Abstract
We previously demonstrated efficient l-valine production by metabolically engineered Corynebacterium glutamicum under oxygen deprivation. To achieve the high productivity, a NADH/NADPH cofactor imbalance during the synthesis of l-valine was overcome by engineering NAD-preferring mutant acetohydroxy acid isomeroreductase (AHAIR) and using NAD-specific leucine dehydrogenase from Lysinibacillus sphaericus. Lactate as a by-product was largely eliminated by disrupting the lactate dehydrogenase gene ldhA. Nonetheless, a few other by-products, particularly succinate, were still produced and acted to suppress the l-valine yield. Eliminating these by-products therefore was deemed key to improving the l-valine yield. By additionally disrupting the phosphoenolpyruvate carboxylase gene ppc, succinate production was effectively suppressed, but both glucose consumption and l-valine production dropped considerably due to the severely elevated intracellular NADH/NAD+ ratio. In contrast, this perturbed intracellular redox state was more than compensated for by deletion of three genes associated with NADH-producing acetate synthesis and overexpression of five glycolytic genes, including gapA, encoding NADH-inhibited glyceraldehyde-3-phosphate dehydrogenase. Inserting feedback-resistant mutant acetohydroxy acid synthase and NAD-preferring mutant AHAIR in the chromosome resulted in higher l-valine yield and productivity. Deleting the alanine transaminase gene avtA suppressed alanine production. The resultant strain produced 1,280 mM l-valine at a yield of 88% mol mol of glucose−1 after 24 h under oxygen deprivation, a vastly improved yield over our previous best.
INTRODUCTION
The branched-chain amino acid (BCAA) l-valine is essential for vertebrates. It is used in dietary products, pharmaceuticals, cosmetics, and as a precursor of antibiotics or herbicides, and it is expected to play a leading role in future feed additives (1, 2). l-Valine has been produced to date by bacterial fermentation, mainly using engineered Corynebacterium glutamicum and Escherichia coli. Several approaches to develop efficient l-valine-producing strains in order to meet the growing world market are well known (2–4). The l-valine synthesis pathway has recently attracted attention, because 2-ketoisovalerate, a direct precursor of l-valine (Fig. 1), is also used as a precursor of isobutanol, the expected mainstay of future renewable biofuels (5), in an Ehrlich pathway (6–8).
Fig 1.
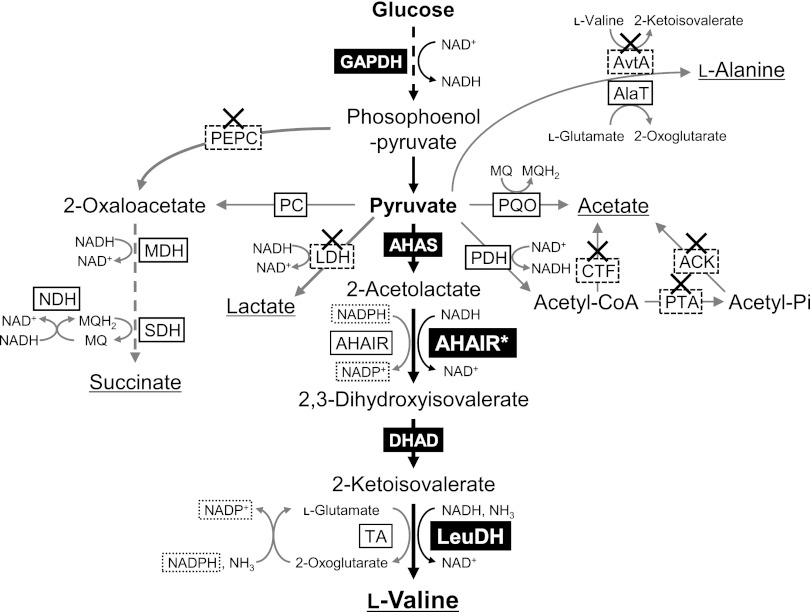
Biosynthetic pathway of l-valine and by-products of C. glutamicum under oxygen deprivation. Enzymes whose genes were overexpressed are highlighted in black boxes, and enzymes whose genes were disrupted are marked with an X. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PEPC, phosphoenolpyruvate carboxylase; PC, pyruvate carboxylase; MDH, malate dehydrogenase; SDH, succinate dehydrogenase; NDH, NADH dehydrogenase; MQ, menaquinone; LDH, lactate dehydrogenase; AlaT, alanine transaminase encoded by alaT; AvtA, alanine transaminase encoded by avtA; PDH, pyruvate dehydrogenase; PQO, pyruvare:quinone oxidoreductase; CTF, CoA transferase; PTA, phosphotransacetylase; ACK, acetate kinase; AHAS, acetohydroxy acid synthase; AHAIR, acetohydroxy acid isomeroreductase (AHAIR*, NAD-preferring mutant); DHAD, dihydroxy acid dehydratase; TA, transaminase B; LeuDH, leucine dehydrogenase (L. sphaericus).
As shown in Fig. 1, l-valine in C. glutamicum is synthesized from pyruvate in a series of four successive reactions catalyzed by ilvBN-encoded acetohydroxy acid synthase (AHAS), ilvC-encoded acetohydroxy acid isomeroreductase (AHAIR), ilvD-encoded dihydroxy acid dehydratase, and ilvE-encoded transaminase B (2, 9). The other BCAAs, l-leucine and l-isoleucine, as well as d-pantothenate, are synthesized through the same pathway (2). The key enzyme regulating synthesis of BCAAs is AHAS, which is subject to feedback inhibition and transcriptional attenuation by all BCAAs (2). Given this knowledge, improved l-valine-producing strains have been constructed by overexpressing the genes encoding l-valine synthesis enzymes, derepressing feedback inhibition of AHAS, and disrupting by-product-synthesizing pathways (1, 2, 4, 10).
Stoichiometrically, 1 mol of l-valine is produced from 1 mol of glucose. However, there exists a cofactor imbalance in the overall l-valine synthesis pathway; 2 mol of NADH is produced via glycolysis (in a glyceraldehyde-3-phosphate dehydrogenase [GAPDH] reaction), whereas 2 mol of NADPH is consumed for l-valine synthesis (AHAIR reaction and the regeneration of l-glutamate [11] as an amino-group donor for the transaminase reaction [12, 13]) (Fig. 1). The resultant surplus NADH and insufficient NADPH are suspected to suppress l-valine production (4, 14, 15). To overcome this, a few strategies, including enhanced pentose phosphate pathway flux to increase NADPH supply (4), introduction of pyridine nucleotide transhydrogenase to regenerate NADPH by hydride ion transfer from NADH (7, 8, 14), and replacement of the native NADP-dependent enzymes with NAD-dependent homologs or engineered enzymes (8, 16), have permitted notably improved l-valine-producing strains.
We have recently attained efficient l-valine production by engineered C. glutamicum strains under oxygen deprivation (16). When deprived of oxygen, C. glutamicum cells sustain metabolic activity despite arrested cell growth and thus are able to catabolize sugars and produce organic acids in order to reoxidize NADH generated via glycolysis (17). This ability underlies several high-productivity and high-yielding production processes of disparate chemicals (7, 18–21). However, in the case of l-valine production under oxygen deprivation, the cofactor imbalance is a critical problem, because generated NADH cannot be oxidized by the synthesis of l-valine, leading to the elevated intracellular NADH/NAD+ ratio that decreases glycolytic flux (18, 22, 23). We circumvented this problem by switching cofactor requirement of AHAIR from NADPH to NADH and introducing NAD-specific leucine dehydrogenase (LeuDH) derived from Lysinibacillus sphaericus (24) instead of transaminase (Fig. 1). As a result, the intracellular redox state of the constructed strain BNGECTMDLD/ΔLDH has significantly improved, and unprecedented l-valine productivity of 1,470 mM after 24 h became possible (16). However, considerable amounts of by-products were still produced, and l-valine yield was suppressed up to 63% mol mol−1 from consumed glucose. The present study describes the modifications that enable suppressed by-product formation without compromising the high l-valine productivity under oxygen-deprived conditions.
MATERIALS AND METHODS
Bacterial strains, media, and cultivation conditions.
Bacterial strains used in this study are listed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani medium (25). C. glutamicum R (JCM 18229) (26) and its recombinants were grown at 33°C in complex medium (A medium) or minimal medium (BT medium) with 4% glucose (18). The final concentrations of kanamycin and chloramphenicol used were 50 μg ml−1 for each for E. coli and 50 and 5 μg ml−1, respectively, for C. glutamicum.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi hsdR17 (rK− mK+) e14− (mcrA) supE44 relA1 Δ(lac-pro0AB)/F′ (traD36 proAB+ lacIq lacZΔM15) | TaKaRa Bio Inc. |
| JM110 | dam dcm supE44 hsdR17 thi leu rpsL1 lacY galK galT ara tonA thr tsx Δ(lac-proAB)/F′ (traD36 proAB+ lacIq lacZΔM15) | 25 |
| C. glutamicum R JCM 18229 | 26 | |
| ΔLP | Markerless ldhA and ppc double deletion mutant | 19 |
| ΔLP_ΔAc | ΔLP with markerless deletion of pta, ackA, and ctfA (acetate synthesis pathway) | This work |
| ΔLP_+GP | ΔLP with induction of gapA, pyk, pfk, pgi, and tpi (glycolytic pathway) | This work |
| ΔLP_ΔAc+GP | ΔLP with markerless deletion of pta, ackA, and ctfA and induction of gapA, pyk, pfkA, pgi, and tpi | This work |
| ΔLP_ΔAc+GP_ilvNGE | ΔLP_ΔAc+GP with replacement of wild-type ilvN with ilvNGE | This work |
| ΔLP_ΔAc+GP_ilvCTM | ΔLP_ΔAc+GP with replacement of wild-type ilvC with ilvCTM | This work |
| ΔLP_ΔAc+GP_ilvNGECTM | ΔLP_ΔAc+GP with replacement of wild-type ilvNC with ilvNGECTM | This work |
| ΔLP_ΔAc+GP_ilvNGECTM_ΔAla | ΔLP_ΔAc+GP_ilvNGECTM with markerless deletion of avtA | This work |
| Val-1 (BNCTMDLD/ΔLP) | ΔLP harboring pCRB-BNCTM and pCRB-DLD | This work |
| Val-2 (BNCTMDLD/ΔLP_ΔAc) | ΔLP_ΔAc harboring pCRB-BNCTM and pCRB-DLD | This work |
| Val-3 (BNCTMDLD/ΔLP_+GP) | ΔLP_+GP harboring pCRB-BNCTM and pCRB-DLD | This work |
| Val-4 (BNCTMDLD/ΔLP_ΔAc+GP) | ΔLP_ΔAc+GP harboring pCRB-BNCTM and pCRB-DLD | This work |
| Val-5 (BNGECTMDLD/ΔLP_ΔAc+GP) | ΔLP_ΔAc+GP harboring pCRB-BNGECTM and pCRB-DLD | This work |
| Val-6 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGE) | ΔLP_ΔAc+GP_ilvNGE harboring pCRB-BNGECTM and pCRB-DLD | This work |
| Val-7 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvCTM) | ΔLP_ΔAc+GP_ilvCTM harboring pCRB-BNGECTM and pCRB-DLD | This work |
| Val-8 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM) | ΔLP_ΔAc+GP_ilvNGECTM harboring pCRB-BNGECTM and pCRB-DLD | This work |
| Val-9 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM_ΔAla) | ΔLP_ΔAc+GP_ilvNGECTM_ΔAla harboring pCRB-BNGECTM and pCRB-DLD | This work |
| Plasmids | ||
| pCRA725 | Ptac-sacR-sacB in pHSG298 | 19 |
| pCRB-BNCTM | PgapA-ilvBNCTM (C. glutamicum) in pCRB21 | 16 |
| pCRB-BNGECTM | PgapA-ilvBNGECTM (C. glutamicum) in pCRB21 | 16 |
| pCRB-DLD | Ptac-ilvD (C. glutamicum) and Ptac-LeuDH gene (L. sphaericus) in pCRB12 | 16 |
ilvNGE, feedback-resistant mutant ilvN (G156E); ilvCTM, NAD-preferring mutant ilvC (S34G, L48E, R49F).
Construction of plasmids and strains.
General DNA manipulations were performed as described earlier (25). Primers, plasmids, and strains used and constructed in this study are listed in Tables 1 and 2. Chromosomal gene deletion, insertion, and substitution were achieved via a markerless system using suicide vector pCRA725 carrying the sacB gene (19, 27, 28). Plasmids were transformed into C. glutamicum by electroporation (29).
Table 2.
Oligonucleotides used in this study
| Primer | Target gene | Sequencea (5′–3′) | Restriction site |
|---|---|---|---|
| Primer1 | SSI1-2 region | CTCTGTCGACTGACCATCACCAATGCTGTG | SalI |
| Primer2 | CTCTGTCGACAAGTTGTCACCGTCTTCGGT | SalI | |
| Primer3 | tpi | GCCCGAATTCATGGCACGTAAGCCAC | EcoRI |
| Primer4 | GCCCGAATTCTTAAGCGACGCTCGCA | EcoRI | |
| Primer5 | ilvNGE | CTCTGCATGCGTTTCCAGATGACCAACCAG | SphI |
| Primer6 | CTCTTCTAGATTAAGCGGTTTCTGCGCGA | XbaI | |
| Primer7 | ilvCTM | CTCTGCATGCGTTTCCAGATGACCAACCAG | SphI |
| Primer8 | CTCTTCTAGATTAAGCGGTTTCTGCGCGA | XbaI | |
| Primer9 | ilvNGECTM | CTCTGCATGCGTTTCCAGATGACCAACCAG | SphI |
| Primer10 | CTCTTCTAGATTAAGCGGTTTCTGCGCGA | XbaI | |
| Primer11 | avtA | CTCTGTCGACAACTTTGGCAGAGCAGCC | SalI |
| Primer12 | CTCTGTCGACTTGTGGATGTGCTCATGGC | SalI | |
| Primer13b | CTCTACTAGTTCTCTTTCCTTGTGTGCTCC | SpeI | |
| Primer14b | CTCTACTAGTAGGCATTGCCACATAATCCC | SpeI |
The restriction site used in the cloning procedure is underlined.
Primers used for inverse amplification to truncate a middle region of avtA.
Measurement of AHAS activity.
Crude extracts of the cells for enzyme assay were prepared using disruption buffer (100 mM potassium phosphate [pH 7.3], 0.5 mM dithiothreitol, and 20% [vol/vol] glycerol) according to previous reports (9, 16). AHAS activity was determined at 30°C by monitoring the pyruvate decrease at 333 nm (ε = 17.5 M−1 cm−1) (30). The reaction mixture contained 100 mM potassium phosphate (pH 7.5), 50 mM sodium pyruvate, 10 mM MgCl2, 0.1 mM thiamine pyrophosphate, 0.1 mM flavin adenine dinucleotide, and crude extract (16).
Conditions for l-valine production.
C. glutamicum strains were aerobically cultivated at 33°C for 14 h in A medium with 4% glucose to an optical density at 610 nm (OD610) of 6.0 to 9.0 from 0.1. Harvested cells were washed with BT medium without urea (BT-U medium) and resuspended in 50 ml BT-U medium containing 200 to 400 mM glucose. This cell suspension was incubated at 33°C with no aeration but with gentle agitation. The pH of the reaction solution was maintained at 7.5 by supplementing with NH3 (18). When necessary, glucose in the reaction solution was replenished before its depletion. Due to the addition of NH3 solution and glucose, the reaction volume significantly increased throughout the reaction for l-valine production. Glucose, l-valine, and by-product concentrations thus were calculated to correct for the increased volume and consequent dilution.
Analytical procedures.
Glucose concentration was measured by an enzyme electrode glucose sensor (BF-4; Oji Scientific Instruments, Hyogo, Japan). Organic acid concentrations were determined by high-performance liquid chromatography (HPLC) (8020; Tosoh Corporation, Tokyo, Japan) equipped with TSKgel OApak-A (Tosoh Corporation). Amino acid concentrations were determined by HPLC (Prominence 20A; Shimazu Corporation, Kyoto, Japan) equipped with Shim-pack Amino-Na (Shimazu Corporation) after derivatization with ο-phthalaldehyde. Intracellular amino acids were extracted by silicone oil centrifugation and sonication as described elsewhere (31). Intracellular metabolites, including NAD(H), were extracted from C. glutamicum cells by using cold ethanol and chloroform according to previously described protocols (16). The resultant extracts were analyzed by LC-tandem mass spectrometry consisting of HPLC (Prominence 20A) and a linear ion trap mass spectrometer (4000 Q TRAP; Applied Biosystems/MDS SCIEX) (32). A factor of 1.8 ml/g cell dry weight was assumed as the cell volume for the calculation of intracellular concentrations. Statistical analysis was performed by using Welch's t test for proper comparison between data measured. Significance was accepted at P < 0.05.
RESULTS
Deletion of the ppc gene leads to suppressed succinate production at the expense of glucose consumption.
Under oxygen deprivation, C. glutamicum produces lactate in the major reaction that reoxidizes the NADH generated via glycolysis (Fig. 1), and disruption of the ldhA gene encoding lactate dehydrogenase (LDH) eliminates almost all of the lactate production (18, 19). However, to compensate for this loss of NADH reoxidizing power in ΔLDH-based l-valine-producing strains, considerable amounts of succinate are produced, permitting NADH reoxidization via reactions catalyzed by malate dehydrogenase (MDH) and succinate dehydrogenase (SDH) in the reductive tricarboxylic acid (TCA) cycle (16). Since succinate is predominantly produced via phosphoenolpyruvate carboxylase (PEPC), which is encoded by the ppc gene, under oxygen-deprived conditions (18, 19), we constructed a ldhA-ppc double deletion mutant, designated ΔLP (Table 1), to suppress by-product formation of succinate in addition to lactate. l-Valine-producing strains were constructed by overexpressing ilvBN (wild-type ilvBN or the G156E mutant [ilvBNGE]), ilvCTM (corresponding to S34G, L48E, and R49F mutations), ilvD, and the LeuDH gene in ΔLP using the respective plasmids (Table 1 and Fig. 1) in the same manner as that used for ΔLDH-based strains constructed previously (16). ilvBNGE and ilvCTM encode feedback-resistant mutant AHAS (IlvBNGE) and NAD-preferring mutant AHAIR (IlvCTM), respectively. LeuDH effectively catalyzes the reversible deamination of l-leucine and the other BCAAs, l-valine and l-isoleucine, using NAD+ as a cofactor (Fig. 1) (24). The inherent cofactor imbalance in the l-valine synthesis pathway (2, 9) is solved by the use of NAD-preferring mutant AHAIR (IlvCTM) and NAD-specific LeuDH (Fig. 1).
Succinate production by the resultant strain Val-1 (BNCTMDLD/ΔLP; Table 1), at 7.1% from consumed glucose after 24 h under oxygen-deprived conditions (Table 3), was 3-fold less than the 23.4% produced by the ΔLDH-based BNCTMDLD/ΔLDH (16). However, glucose consumption and l-valine production of Val-1, at 166 and 106 mM, respectively (Table 3), were less than one-tenth of the 2,160 and 1,170 mM obtained with BNCTMDLD/ΔLDH (16). Nevertheless, l-valine yield improved from 54.1 to 64.0% on the strength of the reduced by-product formation in Val-1.
Table 3.
l-Valine production, l-valine and by-product yield on glucose, and glucose consumption of C. glutamicum strains after 24 h under oxygen deprivation
| Strain | Production of: |
Glucose consumptiona (mM) | |||||
|---|---|---|---|---|---|---|---|
|
l-Valine |
Alanineb (%) | Lactateb (%) | Succinateb (%) | Acetateb (%) | |||
| mMa | %b | ||||||
| ΔLP | 4.4 ± 1.8 | 10.7 ± 1.8 | 21.5 ± 2.3 | 10.1 ± 1.4 | 3.8 ± 0.3 | 6.8 ± 0.4 | 40 ± 10 |
| Val-1 (BNCTMDLD/ΔLP) | 106 ± 26 | 64.0 ± 1.1 | 10.0 ± 0.5 | 1.6 ± 0.5 | 7.1 ± 1.5 | 4.8 ± 0.2 | 166 ± 42 |
| Val-2 (BNCTMDLD/ΔLP_ΔAc) | 332 ± 87 | 70.6 ± 1.1 | 11.7 ± 2.2 | 0.9 ± 0.2 | 8.6 ± 1.2 | 1.6 ± 0.2 | 472 ± 129 |
| Val-3 (BNCTMDLD/ΔLP_+GP) | 956 ± 58 | 75.5 ± 2.1 | 5.8 ± 0.6 | 0.9 ± 0.0 | 9.0 ± 0.5 | 1.5 ± 0.1 | 1,270 ± 70 |
| Val-4 (BNCTMDLD/ΔLP_ΔAc+GP) | 1,120 ± 70 | 74.7 ± 1.0 | 5.5 ± 0.5 | 0.7 ± 0.0 | 10.1 ± 0.7 | 1.0 ± 0.0 | 1,500 ± 80 |
| Val-5 (BNGECTMDLD/ΔLP_ΔAc+GP) | 902 ± 33 | 86.4 ± 0.8 | 2.4 ± 0.3 | 1.1 ± 0.0 | 5.7 ± 0.2 | 0.6 ± 0.0 | 1,040 ± 30 |
| Val-6 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGE) | 822 ± 28 | 85.8 ± 1.0 | 2.7 ± 0.3 | 1.1 ± 0.0 | 5.3 ± 0.2 | 0.6 ± 0.0 | 958 ± 29 |
| Val-7 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvCTM) | 1,210 ± 60 | 84.0 ± 0.8 | 1.8 ± 0.1 | 0.8 ± 0.1 | 4.4 ± 0.2 | 0.6 ± 0.0 | 1,440 ± 80 |
| Val-8 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM) | 1,250 ± 50 | 86.4 ± 1.0 | 2.6 ± 0.3 | 0.8 ± 0.1 | 4.6 ± 0.3 | 0.6 ± 0.0 | 1,440 ± 50 |
| Val-9 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM_ΔAla) | 1,280 ± 100 | 88.4 ± 0.8 | 0.4 ± 0.1 | 0.8 ± 0.1 | 4.6 ± 0.4 | 0.6 ± 0.0 | 1,440 ± 110 |
l-Valine production and glucose consumption values were corrected for dilution caused by the addition of NH3 solution and glucose throughout the reaction.
Yield is expressed as a percentage of the theoretical yield (100% means 1 mol of l-valine or 2 mol of alanine and organic acids produced per 1 mol of glucose consumed). Cell concentration was 40 g cell dry weight liter−1. The data represent an average of three independent experiments.
The intracellular metabolite analysis revealed that metabolites upstream of GAPDH (dihydroxyacetone phosphate [DHAP] and glyceraldehyde-3-phosphate [GAP]) are more abundant in Val-1 (10.72 and 0.85 mM, respectively [Table 4]) than the corresponding ΔLDH strain BNCTMDLD/ΔLDH (2.98 and 0.26 mM [16]). Conversely, metabolites downstream of GAPDH (1,3-bisphosphoglycerate [BPG] and pyruvate) are less abundant in Val-1 (0.05 mM and below the detection limit [Table 4]) than those in BNCTMDLD/ΔLDH (0.17 and 21.2 mM [16]). These results suggest that GAPDH activity in Val-1 was comparatively inhibited. Moreover, the intracellular NADH/NAD+ ratio of 4.14 in Val-1 (Table 5) is an order of magnitude higher than the ratio (0.52) in BNCTMDLD/ΔLDH (16). Therefore, the drastic reduction in glucose consumption and l-valine production of Val-1 under oxygen deprivation must result from the negative effect of a high NADH/NAD+ ratio on GAPDH activity upon deletion of both ldhA and ppc despite the engineered l-valine synthesis pathway in Val-1 to reoxidize NADH.
Table 4.
Intracellular concentration of metabolic intermediates from glycolysis at 2 h during l-valine production under oxygen deprivation
| Strain | Metabolite concna (mM) |
|||
|---|---|---|---|---|
| DHAP | GAP | BPG | Pyruvate | |
| Val-1 (BNCTMDLD/ΔLP) | 10.72 ± 1.86 | 0.85 ± 0.12 | 0.05 ± 0.00 | Not detected |
| Val-2 (BNCTMDLD/ΔLP_ΔAc) | 6.02 ± 0.71 | 0.44 ± 0.07 | 0.17 ± 0.01 | Not detected |
| Val-3 (BNCTMDLD/ΔLP_+GP) | 8.17 ± 0.51 | 0.45 ± 0.02 | 0.21 ± 0.01 | 0.97 ± 0.09 |
| Val-4 (BNCTMDLD/ΔLP_ΔAc+GP) | 6.84 ± 0.33 | 0.52 ± 0.05 | 0.19 ± 0.02 | 18.03 ± 2.75 |
| Val-5 (BNGECTMDLD/ΔLP_ΔAc+GP) | 8.33 ± 0.38 | 0.62 ± 0.05 | 0.13 ± 0.01 | Not detected |
| Val-6 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGE) | 10.77 ± 0.59 | 0.64 ± 0.03 | 0.12 ± 0.03 | Not detected |
| Val-7 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvCTM) | 7.03 ± 1.35 | 0.56 ± 0.07 | 0.19 ± 0.02 | Not detected |
| Val-8 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM) | 7.41 ± 0.75 | 0.83 ± 0.14 | 0.17 ± 0.02 | Not detected |
| Val-9 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM_ΔAla) | 7.40 ± 0.83 | 0.78 ± 0.10 | 0.23 ± 0.01 | 4.91 ± 0.57 |
DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde-3-phosphate; BPG, 1,3-bisphosphoglycerate. The data represented are from five analytical replicates from one experiment.
Table 5.
Intracellular NADH/NAD+ ratio at 2 h during l-valine production under oxygen deprivation
| Strain | NADH/NAD+ ratioa |
|---|---|
| Val-1 (BNCTMDLD/ΔLP) | 4.14 ± 0.27 |
| Val-2 (BNCTMDLD/ΔLP_ΔAc) | 3.49 ± 0.28 |
| Val-3 (BNCTMDLD/ΔLP_+GP) | 2.58 ± 0.30 |
| Val-4 (BNCTMDLD/ΔLP_ΔAc+GP) | 0.84 ± 0.11 |
| Val-5 (BNGECTMDLD/ΔLP_ΔAc+GP) | 1.28 ± 0.15 |
| Val-6 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGE) | 1.94 ± 0.17 |
| Val-7 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvCTM) | 0.93 ± 0.16 |
| Val-8 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM) | 0.95 ± 0.24 |
| Val-9 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM_ΔAla) | 0.98 ± 0.25 |
The data represented are from five analytical replicates from one experiment.
l-Valine production is enhanced by erasing acetate synthesis.
Besides succinate, acetate by-product formation is another important target to suppress in l-valine production. Phosphotransacetylase (PTA), acetate kinase (ACK), and coenzyme A (CoA) transferase (CTF) (Fig. 1), encoded by the pta, ackA, and ctfA genes, respectively, dominate acetate synthesis under oxygen deprivation (28). Their deletion resulted in strain Val-2 (BNCTMDLD/ΔLP_ΔAc; Table 1), of which acetate production decreased 3-fold to 1.6% from consumed glucose compared to that of Val-1 after 24 h under oxygen-deprived conditions (Table 3). l-Valine yield of Val-2 improved to 70.6% from 64.0% for Val-1.
Furthermore, the disruption increased glucose consumption to 472 mM and l-valine production to 332 mM, each representing a 3-fold increase over the corresponding measurements for Val-1 (Table 3). The intracellular NADH/NAD+ ratio in Val-2 was reduced to 3.49 from 4.12 in Val-1 (Table 5), and inhibition of the GAPDH reaction was lessened as evidenced by decreased DHAP and GAP and increased BPG (Table 4). In effect, the higher l-valine productivity and yield of Val-2 relative to Val-1 resulted from the combined effects of suppressed acetate production and increased glucose consumption due to a more enabling intracellular redox state.
Overexpressing glycolytic genes increases glucose consumption and l-valine production.
To address the reduced glucose consumption under oxygen deprivation in the ΔLDHΔPEPC background, we chromosomally integrated and overexpressed glycolytic pathway genes gapA, pyk, pfkA, pgi, and tpi in accordance with our previous findings (20, 27). These genes encode GAPDH, pyruvate kinase, phosphofructokinase, phosphoglucose isomerase, and triosephosphate isomerase, respectively (Table 1). The resultant strain Val-3 (BNCTMDLD/ΔLP_+GP; Table 1) was characterized by enhanced glycolysis. Its glucose consumption and l-valine production increased 7.6-fold to 1,270 mM and 9.0-fold to 956 mM, respectively, over the corresponding measurements for Val-1 after 24 h under oxygen deprivation (Table 3). The l-valine yield of Val-3 improved to 75.5% from 64.0% for Val-1. Its alanine and acetate formations decreased to half and one-third, respectively. In addition, the intracellular NADH/NAD+ ratio in Val-3 was reduced to 2.58 from 4.14 in Val-1 (Table 5). The observed increase in l-valine production of Val-3 thus was attributed not only to the strain's enhanced glycolytic enzyme activities but also to the resultant reduction in the intracellular redox ratio.
We subsequently constructed strain Val-4 (BNCTMDLD/ΔLP_ΔAc+GP; Table 1) and confirmed that the benefits accruing from the enhanced glycolytic pathway and those derived from the deleted acetate synthesis pathway worked simultaneously; the intracellular NADH/NAD+ ratio in Val-4 markedly reduced to 0.84 (Table 5), and glucose consumption increased 9.0-fold to 1,500 mM and l-valine production 10.6-fold to 1,120 mM, respectively, over the corresponding measurements for Val-1 (Table 3). Its l-valine yield of 74.7% was virtually unchanged from that of Val-3.
Feedback-resistant mutant AHAS increases l-valine yield but decreases l-valine production.
Strains Val-1 through Val-4 all overexpress native AHAS that is susceptible to feedback inhibition. As derepression of this feedback inhibition of AHAS has been shown to increase l-valine production (2, 10), a feedback-resistant mutant AHAS (IlvBNGE), which was previously described (16), was overexpressed in strain Val-5 (BNGECTMDLD/ΔLP_ΔAc+GP; Table 1) in place of wild-type AHAS. However, glucose consumption and l-valine production of Val-5 decreased relative to the corresponding measurements for Val-4 (Table 3). On the other hand, the l-valine yield of Val-5 improved to 86.4% from 74.7% for Val-4, and the formation of by-products alanine, acetate, and succinate decreased (Table 3). The intracellular metabolite analysis shows reduced intracellular pyruvate to a level below its detection limit (Table 4), and a raised intracellular NADH/NAD+ ratio to 1.28 from the level in Val-4 (0.84) (Table 5). The effect of mutant AHAS therefore was to efficiently catalyze conversion of pyruvate to 2-acetolactate. This conversion depleted intracellular pyruvate, directly improving the l-valine yield while simultaneously suppressing by-products derived from pyruvate. Conversely, the elevated NADH/NAD+ ratio decreased glucose consumption and, consequently, l-valine production of Val-5.
Replacement of chromosomal wild-type ilvN and ilvC with their respective mutants enhances l-valine production.
Unless expressly stated otherwise, strains Val-1 through Val-5 rely exclusively on plasmids to overexpress target enzymes (Table 1). They all inevitably retain wild-type ilvN and ilvC genes on their chromosome, from which wild-type AHAS and AHAIR may be expressed. Even in strains where mutant versions of both were overexpressed (16), small amounts of the chromosome-based wild-type enzyme activities might unfavorably affect l-valine production. To eliminate such a possibility, the chromosomal wild-type ilvN and ilvC were replaced with feedback-resistant mutant ilvNGE and NAD-preferring mutant ilvCTM, respectively (Table 1).
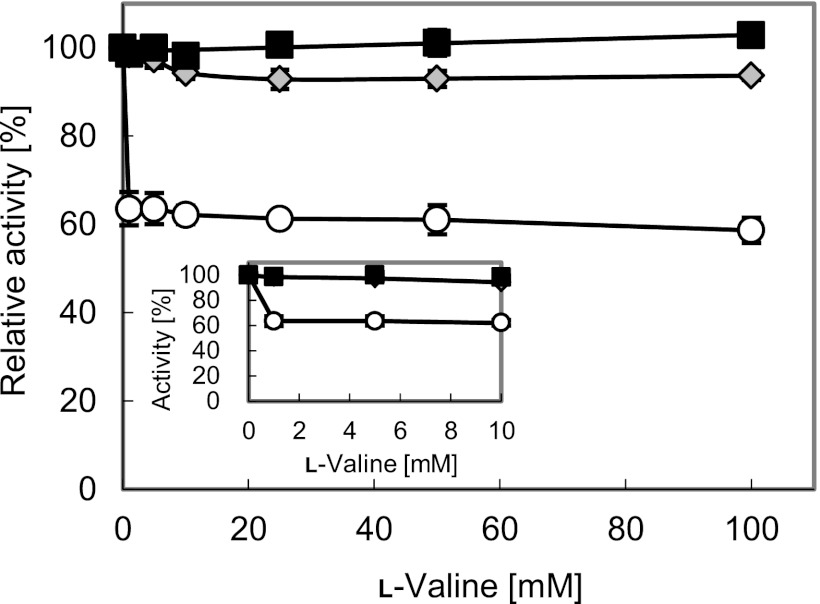
Strain Val-6 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGE; Table 1), in which native chromosomal ilvN was replaced, expressed mutant AHAS only. Figure 2 shows sensitivity to l-valine inhibition of AHAS activities, which were derived from strains expressing the wild type and the mutant in different ratios. While the mutant AHAS activity of Val-6 was barely inhibited by l-valine, the wild-type AHAS activity expressed by Val-4 decreased to ca. 60% even at low l-valine concentrations. The AHAS activity of Val-5, which consisted of overexpressed mutant AHAS via plasmid but also small amounts of the chromosomal wild type, was marginally inhibited by l-valine (activity decreased to ca. 95%). These results confirm that chromosomally replacing wild-type AHAS with the mutant is effective in derepressing product inhibition despite overexpression of the mutant. However, as shown in Table 3, glucose consumption and l-valine production under oxygen deprivation of Val-6 were decreased because of the elevated intracellular NADH/NAD+ ratio to 1.94 from 1.28 for Val-5 (Table 5), as well as Val-5 overexpressing the mutant AHAS. Moreover, unlike Val-5, the l-valine yield of Val-6 was not improved.
Fig 2.
Relative activity of the wild-type AHAS (IlvBN) and the mutant (IlvBNGM) in the presence of l-valine. Open circles, Val-4: IlvBN[plasmid]/IlvBN[chromosome]; gray diamonds, Val-5: IlvBNGE[plasmid]/IlvBN[chromosome]; black squares, Val-6: IlvBNGE[plasmid]/IlvBNGE[chromosome] (the location of ilvBN [wild type or mutant] on each strain is shown in the brackets). The inset shows the relative activity of AHAS in the presence of lower concentrations of l-valine. The data represent averages from three independent experiments.
In strain Val-7 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvCTM; Table 1), in which chromosomal ilvC was replaced, glucose consumption increased 1.4-fold to 1,440 mM and l-valine production 1.3-fold to 1,210 mM over the corresponding measurements for Val-5 after 24 h under oxygen deprivation (Table 3). Added to the reduction in the intracellular NADH/NAD+ ratio from 1.28 to 0.93 (Table 5), the changes suggest that the more weakly expressed chromosomal wild-type AHAIR affected the intracellular redox state. On the other hand, the l-valine yield of Val-7 was slightly decreased to 84.0% from 86.4% for Val-5.
Finally, strain Val-8 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM; Table 1), in which each of chromosomal wild-type ilvN and ilvC was replaced with its respective mutant, was constructed. The intracellular NADH/NAD+ ratio in Val-8 of 0.95 was as low as that in Val-7, and its glucose consumption and l-valine production, at 1,440 and 1,250 mM, respectively, were as high as those of Val-7 (Tables 3 and 5). Moreover, its l-valine yield of Val-8, at 86.4%, was as high as that of Val-5. It follows that mutant ilvNGE and ilvCTM on the chromosome had a cooperative effect, retaining the high l-valine production attributed to ilvCTM while maintaining the high l-valine yield attributed to ilvNGE under oxygen deprivation.
Deficiency of alanine synthesis enzyme suppresses alanine by-product formation, and l-valine productivity and yield are further enhanced.
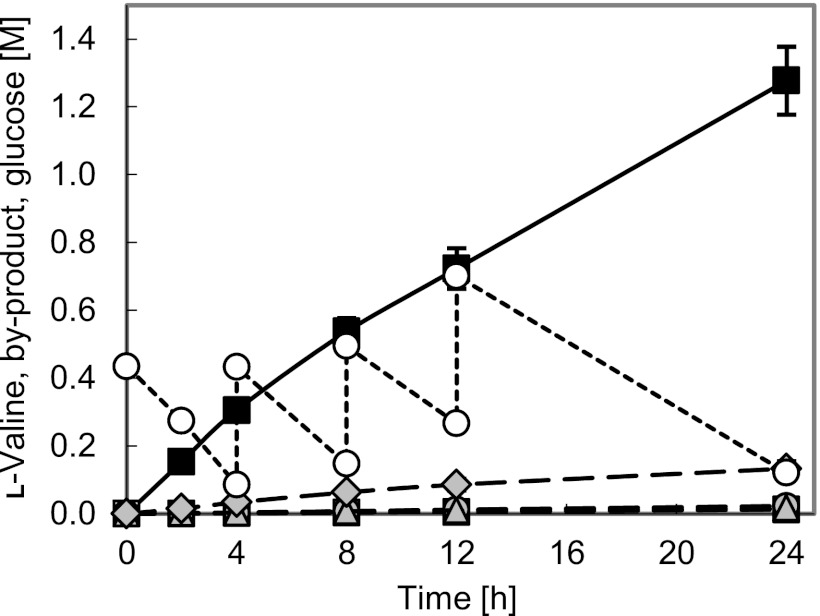
We additionally targeted alanine by-product formation to suppress production and evaluated the effect of deletion of the avtA gene encoding an alanine transaminase AvtA (Fig. 1) on l-valine production under oxygen deprivation. Accordingly, we constructed the avtA-deficient strain Val-9 (BNGECTMDLD/ΔLP_ΔAc+GP_ilvNGECTM_ΔAla; Table 1). As shown in Table 3, its alanine production decreased from 2.6 to 0.4% while its l-valine yield was further improved from 86.4 to 88.4% under oxygen deprivation, demonstrating that AvtA dominates alanine synthesis under the reaction conditions. Intracellularly detected pyruvate in Val-9, unlike that in avtA-active Val-8 (Table 4), would also result from the suppression of alanine synthesis. The l-valine production of Val-9 reached 1,280 mM after 24 h under oxygen-deprived conditions with little by-product formation, except for small amounts of succinate (Fig. 3).
Fig 3.
l-Valine production of Val-9 under oxygen deprivation. Black squares, l-valine; open squares, alanine; gray circles, lactate; gray triangles, acetate; gray diamonds, succinate; open circles, glucose. Each value was corrected for dilution caused by the addition of NH3 solution and glucose throughout the reaction. The data represent averages from three independent experiments.
DISCUSSION
In this study, disruption of ppc in addition to ldhA to suppress succinate production resulted in the steeply elevated intracellular NADH/NAD+ ratio (Table 5), considerably dropping glucose consumption and l-valine production (Table 3) (16). Nonetheless, the strain possesses the NADH-oxidizing l-valine synthesis pathway containing NAD-preferring mutant AHAIR and NAD-specific LeuDH (Fig. 1) (16). Clearly, disrupting lactate and succinate synthesis pathways has a much more pronounced effect on the NADH/NAD+ ratio than the two engineered enzymes. Note that succinate synthesis under oxygen-deprived conditions through oxaloacetate and the reductive TCA cycle consumes an additional 1 mol of NADH; 1 mol of NADH is generated at GAPDH, and a total of 2 mol of NADH is consumed at MDH and SDH per 1 mol of succinate synthesis (Fig. 1) (33). Thus, it cannot be gainsaid that anaerobic succinate synthesis plays a key role in downregulating the intracellular NADH/NAD+ ratio favorably to maintain glycolysis. Taken together, for viable ΔLDHΔPEPC-based l-valine production under oxygen deprivation, we had to compensate for this role of succinate synthesis by the other mechanism(s). We accordingly demonstrated that inactivation of the acetate synthesis pathway, enhancement of the glycolytic pathway, and chromosomal replacement of ilvC with the NAD-preferring mutant ilvCTM combined to reduce the intracellular NADH/NAD+ ratio so much (Table 5) that increased glucose consumption and l-valine production were possible (Table 3).
Inactivation of the acetate synthesis pathway decreased generation of acetate as a by-product and, in addition, reduced the intracellular NADH/NAD+ ratio, leading to increased glucose consumption and l-valine production (Tables 3 and 5). Contrary to succinate synthesis, acetate synthesis via a pyruvate dehydrogenase (PDH) reaction generates one surplus mole of NADH; a total of 2 mol of NADH is generated at GAPDH and PDH per 1 mol of acetate synthesis (Fig. 1). This directly upregulates the intracellular NADH/NAD+ ratio. Therefore, suppressed acetate production led to the improved intracellular redox state.
Overexpressing five glycolytic genes enhanced glucose consumption, improved l-valine production and yield, reduced the intracellular NADH/NAD+ ratio, and suppressed formation of alanine and acetate (Tables 3 and 5). The processes leading to the results were assumed as follows. By overexpressing glycolytic genes, the corresponding enzymes are able to metabolize larger amounts of glucose at the prevailing high intracellular redox ratio. More l-valine can consequently be produced using the overexpressed l-valine synthesis enzymes. Alanine and acetate production is comparatively suppressed, given that the encoding genes would not be expressed strongly enough to handle the enlarged glycolysis flux (27). An increased intracellular ATP pool from substrate-level phosphorylation accruing from the enhanced glycolysis (the intracellular ATP/ADP ratio in Val-3 was higher than that in Val-1; data not shown) may additionally suppress acetate synthesis by inhibiting the ACK reaction (Fig. 1). Less acetate production reduced the intracellular NADH/NAD+ ratio, further accelerating glucose consumption and l-valine synthesis. The improved l-valine yield of Val-3 can be attributed to an overall decrease in total by-products.
Feedback-resistant mutant AHAS (IlvBNGE) played a key role in enhancing l-valine yield by efficiently converting pyruvate into 2-acetolactate (Table 4) and suppressing by-products from pyruvate (Table 3). On the other hand, the mutant also somewhat elevated the intracellular NADH/NAD+ ratio (Table 5), decreasing glucose consumption and l-valine production. Comparison of Val-5 to Val-4 reveals that the decreased amount of NADH-consuming succinate production exceeded that of NADH-generating acetate production by overexpressing the mutant AHAS, resulting in net elevation of the intracellular redox ratio and therefore slower metabolism.
Chromosomal substitution with ilvNGE resulted in the almost total derepression of l-valine inhibition in Val-6 (Fig. 2) but did not improve l-valine yield (Table 3). It is difficult to explain the results, but we suspect that its elevated AHAS activity exceeded the capacity that Val-6 could exploit in l-valine production. Replacement with ilvCTM reduced the intracellular redox ratio of Val-7 (Table 5), thereby improving glucose consumption and l-valine production but slightly decreasing its l-valine yield (Table 3). It is possible that enzymes of l-valine synthesis were expressed in quantities insufficient to handle the enlarged capacity of Val-7 for glucose consumption. Strain Val-8 with ilvNGE and ilvCTM did not suffer such capacity limitations and was able to register increased l-valine production without suppression of yield (Table 3). The resulting 38% increase in l-valine production from Val-5 to Val-8, in keeping with the high l-valine yield of 86%, was much higher than expected, since both mutant AHAS and AHAIR were already overexpressed more than 40-fold compared to corresponding enzymes expressed from the chromosome in both strains (16).
Disruption of the alanine transaminase gene avtA suppressed alanine production and further enhanced l-valine yield. In C. glutamicum, alanine synthesis from pyruvate is catalyzed by the alanine transaminases AlaT and AvtA encoded by alaT and avtA, respectively, and AlaT is thought to be important for alanine supply (12, 13) (Fig. 1). However, in preliminary tests, disrupting avtA significantly reduced alanine by-product formation, whereas disrupting alaT had surprisingly little effect on alanine production (data not shown). Differences in amino-group donors between the two could partially explain the observations; AlaT utilizes mainly l-glutamate, while AvtA uses l-valine (12, 13) (Fig. 1). The intracellular concentration of l-valine determined was as much as 10-fold higher than that of l-glutamate during l-valine production under oxygen deprivation in strain Val-9. Consequently, AvtA-catalyzed alanine synthesis by transamination from l-valine must be the predominant reaction under these conditions (34).
A final l-valine production of 1,280 mM at a yield of 88.4% from consumed glucose after 24 h under oxygen deprivation was recorded (Table 3 and Fig. 3). The yield is the highest recorded to date, and the productivity is only slightly less than the highest reported attained by the ΔLDH-based strain BNGECTMDLD/ΔLDH (16). It is vastly superior to those of all other previous reports; the l-valine concentration of Val-9 even without volume correction (i.e., the actual titer of reaction solution) reached 645 mM after 24 h, compared with 412 mM (75% yield) after 74 h, 240 mM (86% yield) after 46 h (4), and 518 mM (33.8% yield) after 29.5 h (35). The resultant high volumetric productivity of l-valine was, in part, owing to the high cell density, but the production rate per cell of Val-9 was 1.33 mmol g cell dry weight−1 h−1, which was also higher than the highest value of 0.97 mmol g cell dry weight−1 h−1 obtained previously (35). Although small amounts of succinate are still produced, further suppression studies based on analysis of its synthesis pathway (7, 18) may yet reveal more improvements in the yield. The findings obtained from this study should be applicable to effective production of l-valine as well as many kinds of commodity chemicals, like other amino acids and isobutanol, under oxygen deprivation.
ACKNOWLEDGMENTS
We thank Crispinus A. Omumasaba (RITE) for critical reading of the manuscript.
This work was partially supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO), Japan.
Footnotes
Published ahead of print 14 December 2012
REFERENCES
- 1. Park JH, Lee KH, Kim TY, Lee SY. 2007. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl. Acad. Sci. U. S. A. 104:7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park JH, Lee SY. 2010. Fermentative production of branched chain amino acids: a focus on metabolic engineering. Appl. Microbiol. Biotechnol. 85:491–506 [DOI] [PubMed] [Google Scholar]
- 3. Hermann T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104:155–172 [DOI] [PubMed] [Google Scholar]
- 4. Blombach B, Schreiner ME, Bartek T, Oldiges M, Eikmanns BJ. 2008. Corynebacterium glutamicum tailored for high-yield L-valine production. Appl. Microbiol. Biotechnol. 79:471–479 [DOI] [PubMed] [Google Scholar]
- 5. Durre P. 2007. Biobutanol: an attractive biofuel. Biotechnol. J. 2:1525–1534 [DOI] [PubMed] [Google Scholar]
- 6. Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89 [DOI] [PubMed] [Google Scholar]
- 7. Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 77:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bastian S, Liu X, Meyerowitz JT, Snow CD, Chen MM, Arnold FH. 2011. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 13:345–352 [DOI] [PubMed] [Google Scholar]
- 9. Leyval D, Uy D, Delaunay S, Goergen JL, Engasser JM. 2003. Characterisation of the enzyme activities involved in the valine biosynthetic pathway in a valine-producing strain of Corynebacterium glutamicum. J. Biotechnol. 104:241–252 [DOI] [PubMed] [Google Scholar]
- 10. Elišáková V, Pátek M, Holátko J, Nešvera J, Leyval D, Goergen JL, Delaunay S. 2005. Feedback-resistant acetohydroxy acid synthase increases valine production in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71:207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Börmann ER, Eikmanns BJ, Sahm H. 1992. Molecular analysis of the Corynebacterium glutamicum gdh gene encoding glutamate dehydrogenase. Mol. Microbiol. 6:317–326 [DOI] [PubMed] [Google Scholar]
- 12. Marienhagen J, Kennerknecht N, Sahm H, Eggeling L. 2005. Functional analysis of all aminotransferase proteins inferred from the genome sequence of Corynebacterium glutamicum. J. Bacteriol. 187:7639–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marienhagen J, Eggeling L. 2008. Metabolic function of Corynebacterium glutamicum aminotransferases AlaT and AvtA and impact on L-valine production. Appl. Environ. Microbiol. 74:7457–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartek T, Blombach B, Lang S, Eikmanns BJ, Wiechert W, Oldiges M, Nöh K, Noack S. 2011. Comparative 13C metabolic flux analysis of pyruvate dehydrogenase complex-deficient, L-valine-producing Corynebacterium glutamicum. Appl. Environ. Microbiol. 77:6644–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartek T, Blombach B, Zönnchen E, Makus P, Lang S, Eikmanns BJ, Oldiges M. 2010. Importance of NADPH supply for improved L-valine formation in Corynebacterium glutamicum. Biotechnol. Prog. 26:361–371 [DOI] [PubMed] [Google Scholar]
- 16. Hasegawa S, Uematsu K, Natsuma Y, Suda M, Hiraga K, Jojima T, Inui M, Yukawa H. 2012. Improvement of the redox balance increases L-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl. Environ. Microbiol. 78:865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inui M, Suda M, Okino S, Nonaka H, Puskás LG, Vertès AA, Yukawa H. 2007. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504 [DOI] [PubMed] [Google Scholar]
- 18. Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7:182–196 [DOI] [PubMed] [Google Scholar]
- 19. Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8:243–254 [DOI] [PubMed] [Google Scholar]
- 20. Jojima T, Fujii M, Mori E, Inui M, Yukawa H. 2010. Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid L-alanine under oxygen deprivation. Appl. Microbiol. Biotechnol. 87:159–165 [DOI] [PubMed] [Google Scholar]
- 21. Litsanov B, Brocker M, Bott M. 2012. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl. Environ. Microbiol. 78:3325–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Omumasaba CA, Okai N, Inui M, Yukawa H. 2004. Corynebacterium glutamicum glyceraldehyde-3-phosphate dehydrogenase isoforms with opposite, ATP-dependent regulation. J. Mol. Microbiol. Biotechnol. 8:91–103 [DOI] [PubMed] [Google Scholar]
- 23. Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern JL, Cocaign-Bousquet M, Lindley ND. 1998. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur. J. Biochem. 254:96–102 [DOI] [PubMed] [Google Scholar]
- 24. Ohshima T, Misono H, Soda K. 1978. Properties of crystalline leucine dehydrogenase from Bacillus sphaericus. J. Biol. Chem. 253:5719–5725 [PubMed] [Google Scholar]
- 25. Sambrook J, Fritsh EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 26. Yukawa H, Omumasaba CA, Nonaka H, Kós P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M. 2007. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058 [DOI] [PubMed] [Google Scholar]
- 27. Yamamoto S, Gunji W, Suzuki H, Toda H, Suda M, Jojima T, Inui M, Yukawa H. 2012. Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl. Environ. Microbiol. 78:4447–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yasuda K, Jojima T, Suda M, Okino S, Inui M, Yukawa H. 2007. Analyses of the acetate-producing pathways in Corynebacterium glutamicum under oxygen-deprived conditions. Appl. Microbiol. Biotechnol. 77:853–860 [DOI] [PubMed] [Google Scholar]
- 29. Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H. 1993. Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res. Microbiol. 144:181–185 [DOI] [PubMed] [Google Scholar]
- 30. Hill CM, Duggleby RG. 1998. Mutagenesis of Escherichia coli acetohydroxyacid synthase isoenzyme II and characterization of three herbicide-insensitive forms. Biochem. J. 335:653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hermann T, Kramer R. 1996. Mechanism and regulation of isoleucine excretion in Corynebacterium glutamicum. Appl. Environ. Microbiol. 62:3238–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ehira S, Shirai T, Teramoto H, Inui M, Yukawa H. 2008. Group 2 sigma factor SigB of Corynebacterium glutamicum positively regulates glucose metabolism under conditions of oxygen deprivation. Appl. Environ. Microbiol. 74:5146–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bott M, Niebisch A. 2003. The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104:129–153 [DOI] [PubMed] [Google Scholar]
- 34. Hou X, Chen X, Zhang Y, Qian H, Zhang W. 2012. l-Valine production with minimization of by-products' synthesis in Corynebacterium glutamicum and Brevibacterium flavum. Amino Acids 43:2301–2311 [DOI] [PubMed] [Google Scholar]
- 35. Park JH, Jang YS, Lee JW, Lee SY. 2011. Escherichia coli W as a new platform strain for the enhanced production of L-valine by systems metabolic engineering. Biotechnol. Bioeng. 108:1140–1147 [DOI] [PubMed] [Google Scholar]