sequences of anomalously short length and poor quality (i.e., containing N′s) were removed from the data set prior to subjecting sequences to BLAST searches in GenBank (http://www.ncbi.nlm.nih.gov/) to identify and remove non-soxB genes. Taxonomic affinities of soxB genes were determined using a BLAST search that excluded uncultured/environmental microorganisms.
soxB DNA sequences were translated into amino acid sequences. Sequences were manually aligned to both published soxB amino acid alignments (30) and to one another to account for differences not present in previously published alignments. The program CD-HIT was utilized to cluster the soxB DNA sequences and translated amino acid sequences into OTUs using 90, 80, and 70% cutoffs (DNA sequences were not clustered below 80%, because CD-HIT does not allow for DNA clustering below 75%) (47, 48). We chose an 80% cutoff for analysis to preserve soxB gene taxonomic signatures within OTUs and to allow for greater variation among the amino acid sequences comprising the resulting OTUs that could result in functional differences in the SoxB enzymes.
The alignment of translated soxB genes representing OTUs and sequences from NCBI was verified in ClustalW (49) prior to being truncated to the same length (270 residues) and used to reconstruct a phylogenetic tree using maximum likelihood under the WAG +G (1.69) +I (0.01) +F (0.081) amino acid substitution model (50) based on testing different models in Mega5 (51).
Translated sequences that contained nonsense mutations when translated remained in the OTU data set but not the phylogenetic analyses, because these sequences were considered to represent alternative soxB gene sequences since amplification occurred, even if for nonfunctional enzymes or possibly resulting from sequencing error(s). This occurred in approximately 8% of the sequences. Without full-length gene sequences to compare these sequences to, it was difficult to assess what the true reading frame for the gene was or to assess the significance of nonsense mutations. Most of the nonsense mutations were due to a single-base-pair insertion or deletion that caused a frameshift mutation; this could have resulted from sequencing error. Many of these sequences clustered as an OTU containing only one sequence, which had little impact on the full data set.
Statistical analysis.
Abundance data from the pyrosequence and soxB OTU diversity data were normalized for each sample to evaluate correlations of soxB gene diversity and geochemistry. To correct skewness in the environmental data, variables were log10(x + 1) transformed (52). Using PAST software (version 2.14) (53), relationships between major taxonomic groups and environmental factors (e.g., pH, sulfide, etc.) were analyzed by stepwise canonical correspondence analysis (CCA) to sequentially remove the least important variables and maximize correlations between principal axes and linear combinations of environmental variables (54). Permutation tests were carried out. For all analyses, a P value of <0.05 was considered significant.
NCBI sequence submissions and nucleotide sequence accession numbers.
The raw pyrosequence reads are available at the NCBI Sequence Read Archive under study number SRA057273. soxB gene sequences resulting from this study were deposited in GenBank under accession numbers JX471149 to JX471559.
RESULTS
Geochemical zonation along the stream channel.
Rattlesnake Spring (Fig. 1A) was sampled on five different occasions beginning in 2010, and ion concentrations did not vary significantly over the time period (unpublished data). Decreasing dissolved sulfide and increasing dissolved oxygen geochemical profiles were measured along the outflow channel (Fig. 1B). pH continually increased downstream, while alkalinity (as HCO3−) increased to its maximum concentration at 1.6 m and then decreased downstream (Table 1). Temperature increased downstream, likely due to solar heating. In general, anion and cation concentrations were relatively stable, except for ammonium, which increased downstream.
Table 1.
Representative geochemical composition of transect sample locations along an outflow stream from Rattlesnake Spring, Bromide, Oklahomaa
| Sample distance (m) | pH | Temp (°C) | Dissolved O2 (μmol/liter) | Dissolved sulfide (μmol/liter) | Conductivity (mS) | Alkalinity (mmol/liter) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCO3− | Li | Na | NH4+ | K | Mg | Ca | Sr | F | Cl | Br | SO4 | ||||||
| 0 (orifice) | 6.85 | 20.7 | 4.4 | 181.8 | 7.14 | 4.7 | 0.41 | 51.0 | 3.54 | 2.02 | 0.46 | 25.8 | 0.15 | 0.17 | 66.3 | 0.07 | 3.44 |
| 1.6 | 7.16 | 20.7 | 90.9 | 118.7 | 7.60 | 5.0 | 0.41 | 48.9 | 3.51 | 2.04 | 0.46 | 25.8 | 0.15 | 0.08 | 70.8 | 0.09 | 3.44 |
| 5.5 | 7.48 | 21.0 | 149.4 | 19.7 | 7.61 | 4.7 | 0.53 | 48.3 | 3.49 | 2.00 | 0.46 | 25.8 | 0.15 | 0.17 | 66.1 | 0.08 | 3.36 |
| 9.5 | 7.64 | 22.0 | 175.6 | BDb | 7.61 | 4.6 | 0.43 | 49.0 | 4.64 | 2.04 | 0.46 | 25.8 | 0.15 | 0.17 | 66.7 | 0.09 | 3.43 |
| 14 (end) | 7.78 | 25.0 | 156.6 | BD | 6.08 | 4.4 | 0.43 | 48.1 | 4.34 | 2.00 | 0.46 | 25.8 | 0.15 | 0.17 | 65.4 | 0.08 | 3.35 |
Data are the average readings from the spring in July and October 2012.
BD, below the limit of detection for the instrument.
Community composition based on 16S rRNA data.
Presence/absence results at the levels of phyla and class (and genera, where classification was possible) were collated from the 60,279 pyrosequences that comprised 3,092 OTUs (Fig. 1C and Table 2; also see Table S1 in the supplemental material). Although there were some general differences between the July and October pyrosequence data sets, as evidenced from the diversity indices (Table 2) and rarefaction curves for each sample (see Fig. S1), the overall distribution of major taxa at each sampling point along the transect and OTUs formed from pyrosequences were similar from both data sets. Therefore, pyrosequences from July and October were combined for each sampling location and provided a semiquantitative assessment of the alpha diversity for each region of the spring outflow channel. Relative abundances of different microbial communities shifted along the flowpath, and each location was dominated by distinct groups. The orifice was dominated by Chlorobi, Chloroflexi, Gammaproteobacteria, and Epsilonproteobacteria, which are groups that have putative sulfur-based metabolisms. At the genus level (e.g., greater than 94% similarity according to the RDP classifier), 93% of Chlorobi pyrosequences could be identified as belonging to Chlorobium spp., 51% of the gammaproteobacterial sequences could be identified as belonging to Desulfocapsa spp., 28% of gammaproteobacterial pyrosequences could be identified as belonging to Thiothrix spp., and 39% could be identified as belonging to the genus Thiofaba. At ≤94% sequence similarity, 91% of Chloroflexi pyrosequences were related to Chloroflexi spp. and 77% of Epsilonproteobacteria were related to Sulfurovum spp. Among the Cyanobacteria, only 7% were identifiable to the genus level, but at ≤94% sequence similarity, 79% of Cyanobacteria sequences were related to the group I Cyanobacteria (from 21 to 93% sequence similarity) (see Table S1). At 1.6 m, 86% of all pyrosequences were related to Thiothrix spp., which formed 13 prominent OTUs (i.e., having more than 100 sequences) and 46 minor OTUs. At 5.5 m, 36% of all pyrosequences were related to Thiothrix spp., and 27% formed a single OTU related to an unidentified Gammaproteobacteria related (at only 18 to 64% similarity) to the genus Zymobacter. Although Alphaproteobacteria were generally unidentifiable to the genus level at both 9.5 and 14 m, representation among Alphaproteobacteria and Betaproteobacteria increased from 9.5 to 14 m, in addition to sequences identified as eukaryotic chloroplasts (Fig. 1C) primarily from Bacillariophyta (41% of cyanobacterial sequences at 9.5 m and 20% at 14 m) and Streptophyta (47% of cyanobacterial sequences at 9.5 m and 40% at 14 m). The dominant genus from the Betaproteobacteria at 9.5 and 14 m was Thiobacillus.
Table 2.
Separate and combined 16S rRNA and soxB gene sequence data from July and October 2010, including diversity indices used in this study for 16S rRNA genes
| Sample distance (m) | 16S rRNA data |
No. of soxB sequences |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| July 2010 |
October 2010 |
Total | H′ | July | October | Total | |||||||||
| No. of sequences | No. of OTUs | Chao1 estimate (range) | H′ | E | No. of sequences | No. of OTUs | Chao1 estimate (range) | H′ | E | ||||||
| 0 (orifice) | 5,485 | 517 | 894 (783–1,052) | 4.1 | 0.6 | 8,205 | 144 | 193 (168–245) | 1.7 | 0.3 | 13,690 | 3.6 | 35 | 25 | 60 |
| 1.6 | 2,873 | 119 | 156 (136–196) | 2.1 | 0.4 | 9,997 | 167 | 220 (194–271) | 2.4 | 0.5 | 12,870 | 2.6 | 44 | 83 | 127 |
| 5.5 | 3,920 | 197 | 293 (252–364) | 2.3 | 0.4 | 7,886 | 512 | 782 (703–895) | 3.6 | 0.6 | 11,806 | 3.6 | 52 | 71 | 123 |
| 9.5 | 6,065 | 358 | 612 (527–738) | 2.9 | 0.5 | 6,640 | 814 | 1,273 (1,160–1,423) | 5.1 | 0.8 | 12,705 | 4.6 | 22 | 14 | 36 |
| 14 (end) | 3,962 | 777 | 1,383 (1,239–1,572) | 5.2 | 0.8 | 5,246 | 407 | 649 (568–771) | 4.1 | 0.7 | 9,208 | 5.1 | 48 | 17 | 65 |
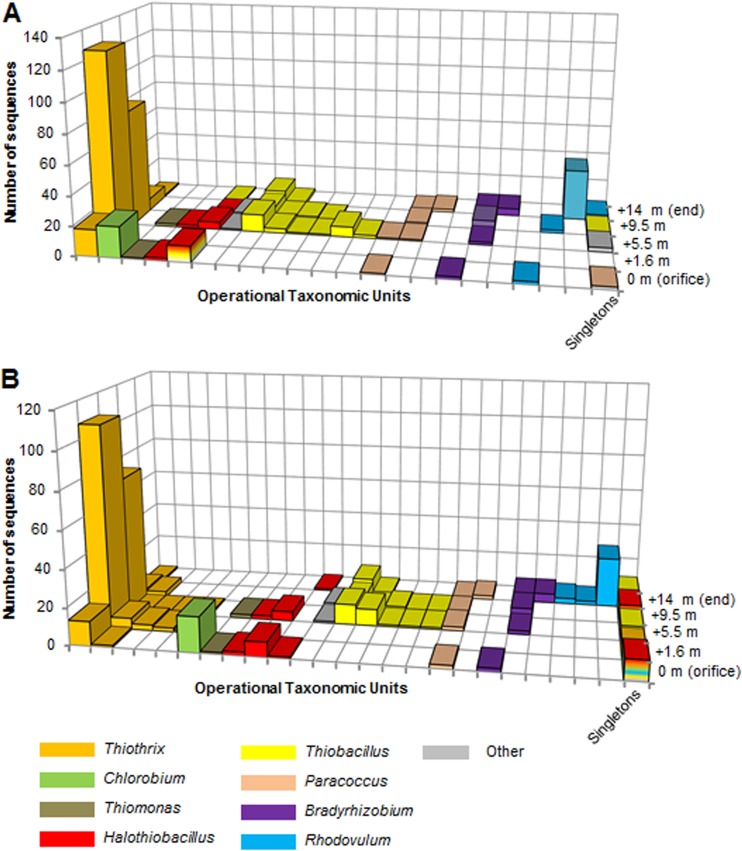
soxB gene sequence clusters.
There were 411 soxB sequences in the final data set (Table 2; also see Fig. S1 in the supplemental material) that ranged in length from 499 to 783 bp (mean, 723 bp) and from 166 to 261 amino acid residues (mean, 241 residues). The percent sequence similarity to the closest soxB relatives varied among identified taxa. Most sequences had 70 to 85% sequence similarity to their closest relatives according to a BLAST search, but some soxB genes related to the genera Thiothrix, Chlorobium, and Bradyrhizobium had higher sequence similarities of 88 to 96%. At 80% sequence identity, 32 OTUs (12 were singletons) formed from soxB DNA sequences (Fig. 2A), but 58 soxB amino acid OTUs were generated (35 OTUs were singletons) (Fig. 2B). DNA-based (Fig. 2A) or amino acid (Fig. 2B) clustering generated similar soxB gene distributions of taxonomic groups along the outflow channel, with only minor differences in the numbers of OTUs between the two data sets.
Fig 2.
soxB gene clustering of DNA-based gene sequences (A) and translated amino acid sequences (B) using an 80% cutoff.
Diversity of soxB gene groups compared to 16S rRNA gene diversity.
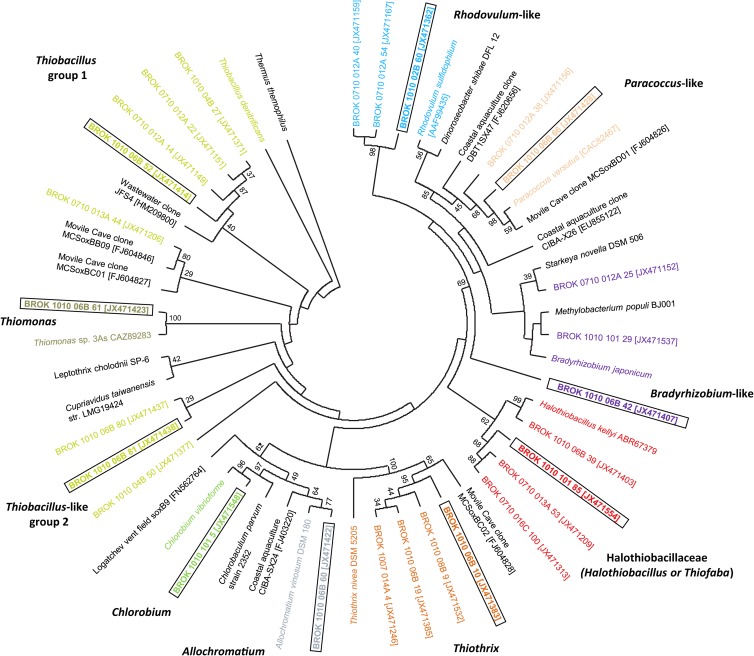
There were distinct phylogenetic associations, some supported by high bootstrap values, to known strains from the genera Thiobacillus, Thiomonas, Chlorobium, Allochromatium, Thiothrix, and Halothiobacillus that utilize the branched pathway (14), and to Rhodovulum, Paracoccus, and Bradyrhizobium that utilize the PSO pathway (14) (Fig. 3). OTUs were affiliated with specific genera associated with a particular Sox pathway and were found at definite locations along the outflow channel, almost to the exclusion of being retrieved from anywhere else along the outflow channel. For example, this is evident for Chlorobium from the orifice or the Rhodovulum-like soxB genes from 14 m (Fig. 2B). All of the retrieved soxB genes from 1.6 m, the location of thick white mats, were affiliated with Thiothrix. Most soxB genes at 5.5 m were related to Gammaproteobacteria, specifically Thiothrix (16S rRNA genes as well) and Halothiobacillus, while soxB genes related to Thiobacillus-like groups were restricted to the downstream portion of the outflow channel (Fig. 2A and B). The largest OTU, comprised of Thiobacillus-like soxB gene sequences, were 84 to 87% similar to previously described Thiobacillus soxB genes (referred to as Thiobacillus group 1), but there were several other OTUs affiliated with Thiobacillus with less sequence similarity (referred to as Thiobacillus group 2) (Fig. 2B).
Fig 3.
Phylogenetic tree of translated soxB sequences from clones that represent distinct OTU groups obtained from Rattlesnake Spring. Taxonomic identification for each of the OTU groups is color coded for comparison to Fig. 2 and 4. Accession numbers for clones from this study, environmental clones previously identified from other investigations, and cultured isolates are shown in brackets. Topology was inferred using maximum likelihood under the WAG+G+I+F method for amino acid substitution (G = 1.69, I = 0.01, and F = 0.081). Bootstrap values of major branch points are shown.
Some soxB gene sequences related to the alphaproteobacterial genera Paracoccus, Bradyrhizobium, and Rhodovulum were retrieved throughout the channel, but most sequences were identified from downstream samples where sulfide was undetectable. 16S rRNA gene pyrosequences affiliated with Alphaproteobacteria were largely unidentifiable to the genus level, so no 16S rRNA gene pyrosequences could be linked to the genera Rhodovulum, Paracoccus, or Bradyrhizobium that would confirm taxonomic identities of the soxB genes. Phylogenetically, the soxB genes from these genera were also weakly related to previously characterized isolates or environmental clones (Fig. 3), which suggested that these OTUs could be related to different, potentially novel, groups. To highlight this taxonomic question, we refer to these OTUs as Rhodovulum-, Paracoccus-, and Bradyrhizobium-like.
There was some discrepancy between the distributions of 16S rRNA and soxB genes for the genera Halothiobacillus, Thiofaba, and Thiobacillus. Both Halothiobacillus and Thiofaba belong to the family Halothiobacillaceae. Phylogenetically, representative soxB genes formed a clade with moderately low bootstrap values with Halothiobacillus (Fig. 3), but this is because there were no Thiofaba soxB genes within the NCBI database for comparison, and we regard this taxonomic assignment as putative. Therefore, phylogenetically, we clustered these sequences as Halothiobacillaceae (Fig. 3).
Additionally, because of poor similarity to previously characterized soxB genes, one soxB DNA OTU was comprised of genes that were affiliated with Halothiobacillus and Thiobacillus, but all translated amino acid sequences in the OTU were closely related to Halothiobacillus. All of the genes in this OTU possessed a 30-bp insertion in the linking loop region that no other soxB sequences retrieved in our study possessed, except for other Halothiobacillus soxB genes. The Thiobacillus soxB genes obtained from the NCBI database do not possess this additional 30-bp insertion in the linking loop; only Halothiobacillus soxB genes do. All of the sequences contained an additional 30-bp insertion and Thiobacillus 16S rRNA sequences were generally confined to the downstream stream reach, but sequences associated with this OTU were found primarily upstream. Because of this, these soxB genes most likely are related to Halothiobacillus.
Distribution of 16S rRNA and soxB gene OTUs in relation to habitat geochemistry.
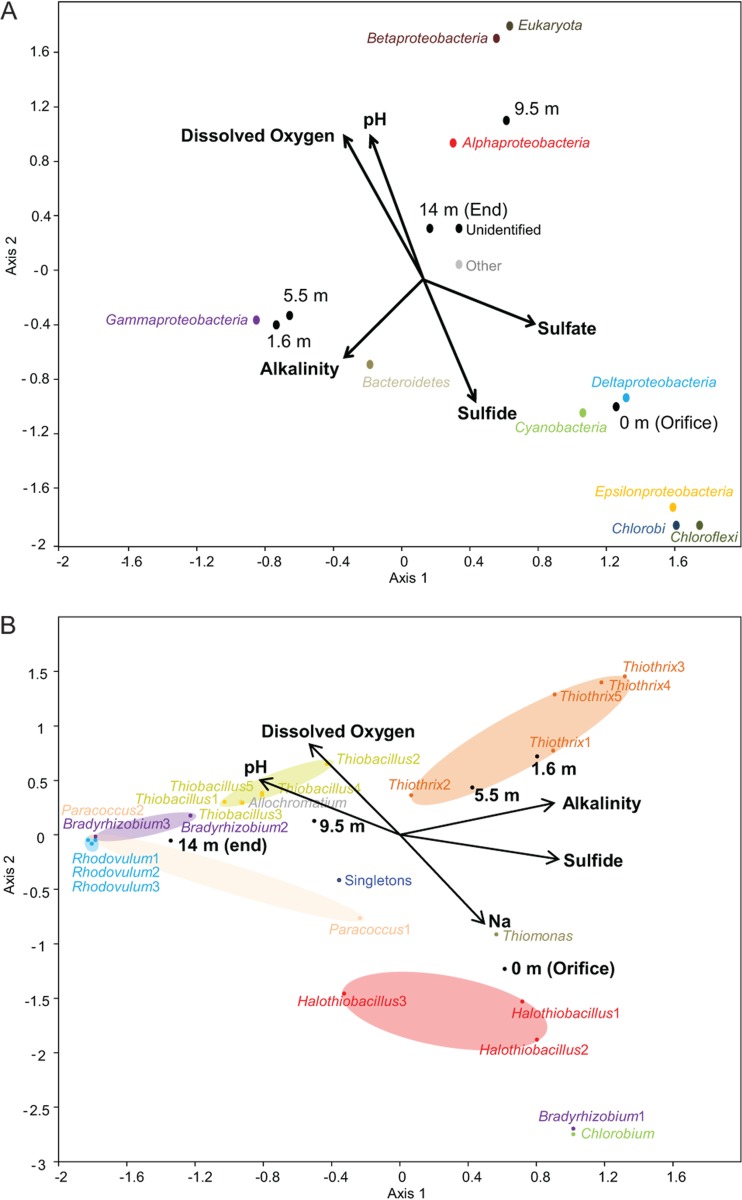
The relationships between geochemistry, particularly related to substrate (e.g., sulfide and oxygen) concentrations, and the distribution of microbial groups along the outflow channel were verified by CCA using 16S rRNA and soxB gene sequence diversity (Fig. 4A and B, respectively). Almost 100% of the variance could be described by two axes of the final CCA, although only a best global P value of 0.06 following 10,000 Monte Carlo permutations could be obtained for any stepwise iteration of the geochemistry and 16S rRNA gene sequence data sets, which would be considered insignificant. CCA axis 1 (58.0% of the variance) was positively correlated with sulfate and negatively correlated with alkalinity, whereas CCA axis 2 (38.1% of the variance) was positively correlated with dissolved oxygen and pH and negatively correlated with sulfide concentrations (Fig. 4A). The CCA triplot verified the notable associations among 16S rRNA genes designated to taxonomic groups and presence/absence results along the outflow channel related to environmental conditions (Fig. 4A). Specifically, Chloroflexi, Chlorobi, Cyanobacteria, Deltaproteobacteria, and Epsilonproteobacteria, found predominately near the orifice, were positively correlated with sulfide and sulfate concentrations. Gammaproteobacteria positively correlated with alkalinity, as well as to the 1.6- and 5.5-m sample locations, but the ordination of sequence groups affiliated with the Alphaproteobacteria, Betaproteobacteria, and Eukaryota from downstream sampling locations (9.5 and 14 m) positively correlated with dissolved oxygen and pH but negatively with sulfide concentrations (Fig. 4A).
Fig 4.
(A) Triplot generated from canonical correspondence analysis of 16S rRNA genes from Rattlesnake Spring showing the relationships among five physicochemical variables, noted as labeled vectors, sampling location along the spring-stream transect, and ordination of 16S rRNA gene sequences (10,000 Monte Carlo permutations; global P value of 0.06). (B) Triplot generated from canonical correspondence analysis of soxB genes from Rattlesnake Spring showing the relationships among five physicochemical variables, noted as labeled vectors, sampling location along the spring-stream transect, and ordination of soxB gene sequence OTUs (10,000 Monte Carlo permutations; global P value of 0.03).
In contrast, nearly all of the relationships among soxB OTU distribution to geochemistry along the stream transect could be significantly explained with two axes (global P value of 0.03 following 10,000 Monte Carlo permutations) (Fig. 4B). CCA axis 1 (57.1% of the variance) strongly and positively correlated with sulfide concentration and alkalinity and strongly but negatively correlated with pH. CCA axis 2 (42.0% of the variance) positively correlated with dissolved oxygen concentration and negatively with sodium concentration, which served as a proxy for conductivity (although adding the conductivity parameter decreased the significance of the CCA when included; data not shown). The ordination of soxB OTUs designated to taxonomy strongly correlated with location along the outflow channel, as well as with environmental variables. For instance, the orifice was correlated with the ordination of Chlorobium, Thiomonas, and the Halothiobacillus OTUs and one Bradyrhizobium-like OTU, which also all correlated with sulfide and sodium concentration vector directions. This Bradyrhizobium-like OTU may be only distantly related to the other Bradyrhizobium-like soxB genes in the spring, because not only was it phylogenetically distinct from the other Bradyrhizobium-like soxB OTUs (Fig. 3), it also did not correlate with the location or environmental parameters of other Bradyrhizobium-like OTUs from the outflow channel on the CCA (Fig. 4). The other Bradyrhizobium-like OTUs were tightly ordinated with other OTUs for PSO pathway taxa (e.g., Rhodovulum-like and Paracoccus-like), as well as with the 14-m sampling location, where these groups were more abundant and negatively correlated with the vector direction for sulfide. Thiothrix and Thiobacillus soxB OTUs each formed their own clusters on the CCA, with Thiothrix correlated with the ordination of the 1.6- and 5.5-m locations where the microbial mats were thickest and dominated by Thiothrix pyrosequences. Similarly, the Thiobacillus OTUs were correlated with the 9.5-m location ordination on the CCA, which was where Thiobacillus pyrosequences were abundant. Both of these soxB OTU groups were strongly correlated with sulfide and dissolved oxygen concentrations, although Thiothrix OTUs were more strongly and positively correlated with sulfide than Thiobacillus OTUs.
soxB gene sequence variations.
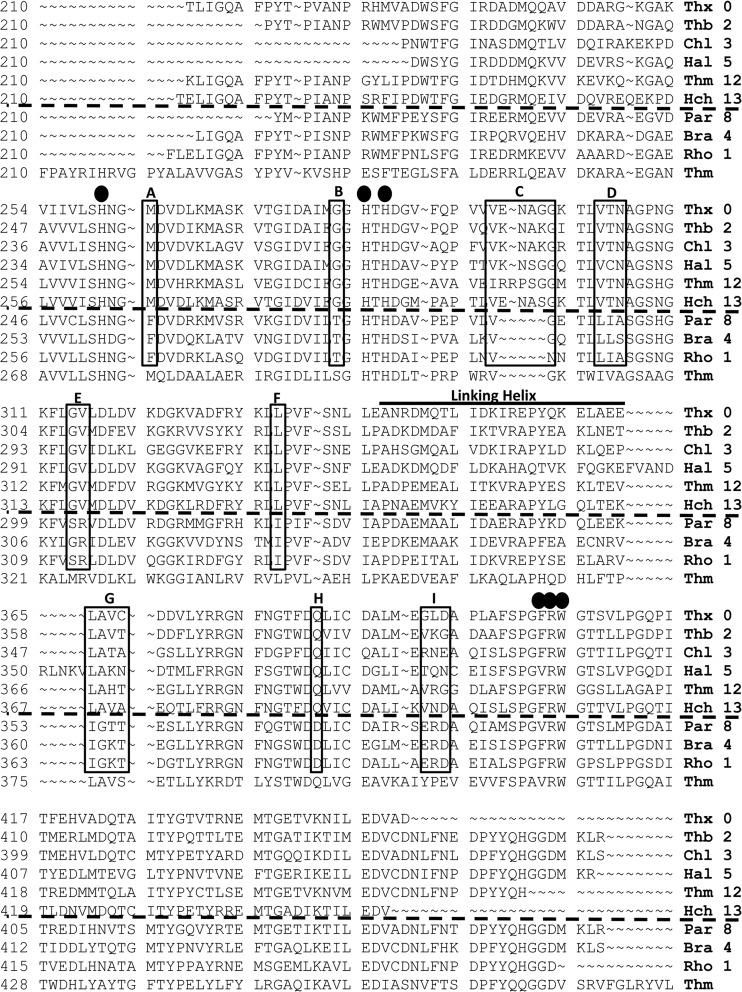
The translated portion of the soxB gene sequenced contained three of the five active-site His residues (His274, His297, and His299) as well as Val415, Arg416, and Trp417. The His residues, Arg416, and Trp417 were almost entirely conserved in all of the translated soxB genes sequenced, with the primary exception being that there were some altered active-site locations caused by frameshift mutations. As shown in Fig. 5, there were compositional differences specific to soxB amino acid sequences associated with the two Sox pathways. In many sequences, a Phe residue replaced the Val415 position. There were also distinct compositional similarities and differences in amino acid variability across OTUs (i.e., Thiothrix versus Chlorobium soxB genes), such that all of the soxB genes whose closest relatives were known to utilize the branched pathway had similar residues that were distinct from those of groups that utilize the PSO pathway (Fig. 5). For example, Fig. 5A shows a residue having a neutral nonpolar amino acid with hydrocarbon or sulfhydral functional groups for the branched pathway taxa compared to a nonpolar amino acid with an aromatic functional group for PSO pathway taxa. Similarly, a neutral amino acid possessing a nonpolar side chain was evident for branched pathway taxa, but different neutral amino acids possessed a polar side chain for PSO pathway taxa (Fig. 5B). Downstream of two of the active-site His residues (297 and 299) (Fig. 5C), all soxB genes associated with the branched pathway possessed four to five more amino acids than those of the PSO pathway (between residues 308 and 309 in T. thermophilus). The soxB gene-linking helix generally varied from OTU to OTU as well, particularly for Halothiobacillus (Fig. 5). Halothiobacillus-like soxB genes possessed 10 additional amino acids not found in other soxB genes retrieved from the spring.
Fig 5.
Representative soxB amino acid sequences from the major OTUs in Rattlesnake Spring: Thiothrix OTU 0 (Thx 0), Thiobacillus OTU 2 (Thb 2), Chlorobium OTU 3 (Chl 3), Halothiobacillus OTU 5 (Hal 5), Thiomonas OTU 12 (Thm 12), Halochromatium OTU 13 (Hch 13), Paracoccus OTU 8 (Par 8), Bradyrhizobium OTU 4 (Bra 4), Rhodovulum OTU 1 (Rho 1), and T. thermophilus HB27 (Thm). The black circles denote amino acids in the active site of the soxB enzyme. The solid black boxes outline key residues between the PSO pathway and branched pathway. The soxB enzymes of the amino acid sequences above the dotted black lines are utilized in the branched pathway, while the soxB enzymes of the sequences below the dotted black line are utilized in the PSO pathway. (A) A conserved methionine in the branched pathway and a phenylalanine in the PSO pathway. (B) A conserved glycine in the branched pathway and either a threonine or serine in the PSO pathway. (C) The branched pathway sequences all possess four to five more amino acids than the PSO pathway soxB genes in this region of the gene. (D) A polar (threonine), polar (asparagine), nonpolar (alanine) motif in the branched pathway and a variable nonpolar (leucine or isoleucine), nonpolar (alanine), polar (serine or threonine) motif in the PSO pathway. (E) A conserved nonpolar glycine residue and a perfectly conserved nonpolar valine residue in branched pathway soxB genes and a conserved, polar serine residue and a perfectly conserved, positively charged arginine residue in the PSO pathway. (F) A conserved leucine in the branched pathway and an isoleucine in the PSO pathway. (G) Conserved leucine and alanine residues, followed by two variable residues in the branched pathway and of isoleucine and glycine, followed by a lysine/threonine in the PSO pathway; however, these residues were not conserved in all of the Bradyrhizobium-like soxB genes. (H) A conserved glutamine residue in the branched pathway and an aspartic acid residue in the PSO pathway. (I) A glutamic acid, arginine, and aspartic acid motif in the PSO pathway that is not conserved in the branched pathway.
DISCUSSION
Microbially mediated geochemical reactions in the sulfur cycle are responsible for the oxidation and reduction of sulfur compounds, production of stored sulfur compounds and volatile phases, and mineralization of organosulfur compounds. Among the microbial groups capable of oxidizing reduced sulfur compounds, multiple oxidation pathways can co-occur in many habitats (for examples, see references 14, 17, 22, and 25). However, investigations of the spatial and functional diversity of a single metabolic gene utilized in different pathways have not been done. Such investigations could uncover controls between functional diversity and niche partitioning among sulfur-oxidizing bacteria within a habitat. This study focused on identifying possible evidence for altered SoxB enzyme function from different groups of putative sulfur-oxidizing bacteria in a single habitat with variable substrate concentrations.
16S rRNA pyrosequencing captured the major groups of sulfur-oxidizing bacteria, occupying the microbial mats and sediments at Rattlesnake Spring, though the taxonomic resolution was poor below the phylum and class levels. Of the known sulfur-oxidizing groups at the orifice, predominately Chlorobi, Chloroflexi, Epsilonproteobacteria, and Gammaproteobacteria, only the 16S rRNA genes of the anoxygenic phototrophs were identifiable to the genus level (i.e., Chlorobium and Chloroflexus). The genera of all other potential sulfur oxidizers at the orifice could not be identified or were very few in number in the whole data set (<50 sequences). The presence of Cyanobacteria at the sulfidic conditions at the orifice was somewhat unexpected but is not without precedence. Cyanobacteria have been shown to switch from oxygenic photosynthesis to anoxygenic photosynthesis in the presence of H2S (55–57). As has been reported by others (15), the primers used for soxB amplification did not amplify soxB genes of Epsilonproteobacteria or Chloroflexi. To our knowledge, Chloroflexi members do not possess Sox genes. Because the soxB diversity among Epsilonproteobacteria remains unknown and this group made up a large portion of the diversity in the microbial mats at the orifice (see Table S1 in the supplemental material), the missing data highlight that there is even more niche partitioning possible than our results currently reveal. At 1.6 and 5.5 m, the dominant putative sulfide-oxidizing group was Thiothrix spp., which is known to possess Sox genes (15). At 9.5 and 14 m, Thiobacillus spp. were the most dominant (i.e., identifiable) putative sulfide oxidizers. Thiobacilli are aerobic and anaerobic sulfide-oxidizing bacteria known to possess Sox genes (58). At 14 m, the gammaproteobacterial pyrosequences were dominated by the genus Aeromonas, whose members generally are not associated with sulfide oxidation and are ubiquitous in aquatic environments (59). Because there were no intact microbial mats growing within the water column between 9.5 and 14 m, these groups likely are associated with the sediments.
The spatial distribution of 16S rRNA-based OTUs, particularly those affiliated with microbial groups capable of oxidizing reduced sulfur compounds, generally correlated with the spatial distribution of soxB-based OTUs from Rattlesnake Spring. Moreover, the spatial distribution of soxB OTUs significantly correlated with geochemical gradients along the outflow channel (Fig. 4), and the closest relatives of the soxB genes, even with low sequence similarities to known microbial groups, offer clues as to how the Sox enzymes are utilized metabolically. As such, soxB genes (and thereby the SoxB enzymes) may be optimized to the geochemical conditions in which the bacterium resides. soxB genes associated with the branched pathway were retrieved throughout the outflow channel, whereas genes for the PSO pathway were more common downstream. There was a distinct divide among branched pathway soxB genes upstream and downstream of 5.5 m (i.e., high H2S correlated with Chlorobium, Thiothrix, Thiomonas, and Halothiobacillus versus low H2S correlated with Thiobacillus and Halochromatium). Only the Thiothrix soxB genes cross this divide (and an OTU comprised of two sequences that are distantly related to Halothiobacillus), but the numbers are low and likely due to pieces of Thiothrix-dominated white mats being dislodged and carried downstream.
Comparison of soxB genes from Rattlesnake Spring to the soxB gene of T. thermophilus, which has a crystalized SoxB enzyme, indicated where differences in the amino acid sequences were occurring within the SoxB enzyme. The translated soxB amino acid compositional differences for all Rattlesnake Spring OTUs (e.g., Thiothrix OTU 0 versus Thiothrix OTU 7, Thiothrix OTU 0 versus Chlorobium OTU 3, etc.) were too numerous to list, but notable amino acid compositional differences were predominately related to whether the taxa utilize the branched versus PSO Sox pathway. Because T. thermophilus possesses a soxC and soxD gene, it would be expected that this organism uses the PSO pathway. However, the T. thermophilus soxB gene shares similarities with both the branched pathway and PSO pathway (Fig. 5), and phylogenetically it is only distantly related to other PSO pathway bacteria (15). Most notably, T. thermophilus does not possess the additional four to five amino acids that the branched pathway genes have, but it does share many compositional differences with other branched pathway soxB genes (Fig. 5). When mapped onto the SoxB enzyme structure of T. thermophilus, most of the amino acid sequence differences in the soxB genes from Rattlesnake Spring are found on the outer portion of the enzyme, generally away from the active site. Although changes in the outer structure of the enzyme might seem unlikely to induce a functional change in an enzyme, previous findings suggest that it is possible (60, 61) and that even a single-amino-acid change can alter an enzyme's capabilities (62–64). For instance, the substitution of six amino acids (Ala, Asp, Cys, Lys, Phe, and Ser) was found to lead to improved environmental stability in the serine protease from Bacillus amyloliquefaciens that allowed the proteases to resist inactivation under higher temperatures and alkalinity (65).
The increased presence of the soxB genes associated with the PSO pathway downstream indicate a preference for, and availability of, thiosulfate in this region of the spring, because thiosulfate has been shown as a substrate for this pathway (14, 17), and the sulfide concentration in this region of the spring was undetectable (Table 1). Many laboratory studies of bacterial genera that utilize the PSO pathway have been noted to prefer thiosulfate in laboratory studies over other reduced sulfur compounds (66–68). However, organisms having the branched pathway can also oxidize thiosulfate, and it is likely that the Thiobacillus soxB genes found downstream alongside the PSO pathway soxB genes could be utilizing thiosulfate as a substrate. Upstream, specifically from the orifice to 5.5 m, thiosulfate is likely available, but the H2S concentration is high enough that it is likely being utilized as a substrate. All of the bacterial genera affiliated with the soxB genes of the branched pathway in Rattlesnake Spring consist of strains that have been reported to oxidize sulfide as well as thiosulfate (4, 34, 69–72). The mechanism whereby SoxB could oxidize a substrate such as sulfide is unclear, because the current model relies on thiosulfate as a substrate and requires a sulfone sulfur, but mechanisms have been proposed (30). Because reconstituted Sox systems can oxidize sulfide or other reduced sulfur compounds (19–21), this suggests that the Sox system does so in natural systems as well, regardless of the presence of other sulfide-oxidizing metabolic systems (i.e., sulfide-quinone reductase). Actual protein expression experiments need to be done to verify these findings to determine if the different soxB gene variations found in Rattlesnake Spring do encode functionally different enzymes. We hypothesize that many of the amino acid compositional differences (particularly insertions/deletions) found in the soxB genes from Rattlesnake Spring could induce structural and thereby functional changes in the SoxB enzymes. The functional differences may only be minor, such that there is a slightly stronger preference for one substrate over another or a difference in rate of reactions, but these differences could be utilized to explain the distribution of bacterial groups within Rattlesnake Spring (i.e., niche partitioning) as well as lead to an improved understanding of the Sox system in natural systems. The version of a soxB gene that a bacterium possesses does not necessarily govern the distribution of that bacterium, because numerous biological and geochemical parameters probably influence the biogeographic location and distribution of bacteria in the environment. However, this does not preclude the soxB genes (and thereby the SoxB enzymes) from being optimized to the geochemical conditions in which the bacterium resides.
Collectively, our results indicate that the SoxB enzymes are optimized to a range of specific geochemical conditions, and the spatial distribution of the different soxB gene variations along the outflow channel at Rattlesnake Spring could be indicative of niche partitioning among the bacteria that oxidize reduced sulfur compounds. The degree to which SoxB enzymes may be functionally different is the subject of our ongoing and future work, specifically to isolate and express full-length soxB, soxA, soxX, soxY, and soxZ genes to test the functionality of the different variations of Sox genes within the spring outflow channel under various geochemical conditions. The functionality of all of the Sox enzymes is particularly important to resolve, as the individual Sox enzymes are heavily reliant on the other enzymes in the pathway (e.g., SoxYZ requires the assistance of SoxXA, and SoxYZ serves to provide the reduced sulfur substrate to SoxB and SoxCD when present). We hypothesize that the distribution of the other Sox genes within Rattlesnake Spring will be similar to the distribution of the soxB genes, and that they will encode functionally different enzymes that are optimized to different, and specific, geochemical conditions. It seems improbable that the most efficient enzyme systems have been characterized in laboratory-grown strains to date, and that it is more likely that nature has a full spectrum of enzyme systems for all biologically compatible geochemical conditions. The identification and characterization of gene variations that encode functionally different enzymes could lead to bioremediation and bioengineering advances.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Murphy and K. McDonough for allowing access to the springs and for field support. T. Brown and C. Liu assisted with sample collection, and B. Maas helped with field sampling and technical laboratory analyses. The manuscript benefited from commentary provided by B. J. Campbell and A. Layton.
The research was supported by grants from the American Association of Petroleum Geologists (Edward B. Picou, Jr., named grant), the Geological Society of America (Student Research grant 9582-11) to B.H., and from the National Science Foundation (DEB-0640835) and Jones Endowment at the University of Tennessee to A.S.E.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02812-12.
REFERENCES
- 1. Edwards KJ, Bond PL, Druschel GK, McGuire MM, Hamers RJ, Banfield JF. 2000. Geochemical and biological aspects of sulfide mineral dissolution: lessons from Iron Mountain, California. Chem. Geol. 169:383–397 [Google Scholar]
- 2. Baker BJ, Banfield JF. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139–152 [DOI] [PubMed] [Google Scholar]
- 3. Engel AS, Porter ML, Stern LA, Quinlan S, Bennett PC. 2004. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic spring dominated by chemolithoautotrophic Epsilonproteobacteria. FEMS Microbiol. Ecol. 51:31–53 [DOI] [PubMed] [Google Scholar]
- 4. Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, Baciu M, Lu Y, Murrell JC. 2009. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 3:1093–1104 [DOI] [PubMed] [Google Scholar]
- 5. Engel AS, Meisinger DB, Porter ML, Payn RA, Schmid M, Stern LA, Schleifer KH, Lee NM. 2010. Linking phylogenetic and functional diversity to nutrient spiraling in microbial mats from Lower Kane Cave (U. S. A.). ISME J. 4:98–110 [DOI] [PubMed] [Google Scholar]
- 6. Kelly DP. 2010. Global consequences of the microbial production and consumption of inorganic and organic sulfur compounds, p 3087–3095 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology Biomedical and Life Sciences, part 28. Springer, Heidelberg, Germany [Google Scholar]
- 7. Henckel T, Friedrich M, Conrad R. 1999. Molecular analysis of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane, monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Segawa T, Miyamoto K, Ushida K, Agata K, Okada N, Kohshima S. 2005. Seasonal change in bacterial flora and biomass in mountain snow from the Tateyama Mountains, Japan, analyzed by 16S rRNA gene sequencing and real-time PCR. Appl. Environ. Microbiol. 71:123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansel CM, Fendorf S, Jardine PM, Francis CA. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74:1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krishnani KK, Gopikrishna G, Pillai SM, Gupta BP. 2010. Abundance of sulphur-oxidizing bacteria in coastal aquaculture using soxB gene analysis. Aquaculture Res. 41:1290–1301 [Google Scholar]
- 11. Bannert A, Kleineidam K, Wissing L, Mueller-Niggemann C, Vogelsang V, Welzl G, Cao Z, Schloter M. 2011. Changes in diversity and functional gene abundances of microbial communities involved in nitrogen fixation, nitrification, and denitrification in a tidal wetland versus paddy soils cultivated for different time periods. Appl. Environ. Microbiol. 77:6109–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo JF, Lin WT, Guo Y. 2011. Functional gene based analysis of sulfur-oxidizing bacteria community in sulfide removing bioreactor. Appl. Microbiol. Biotechnol. 90:769–778 [DOI] [PubMed] [Google Scholar]
- 13. Galand PE, Bourrain M, De Maistre E, Catala P, Desdevises Y, Elifantz H, Kirchman DL, Lebaron P. 2012. Phylogenetic and functional diversity of Bacteria and Archaea in a unique stratified lagoon, the Clipperton atoll (N Pacific). FEMS Microbiol. Ecol. 79:203–217 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh W, Dam B. 2009. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33:999–1043 [DOI] [PubMed] [Google Scholar]
- 15. Meyer B, Imhoff JF, Kuever J. 2007. Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria–evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9:2957–2977 [DOI] [PubMed] [Google Scholar]
- 16. Gregersen LH, Bryant DA, Frigaard NU. 2011. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front. Microbiol. 2:116–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer J. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67:2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelly DP, Shergill JK, Lu WP, Wood AP. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71:95–107 [DOI] [PubMed] [Google Scholar]
- 19. Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, Prinz H. 2000. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182:4677–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rother D, Henrich HJ, Quentmeier A, Bardischewsky F, Friedrich CG. 2001. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein soxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J. Bacteriol. 183:4499–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rother D, Friedrich CG. 2002. The cytochrome complex SoxXA of Paracoccus pantotrophus is produced in Escherichia coli and functional in the reconstituted sulfur-oxidizing enzyme system. Biochim. Biophys. Acta 1598:65–73 [DOI] [PubMed] [Google Scholar]
- 22. Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. 2005. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8:253–259 [DOI] [PubMed] [Google Scholar]
- 23. Hensen D, Sperling D, Truper HG, Brune DC, Dahl C. 2006. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62:794–810 [DOI] [PubMed] [Google Scholar]
- 24. Petri R, Podgorsek L, Imhoff JF. 2001. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol. Lett. 197:171–178 [DOI] [PubMed] [Google Scholar]
- 25. Anandham R, Indiragandhi P, Madhaiyan M, Ryu KY, Jee HJ, Sa TM. 2008. Chemolithoautotrophic oxidation of thiosulfate and phylogenetic distribution of sulfur oxidation gene (soxB) in rhizobacteria isolated from crop plants. Res. Microbiol. 159:579–589 [DOI] [PubMed] [Google Scholar]
- 26. Kilmartin JR, Maher MJ, Krusong K, Noble CJ, Hanson GR, Bernhardt PV, Riley MJ, Kappler U. 2011. Insights into the structure and function of the active site of SoxAX cytochromes. J. Biol. Chem. 286:24872–24881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quentmeier A, Friedrich CG. 2001. The cysteine residue of the SoxY protein as the active site of the protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett. 503:168–172 [DOI] [PubMed] [Google Scholar]
- 28. Quentmeier A, Hellwig P, Bardischewsky F, Grelle G, Kraft R, Friedrich CG. 2003. Sulfur oxidation in Paracoccus pantotrophus: interaction of the sulfur-binding protein SoxYZ with the dimanganese SoxB protein. Biochem. Biophys. Res. Commun. 312:1011–1018 [DOI] [PubMed] [Google Scholar]
- 29. Sauve V, Bruno S, Berks BC, Hemmings AM. 2007. The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J. Biol. Chem. 282:23194–23204 [DOI] [PubMed] [Google Scholar]
- 30. Sauve V, Roversi P, Leath KJ, Garman EF, Antrobus R, Lea SM, Berks BC. 2009. Mechanism for the hydrolysis of a sulfur-sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J. Biol. Chem. 284:21707–21718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Epel B, Schäfer KO, Quentmeier A, Friedrich C, Lubitz W. 2005. Multifrequency EPR analysis of the dimanganese cluster of the putative sulfate thiohydrolase SoxB of Paracoccus pantotrophus. J. Biol. Inorg. Chem. 10:636–642 [DOI] [PubMed] [Google Scholar]
- 32. Bagchi A, Ghosh TC. 2005. A structural study towards the understanding of interactions of SoxY, SoxZ, and SoxB, leading to the oxidation of sulfur anions via the novel global sulfur oxidizing (sox) operon. Biochem. Biophys. Res. Commun. 335:609–615 [DOI] [PubMed] [Google Scholar]
- 33. Ogawa T, Furusawa T, Shiga M, Seo D, Sakurai H, Inoue K. 2010. Biochemical studies of a soxF-encoded monomeric flavoprotein purified from the green sulfur bacterium Chlorobaculum tepidum that stimulates in vitro thiosulfate oxidation. Biosci. Biotechnol. Biochem. 74:771–780 [DOI] [PubMed] [Google Scholar]
- 34. Azai C, Tsukatani Y, Harada J, Oh-oka H. 2009. Sulfur oxidation in mutants of the photosynthetic green sulfur bacterium Chlorobium tepidum devoid of cytochrome c-554 and SoxB. Photosynth. Res. 100:57–65 [DOI] [PubMed] [Google Scholar]
- 35. Welte C, Swetlana H, Kratzer C, Quentmeier A, Friedrich CG, Dahl C. 2009. Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett. 583:1281–1286 [DOI] [PubMed] [Google Scholar]
- 36. Oakley BB, Carbonero F, van der Gast CJ, Hawkins RJ, Purdy KJ. 2010. Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME J. 4:488–497 [DOI] [PubMed] [Google Scholar]
- 37. Hanson RL, Cates SW. 1994. Hydrology of the Chickasaw National Recreation Area, Murray County, Oklahoma: USGS Water-Resources Investigations Report 94–4102. U.S. Geological Survey, Reston, VA [Google Scholar]
- 38. Christenson S, Hunt AG, Parkhurst DL. 2009. Geochemical investigation of the Arbuckle-Simpson aquifer, south-central Oklahoma, 2004–06: U.S. Geological Survey Scientific Investigations Report 2009-5036. U.S. Geological Survey, Reston, VA [Google Scholar]
- 39. Christenson S, Hunt AG, Parkhurst DL, Osborn NI. 2009. Geochemistry of the Arbuckle-Simpson Aquifer: U.S. Geological Survey Fact Sheet 2009-3013. U.S. Geological Survey, Reston, VA [Google Scholar]
- 40. APHA, AWWA, WEF 2005. Standard methods for the examination of water and wastewater, 21st ed American Public Health Association, Washington, DC [Google Scholar]
- 41. Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43 doi:10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125 doi:10.1186/1471-2180-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 45. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed] [Google Scholar]
- 48. Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18:691–699 [DOI] [PubMed] [Google Scholar]
- 51. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony. Methods Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramette A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4 http://palaeo-electronica.org/2001_1/past/issue1_01.htm [Google Scholar]
- 54. Legendre P, Legendre L. 1998. Numerical ecology, 2nd ed Elsevier, Amsterdam, Netherlands [Google Scholar]
- 55. Cohen Y, Padan E, Shilo M. 1975. Facultative anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. J. Bacteriol. 123:855–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Padan E. 1979. Facultative anoxygenic photosynthesis in Cyanobacteria. Annu. Rev. Plant Physiol. 30:27–40 [Google Scholar]
- 57. Cohen Y, Jørgensen Revsbech BBNP, Poplawski R. 1986. Adaptation to hydrogen sulfide of oxygenic and anoxygenic photosynthesis among Cyanobacteria. Appl. Environ. Microbiol. 51:398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beller HR, Chain PSG, Letain TE, Chakicherla A, Larimer FW, Richardson PM, Coleman MA, Wood AP, Kelly DP. 2006. The genome sequences of the obligatory chemolithoautotrophic facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 188:1473–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huddleston JR, Zak JC, Jeter RM. 2006. Antimicrobial susceptibilities of Aeromonas spp. isolated from environmental sources. Appl. Environ. Microbiol. 72:7036–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oue S, Okamoto A, Yano T, Kagamiyama H. 1999. Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues. J. Biol. Chem. 274:2344–2349 [DOI] [PubMed] [Google Scholar]
- 61. Watney JB, Agarwal PK, Hammes-Schiffer S. 2003. Effect of mutation on enzyme motion in dihydrofolate reductase. J. Am. Chem. Soc. 125:3745–3750 [DOI] [PubMed] [Google Scholar]
- 62. Griesbeck C, Schütz M, Schödl T, Bathe S, Nausch L, Mederer N, Vielreicher M, Hauska G. 2002. Mechanism of sulfide-quinone reductase investigated using site-directed mutagenesis and sulfur analysis. Biochemistry 41:11552–11565 [DOI] [PubMed] [Google Scholar]
- 63. Otten LG, Sio CF, van der Sloot AM, Cool RH, Quax WJ. 2004. Mutational analysis of a key residue in the substrate specificity of a cephalosporin acylase. Chem. BioChem. 5:820–825 [DOI] [PubMed] [Google Scholar]
- 64. Swanwick RS, Shrimpton PJ, Allemann RK. 2004. Pivotal role of Gly 121 in dihydrofolate reductase from Escherichia coli: the altered structure of a mutant enzyme may be the basis of its diminished catalytic performance. Biochemistry 43:4119–4127 [DOI] [PubMed] [Google Scholar]
- 65. Pantoliano MW, Whitlow M, Wood JF, Dodd SW, Hardman KD, Rollence ML, Bryan PN. 1989. Large increases in general stability for Subtilisin BPN′ through incremental changes in the free energy of unfolding. Biochemistry 28:7205–7213 [DOI] [PubMed] [Google Scholar]
- 66. Friedrich CG, Mitrenga G. 1981. Oxidation of thiosulfate by Paracoccus denitrificans and other hydrogen bacteria. FEMS Microbiol. Lett. 10:209–212 [Google Scholar]
- 67. Appia-Ayme C, Little PJ, Matsumoto Y, Leech AP, Berks BC. 2001. Cytochrome complex essential for photosynthetic oxidation of both thiosulfate and sulfide in Rhodovulum sulfidophilum. J. Bacteriol. 183:6107–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Masuda S, Eda S, Ikeda S, Mitsui H, Minamisawa K. 2010. Thiosulfate-dependent chemolithoautotrophic growth of Bradyrhizobium japonicum. Appl. Environ. Microbiol. 76:2401–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Larkin JM, Shinabarger DL. 1983. Characterization of Thiothrix nivea. Int. J. Syst. Bacteriol. 33:841–846 [Google Scholar]
- 70. Imhoff JF, Suling J, Petri R. 1998. Phylogenetic relationships among the Chromatiaceae, their taxonomic reclassification and description of the new genera Allochromatium, Halochromatium, Isochromatium, Marichromatium, Thiococcus, Thiohalocapsa and Thermochromatium. Int. J. Syst. Bacteriol. 48:1129–1143 [DOI] [PubMed] [Google Scholar]
- 71. Sievert SM, Heidorn T, Kuever J. 2000. Halothiobacillus kellyi sp. nov., a mesophilic, obligately chemoautotrophic, sulfur-oxidizing bacterium isolated from a shallow-water hydrothermal vent in the Aegean Sea, and emended description of the genus Halothiobacillus. Int. J. Syst. Bacteriol. 50:1229–1237 [DOI] [PubMed] [Google Scholar]
- 72. Chen XG, Geng AL, Yan R, Gould WD, Ng YL, Liang DT. 2004. Isolation and characterization of sulphur-oxidizing Thiomonas sp. and its potential application in biological deodorization. Int. J. Syst. Bacteriol. 39:495–503 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.