Abstract
White-nose syndrome (WNS) is an emerging disease of hibernating bats caused by the recently described fungus Geomyces destructans. First isolated in 2008, the origins of this fungus in North America and its ability to persist in the environment remain undefined. To investigate the correlation between manifestation of WNS and distribution of G. destructans in the United States, we analyzed sediment samples collected from 55 bat hibernacula (caves and mines) both within and outside the known range of WNS using a newly developed real-time PCR assay. Geomyces destructans was detected in 17 of 21 sites within the known range of WNS at the time when the samples were collected; the fungus was not found in 28 sites beyond the known range of the disease at the time when environmental samples were collected. These data indicate that the distribution of G. destructans is correlated with disease in hibernating bats and support the hypothesis that the fungus is likely an exotic species in North America. Additionally, we examined whether G. destructans persists in infested bat hibernacula when bats are absent. Sediment samples were collected from 14 WNS-positive hibernacula, and the samples were screened for viable fungus by using a culture technique. Viable G. destructans was cultivated from 7 of the 14 sites sampled during late summer, when bats were no longer in hibernation, suggesting that the fungus can persist in the environment in the absence of bat hosts for long periods of time.
INTRODUCTION
White-nose syndrome (WNS) is an emerging wildlife disease that by one estimate (http://www.whitenosesyndrome.org/news/north-american-bat-death-toll-exceeds-55-million-white-nose-syndrome) has killed approximately 5.5 million hibernating bats in North America since its discovery in 2007. The disease results from cutaneous infection by the recently described fungus Geomyces destructans (1, 2) and has been implicated in population declines of 72 to 88% for hibernating bat species inhabiting the northeastern United States (3–5). Since 2007, the disease has spread across the eastern United States and Canada, threatening the future of North American bat populations (5, 6). The sudden emergence and rapid spread of WNS has led to questions regarding the origin of G. destructans in North America and how the life cycle of the fungus allows it to exert such significant impacts on hibernating bat populations.
Laboratory experiments have demonstrated that G. destructans is the causative agent of WNS in a North American bat species, eliciting the disease in apparently healthy animals (7, 8). In addition, G. destructans has been found to occur on hibernating bats throughout most of Europe, but it has not been associated with unusual bat mortality on the European continent (9–12). Together, these findings prompt two hypotheses regarding the origin of G. destructans in North America that are consistent with the emergence of a novel infectious disease (13): (i) Geomyces destructans is endemic to North America, but a pathogenic strain spontaneously emerged and is spreading across the landscape (8), or (ii) Geomyces destructans was recently introduced into North America, where it is behaving as an exotic pathogen among naïve populations of bats (8, 9, 11).
Recent research supports the exotic-species hypothesis. For example, a European isolate of G. destructans has been found to induce lesions diagnostic for WNS and mortality in an experimentally infected North American bat species (8). This demonstrates that a European isolate of the fungus is highly pathogenic to North American bats despite having no apparent effects on wild-bat populations of Europe and implicates Europe as the possible source for the introduction of G. destructans into North America (12). In addition, isolates of G. destructans from the eastern United States appear to be genetically identical (14), suggesting that G. destructans in North America is derived from a single isolate that may have been introduced into this continent.
While evidence is mounting to support the hypothesis that G. destructans was introduced into North America, it remains unclear why the WNS-related mortality rate varies between bats of North America and Europe (12) and why some North American species appear to be more vulnerable than others (5). Environmental effects, genetic composition, and behaviors differ among bat species and likely play a role in facilitating infection, disease progression, and mortality, but such factors are difficult to tease apart without a basic understanding of whether the presence of G. destructans in a hibernaculum correlates with manifestation of WNS in bats. Furthermore, the current assumption that G. destructans is limited to areas where WNS has been observed may be biased by the primary means of detecting the fungus through diagnostic analysis of samples derived from sick or dead bats. A previous study addressed these issues by screening sediment samples from bat hibernacula to determine whether G. destructans was indeed restricted to areas where WNS has been observed in bats (15). While nucleic acid from G. destructans was found to occur at three sites within the known range of WNS and no sites outside the range of WNS, the number of sites sampled was too small given the low detection rate to conclude that the distribution of the fungus was correlated to that of the disease. In addition, the method utilized in that study lacked specificity, because similar species of Geomyces cross-reacted with primers of the conventional PCR-based method (16) and may have masked the presence of G. destructans if it was in low abundance. Screening of a greater number of sediment samples from hibernacula using a more specific and sensitive technique, such as a recently described real-time PCR assay (17), may serve to better determine whether the distribution of G. destructans is limited to areas where WNS occurs or whether the fungus is more widespread in North America than currently thought.
The ability to detect G. destructans in environmental samples using PCR-based methods could also reveal important information about WNS disease dynamics. Current determination of whether G. destructans is present in a hibernaculum usually relies upon first detecting the fungus on sick bats, which makes it difficult to address questions such as, “When did the fungus arrive at a given site,” “How long does it take for disease to manifest after the arrival of G. destructans at a new site,” and “Is disease an inevitable outcome of the fungus' presence in a hibernaculum?” An understanding of these aspects of the pathogen's interaction with the environment and its host will facilitate disease surveillance of bat hibernacula and potentially enable earlier deployment of interventional strategies to more effectively limit the spread of and reduce mortality caused by WNS.
Most fungi pathogenic to mammals can persist in the environment in the absence of a host (18, 19). Given the temperature requirements for growth of G. destructans (i.e., it does not grow at or above approximately 20°C [20]), caves and mines have environmental characteristics consistent with potential long-term reservoirs for the fungus, as they remain cool throughout the year, even when bats are absent during summer months. While follow-up culture analyses of sediment samples that contained DNA from G. destructans from a previous study (15) proved that viable G. destructans was present (21), these samples were collected during the hibernation season and may have represented only short-term survival of the fungus after it detached from a bat host. Similarly, G. destructans was cultured from the wall of a cave in Estonia where an infected bat had been observed 9 days prior (12), again demonstrating only temporary persistence. Thus, the ability of G. destructans to survive long term in the environment in the absence of its bat hosts remains uncertain.
The objectives of this study were to (i) determine the distribution of G. destructans in underground bat hibernacula of eastern North America and examine whether the presence of the fungus strictly correlates with the occurrence of WNS and (ii) establish whether hibernacula can serve as reservoirs for G. destructans during the summer months, when bats are largely absent. To address the first objective, we screened sediment samples collected from bat hibernacula across the eastern United States for the presence of G. destructans using a real-time PCR test (17). To address the second objective, sediment samples collected from WNS-affected hibernacula were screened for viable G. destructans, using a previously described culture technique (21), during seasons of both bat hibernation (when bats are present in hibernacula) and activity (when bats are largely absent from hibernacula). By demonstrating the utility of environmental sampling as a noninvasive tool for detecting G. destructans, results from these investigations offer the potential to refine WNS surveillance and management.
MATERIALS AND METHODS
Sample collection.
Sediment samples used for fungal distribution analysis were collected from the eastern United States by volunteers during the winter of 2008 to 2009. For each sample, clean latex gloves were worn, and sterile wooden splints were used to transfer sediment into sterile, labeled sampling bags. A minimum of five samples were collected from the floor of each cave or mine (here referred to as a “site”) and immediately shipped on ice to the U.S. Geological Survey National Wildlife Health Center (Madison, WI), where they were stored at −80°C. The samples included in this study represented a total of 56 sites from 22 states east of the 95th meridian west (95°W longitude), including 8 states within and 14 states outside the known range of WNS at the time when the samples were collected (Fig. 1 and Table 1). Exact locations of the sampled sites are not provided due to the sensitive nature of bat hibernacula.
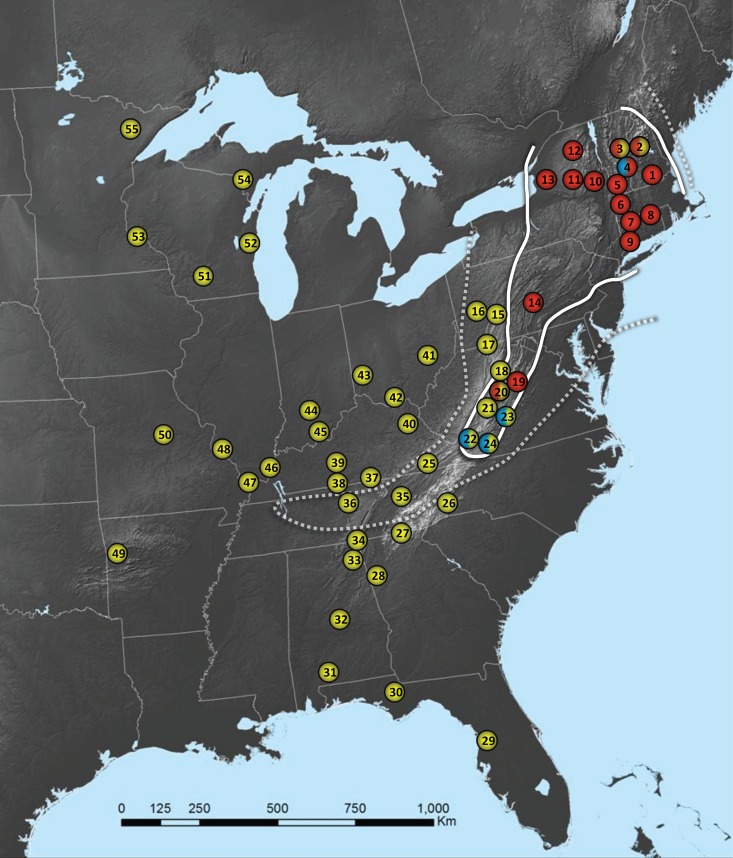
Fig 1.
Map of the eastern United States showing the sampling locations described in Table 1. The left halves of the circles depict the disease status of individual bat hibernacula at the time when the samples were collected (red, diseased; blue, buffer; yellow, clean). The right halves of the circles represent the PCR results (red, G. destructans detected; yellow, G. destructans not detected). The solid white line marks the general geographic range of WNS (i.e., WNS zone) at the time of sample collection; the dotted white line marks the general geographic range of WNS during the following winter (i.e., buffer zone) (http://www.whitenosesyndrome.org/resources/map). Geomyces destructans was detected only in bat hibernacula that were situated within the known range of WNS as of the winter of 2008 to 2009. The PCR results for sites 20, 21, 22, and 23 represent only those for samples collected in the winter of 2008 to 2009 and not those collected in the winter of 2010 to 2011.
Table 1.
Bat hibernacula in the eastern United States from which sediment samples were collected in the winter of 2008 to 2009 to test for the presence of G. destructans by real-time PCRa
| Site | State | Geographic zone | Cave or mine status | PCR result |
|---|---|---|---|---|
| 1 | NH | WNS | Diseased | + |
| 2 | NH | WNS | Diseased | − |
| 3 | VT | WNS | Diseased | − |
| 4 | VT | WNS | Buffer | + |
| 5 | VT | WNS | Diseased | + |
| 6 | MA | WNS | Diseased | + |
| 7 | MA | WNS | Diseased | + |
| 8 | MA | WNS | Diseased | + |
| 9 | CT | WNS | Diseased | + |
| 10 | NY | WNS | Diseased | + |
| 11 | NY | WNS | Diseased | + |
| 12 | NY | WNS | Diseased | + |
| 13 | NY | WNS | Diseased | + |
| 14 | PA | WNS | Diseased | + |
| 15 | PA | Buffer | Clean | − |
| 16 | PA | Buffer | Clean | − |
| 17 | PA | Buffer | Clean | − |
| 18 | WV | WNS | Clean | − |
| 19 | WV | WNS | Diseased | + |
| 20 | WV | WNS | Diseased | − |
| 20* | WV | WNS | Diseased | + |
| 21 | WV | WNS | Clean | − |
| 21* | WV | WNS | Diseased | + |
| 22 | WV | WNS | Buffer | − |
| 22* | WV | WNS | Diseased | + |
| 23 | VA | WNS | Buffer | − |
| 23* | VA | WNS | Diseased | + |
| 24 | VA | WNS | Buffer | − |
| 25 | VA | Buffer | Clean | − |
| 26 | NC | Outside | Clean | − |
| 27 | NC | Outside | Clean | − |
| 28 | GA | Outside | Clean | − |
| 29 | FL | Outside | Clean | − |
| 30 | FL | Outside | Clean | − |
| 31 | AL | Outside | Clean | − |
| 32 | AL | Outside | Clean | − |
| 33 | AL | Outside | Clean | − |
| 34 | TN | Outside | Clean | − |
| 35 | TN | Buffer | Clean | − |
| 36 | TN | Buffer | Clean | − |
| 37 | KY | Outside | Clean | − |
| 38 | KY | Outside | Clean | − |
| 39 | KY | Outside | Clean | − |
| 40 | KY | Outside | Clean | − |
| 41 | OH | Outside | Clean | − |
| 42 | OH | Outside | Clean | − |
| 43 | OH | Outside | Clean | − |
| 44 | IN | Outside | Clean | − |
| 45 | IN | Outside | Clean | − |
| 46 | IL | Outside | Clean | − |
| 47 | IL | Outside | Clean | − |
| 48 | IL | Outside | Clean | − |
| 49 | AR | Outside | Clean | − |
| 50 | MO | Outside | Clean | − |
| 51 | WI | Outside | Clean | − |
| 52 | WI | Outside | Clean | − |
| 53 | WI | Outside | Clean | − |
| 54 | MI | Outside | Clean | − |
| 55 | MN | Outside | Clean | − |
Asterisks indicate sites resampled in winter 2010 to 2011. “Geographic zone,” “cave or mine status,” and “PCR result” depict results from winter 2010 to 2011. Sites with the same number but lacking this symbol represent results from winter 2008 to 2009 for these same sites.
Sediment samples for environmental persistence analysis were collected from 14 bat hibernacula in which bats with WNS had been previously identified. These consisted of 4 sites in New Hampshire, 2 sites in Vermont, 3 sites in Virginia, and 5 sites in West Virginia (Table 2). Five locations within each site were marked, and samples were serially collected within 30 cm of the markers on three separate occasions: once in February to March 2011 (during the bat hibernation period; here referred to as winter 2010 to 2011), once in late July to late August 2011 (near the end of the active season and just prior to large congregations of bats returning to the hibernacula; here referred to as summer 2011), and again in October 2011 to March 2012 (during the next consecutive hibernation period; here referred to as winter 2011 to 2012). Exceptions were as follows: samples were not obtained from sites C6, C7, and C8 during the third sampling period (i.e., winter 2011 to 2012), several sampling markers could not be relocated within sites C7 and C9 during the second visit (i.e., summer 2011), and samples were collected from approximate locations determined by collectors. Additionally, site C4 flooded in September 2011; the markers were relocated during the winter 2011 to 2012 visit, although 2.5 to 20 cm of sediment had been deposited on top of the previously sampled sediment.
Table 2.
Bat hibernacula within the WNS-affected area of the United States in which sediment samples were tested for the presence of viable G. destructans using a culture technique
| Site | State | Sampling location | Culture result |
||
|---|---|---|---|---|---|
| Winter 2010–2011 | Summer 2011 | Winter 2011–2012a | |||
| C1 | VA | Total | − | + | + |
| 1 | − | − | − | ||
| 2 | − | + | − | ||
| 3 | − | − | − | ||
| 4 | − | − | + | ||
| 5 | − | − | − | ||
| C2 | VA | Total | − | − | − |
| 1 | − | − | − | ||
| 2 | − | − | − | ||
| 3 | − | − | − | ||
| 4 | − | − | − | ||
| 5 | − | − | − | ||
| C3 | VA | Total | − | − | − |
| 1 | − | − | − | ||
| 2 | − | − | − | ||
| 3 | − | − | − | ||
| 4 | − | − | − | ||
| 5 | − | − | − | ||
| C4 | VT | Total | + | + | + |
| 1 | − | + | − | ||
| 2 | + | − | − | ||
| 3 | + | − | − | ||
| 4 | − | − | + | ||
| 5 | + | + | − | ||
| C5 | VT | Total | + | − | + |
| 1 | + | − | + | ||
| 2 | − | − | − | ||
| 3 | − | − | + | ||
| 4 | − | − | + | ||
| 5 | − | − | − | ||
| C6 | NH | Total | + | + | NA |
| 1 | + | − | NA | ||
| 2 | − | + | NA | ||
| 3 | − | − | NA | ||
| 4 | − | − | NA | ||
| 5 | − | − | NA | ||
| C7 | NH | Total | + | − | NA |
| 1 | − | − | NA | ||
| 2 | + | − | NA | ||
| 3 | − | − | NA | ||
| 4 | − | − | NA | ||
| 5 | − | − | NA | ||
| C8 | NH | Total | + | + | NA |
| 1 | − | + | NA | ||
| 2 | + | + | NA | ||
| 3 | + | − | NA | ||
| 4 | − | − | NA | ||
| 5 | − | − | NA | ||
| C9 | NH | Total | − | + | + |
| 1 | − | − | − | ||
| 2 | − | − | − | ||
| 3 | − | − | + | ||
| 4 | − | + | + | ||
| 5 | − | − | − | ||
| C10 | WV | Total | − | − | − |
| 1 | − | − | − | ||
| 2 | − | − | − | ||
| 3 | − | − | − | ||
| 4 | − | − | − | ||
| 5 | − | − | − | ||
| C11 | WV | Total | − | + | − |
| 1 | − | + | − | ||
| 2 | − | − | − | ||
| 3 | − | − | − | ||
| 4 | − | − | − | ||
| 5 | − | − | − | ||
| C12 | WV | Total | + | − | − |
| 1 | − | − | − | ||
| 2 | + | − | − | ||
| 3 | − | − | − | ||
| 4 | − | − | − | ||
| 5 | − | − | − | ||
| C13 | WV | Total | + | − | − |
| 1 | − | − | − | ||
| 2 | − | − | − | ||
| 3 | − | − | − | ||
| 4 | + | − | − | ||
| 5 | − | − | − | ||
| C14 | WV | Total | + | + | − |
| 1 | − | − | − | ||
| 2 | + | − | − | ||
| 3 | − | − | − | ||
| 4 | − | + | − | ||
| 5 | − | − | − | ||
NA, no sample collected.
DNA extraction and PCR analysis.
DNA was extracted from the sediment samples for the distribution study using the PowerSoil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. If more than five samples were collected from a given site, five were chosen at random for inclusion in the study. All extracted DNA samples were stored at −20°C.
Real-time PCR targeting the intergenic spacer (IGS) region of the rRNA gene complex of G. destructans was performed on an Applied Biosystems 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA), as described previously (17). Five microliters of each DNA extraction from sediment (diluted 1:1 and 1:10) was added to each 25-μl PCR mixture. All plates included at least two positive-control samples (3.3 pg G. destructans genomic DNA [gDNA]) and one negative-control sample (water added in place of the template). Individual samples that crossed the cycle threshold (CT) (set at 10% of the maximum fluorescence of the positive-control sample for each plate [17, 22]) within 40 cycles were considered positive for the presence of G. destructans. Furthermore, a sample was identified as positive if either or both template dilutions (1:1 or 1:10) crossed the cycle threshold, as described above; a site was considered positive when at least one sample from that site was PCR positive.
Sediment often contains humic acid and other substances that can inhibit DNA amplification. Thus, prior to conducting the real-time PCR assay, all samples were screened for PCR inhibition to reduce the chance of false-negative results. Conventional PCR targeting the internal transcribed spacer (ITS) region of the rRNA gene was performed with primers ITS4 and ITS5 (23), using GoTaq Flexi DNA polymerase (Promega Corporation, Madison, WI) according to the manufacturer's instructions. Five microliters of the 1:10-diluted sediment DNA extraction was used as the template. Cycling conditions were as follows: 98°C for 2 min and then 30 cycles of 98°C for 10 s, 50°C for 30 s, and 72°C for 1 min, followed by a final extension step for 7 min at 72°C. Amplification products were analyzed by using an agarose gel. Control reaction mixtures containing 33 fg gDNA isolated from pure cultures of G. destructans (positive control) or without a template added (negative control) were also included. Samples failing to yield bands by the ITS PCR were subsequently spiked with gDNA from G. destructans and used as the template in a modified version of the real-time PCR assay (17) adapted for use with conventional PCR. Reagents used were the same as those described above for the ITS PCR. Five microliters of the 1:10-diluted sediment DNA extraction, 5 μl (containing 33 fg) gDNA, and 1.25 μl of each primer used in the real-time PCR were included in the 25-μl total reaction mixture volume. No probe was added. Control reaction mixtures, as described above for the ITS PCR, were also included. Cycling conditions were identical to those used for the real-time PCR assay (17). Spiked samples that did not yield amplification products were considered inhibitory. If one or more samples showed evidence of inhibition, the entire site was excluded from the data set. To ensure that the qualitative results of the inhibition screen using conventional PCR were consistent with the more quantitative real-time PCR, a subset of samples was also screened for inhibition on the real-time PCR platform. Single samples from 45 individual sites that tested negative for the presence of G. destructans by real-time PCR were randomly selected, spiked with 33 fg of G. destructans gDNA, and used as the template in the real-time PCR assay. Samples within one CT value of the positive-control well containing 33 fg G. destructans gDNA were considered noninhibitory.
PCR amplicons from each PCR-positive site were subjected to cloning and sequencing to confirm an exact sequence match to G. destructans. PCR products were cloned as described previously (24) and prepared for sequencing according to preestablished methods (15), using primers SP6 (TATTTAGGTGACACTATA) and T7 (TAATACGACTCACTATAG), which target pGEM-T (Promega, Madison, WI) up- and downstream of the insert. Because of short size of the amplicons (103 bp), blue/white screening of bacterial colonies was not possible, so approximately 8 to 16 random colonies were chosen for screening by PCR amplification, and those yielding amplification products were further characterized by DNA sequencing.
Culture analysis.
For the environmental persistence analysis, approximately 200 mg of each thawed sediment sample was placed into a sterile microcentrifuge tube; suspended in 0.5 ml sterile, deionized water; and serially diluted, as previously described (21). Sabouraud dextrose agar plates containing chloramphenicol and gentamicin (BD Diagnostic Systems, Sparks, MD) were inoculated by spreading 150 μl of the 10−1, 10−2, and 10−3 dilutions onto the medium. Each dilution was plated in duplicate. The plates were incubated at 7°C and checked at 30 days and once weekly thereafter for a total of 60 days. Colonies of G. destructans were initially identified by examining tape lifts of suspect colonies using a 40× objective to identify characteristic crescent-shaped conidia borne at the end of verticillately branching conidiophores (2). At least one colony of G. destructans from each site at each time point, when present, was isolated in pure culture. The ITS region of the rRNA gene of each of these isolates was then sequenced to confirm identification of G. destructans, using primers ITS1-F and ITS4 (25) and PCR conditions described previously (21).
Data analysis.
Hibernacula for the fungal distribution analysis were categorized as occurring within one of three zones based on WNS distribution at the time of sample collection: (i) the WNS zone (general geographic area within which the disease had been documented), (ii) the buffer zone (general geographic area within which WNS was documented the year following sample collection), and (iii) the outside zone (general geographic area within which WNS was not documented until at least 2 years after sample collection) (Table 1 and Fig. 1). Not all hibernacula falling within the WNS zone had been confirmed to contain bats exhibiting signs of the disease; similarly, not all hibernacula within the buffer zone were confirmed to contain bats with WNS by the following year. For this reason, individual sites were designated either diseased sites (WNS documented at the site prior to or at the time when the samples were collected), buffer sites (WNS documented 1 year after the samples were collected), or clean sites (WNS not documented to date or documented more than 1 year after the samples were collected), based on interviews with individuals from state and federal wildlife agencies (Table 1 and Fig. 1).
To test the null hypothesis that the distribution of G. destructans is not associated with WNS in North America, PCR results (i.e., the number of PCR-positive and PCR-negative bat hibernacula) for sites that occurred within and outside the known range of WNS (WNS and outside zones) at the time when samples were collected were compared by using Fisher's exact test in SigmaPlot 11.2 (Systat Software, Inc., San Jose, CA). Sites that occurred within the geographic buffer zone were excluded from this analysis because it was equally plausible that G. destructans could be present or absent from those sites.
Estimated probability of detection.
To determine the probability of PCR and culture analysis to detect G. destructans, the results of each test were formulated as a binomial variable (1 = G. destructans detected; 0 = G. destructans not detected). A detection history for each sampled site was then created as a series of zeros and ones. For example, a detection history of 101 for the environmental persistence analysis indicated that G. destructans was detected on the first and last surveys but not the second survey. The probability of detecting the fungus, if present, was then estimated in the following manner. The observed values at site i and replicate (spatial or temporal) t (yi,t), were the detection histories and represent the imperfect observation process uncorrected for the ability of the diagnostic test to detect the fungus. These observations were modeled as Bernoulli trials, where the probability of success (p.effi,t, or the probability that yi,t = 1), was the observed detection at site i and replicate t.
Mathematically, yi,t ∼ Bern(p.effi,t), and the observed detections of the fungus were a function of the true infection status of the site (infected/clean) and the probability of the test correctly detecting the fungus, if present. Therefore, p.effi,t = zi × pi,t, where z represented the true state of the site (infected or clean) and p was the true detection probability. The observations, therefore, were imperfect reflections of the true state due to imperfect detection probabilities. Furthermore, when z was unknown, z was formulated as Bernoulli trials, where the probability of success equals ψ, the true proportion of sites that were infected (as opposed to observed). The analysis was conducted with R (R Foundation for Statistical Computing, Vienna, Austria) (26), using the R2WinBUGS library (27) according to preestablished methods (28). To aid in convergence, logit(pi,t) < −α1 + β1X was formulated, where X was a matrix of covariate values [similarly, logit(ψ) < −α2 + β2X]. The models for both analyses were checked for convergence using the Gelman-Rubin diagnostics function (gelman.diag) in the CODA package in R (29).
For the PCR analysis, it was assumed that the true infection status of buffer sites was unknown and that the true infection status of diseased and clean sites was known. Thus, z was set to 1 for diseased sites and 0 for clean sites. Priors for the estimates of α's and β's were uniform (−5, 5). Ideally, detection and occupancy would have been estimated separately for buffer and infected sites, but there were not enough buffer sites (n = 4) for this analysis to converge. Therefore, a pooled occupancy and detection probability was estimated separately for buffer and infected sites.
For the culture analysis, it was assumed that the true infection status for each site was known as infected, and z was set to 1. Since multiple samples were collected within each cave on 3 different sampling occasions, two models were run, one with an effect of time and one including a random effect for cave. Random effects were formulated as Ν (μ,τ), where τ = 1/(σ × σ). Priors on μ were uniform (−5, 5); priors on σ were also uniform (0, 10).
RESULTS
PCR analysis.
One of the 56 sites tested for the distribution portion of the study exhibited PCR inhibition and was excluded from further analysis. The subset of samples tested for inhibition by using real-time PCR yielded results identical to those of the conventional PCR, indicating that the conventional PCR method used to screen samples for inhibition was accurate. Sequences of PCR amplicons from all sediment samples that were PCR positive for G. destructans were 100% identical to the 103-nucleotide IGS region of the type isolate of G. destructans (GenBank accession no. JX415267 [17]).
Distribution and environmental detection of G. destructans.
Nucleic acid from G. destructans was detected by real-time PCR in 47 samples collected during the winter of 2008 to 2009, representing 13 different sites (Table 1). Seven sites that were initially sampled in winter 2008 to 2009 were resampled in winter 2010 to 2011 as part of the environmental persistence study (see above). Four of these sites (sites C2, C10, C13, and C14) that were PCR negative for G. destructans in winter 2008 to 2009 were subsequently reanalyzed by real-time PCR using DNA extractions from samples collected during winter 2010 to 2011 (sites designated 23*, 22*, 20*, and 21*, respectively, in Table 1). Bats from one of the four sites (site 20) had been diagnosed with WNS in winter 2008 to 2009, but bats from the remaining three sites (sites 21, 22, and 23) did not show signs of the disease until the winter of 2009 to 2010 or the winter of 2010 to 2011. PCR analysis of these samples (collected during winter 2010 to 2011) showed that DNA from G. destructans was present in 9 of 20 samples, representing all four sites.
The occurrence of G. destructans as detected in the environmental samples was synonymous with the known range of WNS at the time when samples were collected (Fig. 1), with hibernacula within the known range of WNS having significantly higher detection rates for the fungus than hibernacula outside the range of the disease (P < 0.0001). Sixteen of the 17 hibernacula in which the fungus was detected were diseased sites; the remaining hibernaculum was a buffer site in which bats with WNS were observed the following year. Of the 12 WNS-positive hibernacula from which G. destructans was detected in winter 2008 to 2009, 5 had been designated WNS positive in winter 2007 to 2008, and 7 were identified as WNS positive in winter 2008 to 2009. Site 6 had not been officially monitored since 1985, but the cave was considered WNS positive based upon observations of clinical signs in bats suggestive of the disease at the time when the samples were collected. The four hibernacula from which G. destructans was detected by PCR in winter 2010 to 2011 included one site known to harbor WNS-positive bats in winter 2008 to 2009, two in winter 2009 to 2010, and one in winter 2010 to 2011.
Environmental persistence of G. destructans.
Geomyces destructans was cultured from 27 of the 195 sediment samples collected from bat hibernacula in 2011 to 2012, with viable fungus detected in 11 of the 14 sites during at least one sampling interval (Table 2). Seven of the 14 sites were found to harbor viable G. destructans in late summer, when bats were either absent from the hibernacula or present in only low numbers. Sequences of the rRNA gene ITS regions of isolates from each site were 100% identical to the ITS region of the type isolate of G. destructans (GenBank accession no. EU884921 [2]).
Estimated probability of detection.
All samples from the 36 caves designated “clean” (i.e., WNS free) tested negative for G. destructans. The remaining 23 samples (representing diseased and buffer sites) were combined into one data set, and probability of detection for the PCR assay was estimated. Estimated probabilities of detection for a single sample were 0.56 (95% confidence interval [CI], 0.47 to 0.67) for diseased sites and 0.11 (95% CI, 0.02 to 0.29) for buffer sites. This indicated that with five samples from a given diseased site, the probability of detecting the fungus was 0.98 and that 4 samples are sufficient to obtain a mean estimated probability of detection of >0.95. For buffer sites, the probability of detecting G. destructans with 5 samples was 0.44, and at least 26 samples would be needed from each site for the mean estimated probability of detection to be >0.95.
The overall probability of detection using the culture technique was 0.14 (95% CI, 0.10 to 0.19). There was no difference between detection probabilities by time period (0.16 [95% CI, 0.080 to 0.25] in winter 2010 to 2011, 0.13 [95% CI, 0.062 to 0.22] in summer 2011, and 0.13 [95% CI, 0.054 to 0.23] in winter 2011 to 2012). However, the random-effects model indicated unexplained variation due to the effect of site (i.e., cave/mine). More than 20 samples would be needed to have a mean estimated probability of detection of ≥0.95.
DISCUSSION
The sudden emergence and spread of WNS in North America have led to speculation that G. destructans is an exotic species and may have been recently introduced from Europe (8, 9, 11). If this hypothesis is valid, the distribution of G. destructans would be expected to mirror that of the disease. We screened a total of 295 sediment samples collected from 55 caves and mines in the eastern United States using a real-time PCR assay specific for G. destructans (17) and detected the fungus in 17 bat hibernacula. All 17 of these sites were situated within the known range of WNS at the time when the samples were collected, and G. destructans was not found to occur outside that area (Fig. 1). Furthermore, the real-time PCR findings paralleled WNS manifestations on a temporal scale. G. destructans was not detected in three sites that were unaffected by WNS in the winter of 2008 to 2009, but the fungus was later detected in sediment samples collected from those same hibernacula subsequent to the appearance of the disease in bats at those sites. These findings suggest that an endemic, less virulent strain of G. destructans likely did not occur in eastern North America prior to the arrival of WNS and offer further support for the exotic-species hypothesis to explain the emergence of G. destructans as a novel pathogen in North America (8, 13).
Sixteen of the 17 sites in which G. destructans was detected by real-time PCR in this study contained bats showing signs of WNS prior to, or at the time of, sample collection. In the remaining site, WNS was observed the following winter. While these results seem to suggest that WNS may be an inevitable outcome once G. destructans is introduced into a hibernaculum, it is important to interpret these results cautiously because a relatively small number of bat hibernacula were sampled, and all positive sites were located within the same geographic area. Thus, it is unknown how clinical signs of WNS and disease severity may vary as G. destructans spreads to new regions of North America with different environmental conditions and host species. The potential importance of site-specific factors in their relation to WNS may be highlighted by the detection of G. destructans in only one of the four buffer sites, which may suggest that different hibernacula have different latency periods between the arrival of G. destructans and the manifestation of the disease in bats or that fungal abundance thresholds that result in the appearance of WNS (i.e., infective doses of the fungus) vary between sites.
This study represents the first application of a high-throughput PCR technique for direct detection of G. destructans in the environment. A previously described PCR assay (16) utilized in a prior study to detect G. destructans in the environment lacked specificity and required cloning and sequencing procedures to differentiate DNA of G. destructans from that of other closely related Geomyces spp. common in cave sediment (15, 21). Additionally, a real-time PCR assay that targets the alpha-l-rhamnosidase gene of G. destructans (30) was not tested against environmental samples but may lack the sensitivity necessary to detect the fungus in sediment, given that the alpha-l-rhamnosidase gene likely exists at a low copy number within the genome of G. destructans (17). The work described here confirms the specificity and sensitivity of a previously developed PCR method that targets the IGS region of G. destructans (17) and supports its application for use with environmental samples. Furthermore, the detection of the fungus in a buffer site suggests that PCR screening of sediment samples within caves may allow for early detection of G. destructans prior to manifestations of visible signs of disease in bats inhabiting a hibernaculum. The relatively low estimated detection probability for samples collected from buffer sites relative to diseased sites may have been an effect of our inability to estimate infection status and detection probability separately for buffer and infected sites. Thus, we may have underestimated detection probability by overestimating the proportion of buffer sites that were infected. Additionally, the probability of detecting the fungus at buffer sites prior to disease onset might be enhanced by collecting sediment samples in early fall instead of midwinter (i.e., months before a hibernaculum might become diseased, as opposed to a full year before manifestation of WNS). Also, future work to determine whether certain types of environmental samples or specific locations within caves and mines are more likely to harbor G. destructans may further enhance sensitivity of detection.
While the use of PCR to detect G. destructans can provide important information about certain aspects of WNS disease ecology, the method is limited in that it cannot discriminate between viable and nonviable fungus. This is of particular importance in determining what role the environment plays in maintaining infectious populations of G. destructans. Detection of live G. destructans in 7 of the 14 caves and mines in late summer provides the first evidence that G. destructans is capable of surviving in bat hibernacula when bats are either absent or at low densities and that caves and mines serve as likely infection sources when bats return for hibernation in early autumn. However, our ability to culture viable G. destructans from sediment samples collected in 2011 from sites C5 and C9 suggests that the fungus can survive much longer than a few months in the environment in the absence of a bat host. Specifically, bats had not been observed in one of the sites (site C9) for approximately 1 year prior to sample collection, and site C5 had been sealed such that bats were excluded from the hibernaculum for approximately 2 years prior to sample collection. Demonstration that sediments from these two mines contained live G. destructans 1 to 2 years after bats had been extirpated/excluded indicates that the fungus can persist long term in caves and mines.
The culture technique used for this experiment lacked the sensitivity of the molecular detection technique for sites known to be infested with G. destructans. Specifically, there was a lack of correlation in the detection of viable G. destructans across replicate, serially diluted, and spatially and temporally separated samples collected within the same sites. The mean probability of detecting G. destructans from contaminated sediment was 0.14, with at least 20 samples being required from an average site to have a 95% chance of detection of the fungus using the described culture technique. However, detection probabilities varied greatly by site, with some sites still not reaching a 50% detection probability with 15 samples. Clumping or aggregation of G. destructans within sediment, competition or inhibition by other fungi on the artificial culture medium, low abundance of G. destructans in environmental samples relative to the abundance of other fungi, differences in abundance of G. destructans between sites, and/or differences in abundance between locations within the same site may account for these discrepancies. Whatever the reason, the described culture-based technique is valuable to demonstrate that viable G. destructans is present in a tested sample. However, the technique is currently suitable neither for quantifying the abundance of G. destructans nor for proving the absence of the fungus in environmental samples. Future work focusing on developing a medium that is more selective for G. destructans may serve to improve the utility of culture-based methods for addressing research questions such as how long the fungus remains viable in different environments, what portions of hibernacula are most conducive to supporting G. destructans (including cave ceilings where bats roost and therefore which may be most likely to come into contact with the fungus), whether G. destructans can propagate (as opposed to simply persist) in hibernacula without bats, and how abundance of the fungus changes spatially or temporally within sites.
Disease ecology is often represented by a triad that involves interactions between a host, a pathogen, and an environment. To date, research on WNS has focused primarily on bats, G. destructans, and interactions between the two. Relatively little information is available regarding the interplay between the pathogen and the environment. This work demonstrates the utility of environmental sampling for enhancing WNS surveillance and furthering research on WNS epidemiology. Specifically, the results of this study show that the presence of G. destructans in environments where bats hibernate is strongly correlated with disease manifestation, the fungus may be detectable in the environment prior to disease manifestation, and the fungus can persist in the sediment of bat hibernacula for long periods of time in the absence of bat hosts. Additional studies to more fully elucidate the role that the environment plays in supporting the proliferation of G. destructans and facilitating the development and progression of WNS will reveal important factors related to the epidemiology of WNS and may provide information useful for WNS disease management.
ACKNOWLEDGMENTS
This work was supported by the National Speleological Society, the U.S. Fish and Wildlife Service, the U.S. Forest Service, and the U.S. Geological Survey.
We thank Thomas H. Kunz (BU) for his early role in the development of this project and Peter Youngbaer (NSS), Mike Warner (Speleobooks, Inc.), and Alan Hicks (NY DEC) for their assistance in coordinating the collection of sediment samples. We are indebted to Emily Brunkhurst (NH FGD), Scott Darling (VT FWD), Joel Flewelling (VT FWD), Tom French (MA DFW), Jeff Hajenga (WV DNR), Carl Herzog (NY DEC), Christina Kocer (FWS), Rick Reynolds (VA DGIF), Craig Stihler (WV DNR), Greg Turner (PA GC), and Susi VonOettingen (FWS) for providing disease surveillance information and/or assisting in repeated collection of sediment samples. We also thank the many individuals who volunteered to collect sediment samples. We are grateful to Andrew Minnis, Michelle Jusino, and Mark Banik for assistance with sequencing of clones and to Paul Cryan for aiding in the design of Fig. 1.
The use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 14 December 2012
REFERENCES
- 1. Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, Coleman JTH, Darling SR, Gargas A, Niver R, Okoniewski JC, Rudd RJ, Stone WB. 2009. Bat white-nose syndrome: an emerging fungal pathogen? Science 323:227 doi:10.1126/science.1163874 [DOI] [PubMed] [Google Scholar]
- 2. Gargas A, Trest MT, Christensen M, Volk TJ, Blehert DS. 2009. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon 108:147–154 [Google Scholar]
- 3. Dzal Y, McGuire LP, Veselka N, Fenton MB. 2011. Going, going, gone: the impact of white-nose syndrome on the summer activity of the little brown bat (Myotis lucifugus). Biol. Lett. 7:392–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brooks RT. 2011. Declines in summer bat activity in central New England 4 years following the initial detection of white-nose syndrome. Biodivers. Conserv. 20:2537–2541 [Google Scholar]
- 5. Turner GG, Reeder DM, Coleman JTH. 2011. A five-year assessment of mortality and geographic spread of white-nose syndrome in North American bats and a look to the future. Bat Res. News 52:13–27 [Google Scholar]
- 6. Frick WF, Pollock JF, Hicks AC, Langwig KE, Reynolds DS, Turner GG, Butchkoski CM, Kunz TH. 2010. An emerging disease causes regional population collapse of a common North American bat species. Science 329:679–682 [DOI] [PubMed] [Google Scholar]
- 7. Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, Ballmann AE, Coleman JTH, Redell DN, Reeder DM, Blehert DS. 2011. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480:376–378 [DOI] [PubMed] [Google Scholar]
- 8. Warnecke L, Turner JM, Bollinger TK, Lorch JM, Misra V, Cryan PM, Wibbelt G, Blehert DS, Willis CKR. 2012. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc. Natl. Acad. Sci. U. S. A. 109:6999–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Puechmaille SJ, Verdeyroux P, Fuller H, Gouilh MA, Bekart M, Teeling EC. 2010. White-nose syndrome fungus (Geomyces destructans) in bat, France. Emerg. Infect. Dis. 16:290–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martínková N, Bačkor P, Bartonička T, Blažková P, Cervený J, Falteisek L, Gaisler J, Hanzal V, Horáček D, Hubálek Z, Jahelková H, Kolaøík M, Korytár L, Kubátová A, Lehotská B, Lehotský R, Lučan RK, Májek O, Matějù J, Rehák Z, Šafář J, Tájek P, Tkadlec E, Uhrin M, Wagner J, Weinfurtová D, Zima J, Zukal J, Horáček I. 2010. Increasing incidence of Geomyces destructans fungus in bats from the Czech Republic and Slovakia. PLoS One 5:e13853 doi:10.1371/journal.pone.0013853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wibbelt G, Kurth A, Hellmann D, Weishaar M, Barlow A, Veith M, Prüger J, Görföl T, Grosche L, Bontadina F, Zöphel U, Seidl HP, Cryan PM, Blehert DS. 2010. White-nose syndrome fungus (Geomyces destructans) in bats, Europe. Emerg. Infect. Dis. 16:1237–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Puechmaille SJ, Wibbelt G, Korn V, Fuller H, Forget F, Mühldorfer K, Kurth A, Bogdanowicz W, Borel C, Bosch T, Cherezy T, Drebet M, Görföl T, Haarsma AJ, Herhaus F, Hallart G, Hammer M, Jungmann C, Le Bris Y, Lutsar L, Masing M, Mulkens B, Passior K, Starrach M, Wojtaszewski A, Zöphel U, Teeling EC. 2011. Pan-European distribution of white-nose syndrome fungus (Geomyces destructans) not associated with mass mortality. PLoS One 6:e19167 doi:10.1371/journal.pone.0019167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rachowicz LJ, Hero J-M, Alford RA, Taylor JW, Morgan JAT, Vredenburg VT, Collins JP, Briggs CJ. 2005. The novel and endemic pathogen hypothesis: competing explanations for the origin of emerging infectious diseases of wildlife. Conserv. Biol. 19:1441–1448 [Google Scholar]
- 14. Ren P, Haman KH, Last LA, Rajkumar SS, Keel MK, Chaturvedi V. 2012. Clonal spread of Geomyces destructans among bats, Midwestern and Southern United States. Emerg. Infect. Dis. 18:883–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lindner DL, Gargas A, Lorch JM, Banik MT, Glaeser J, Kunz TH, Blehert DS. 2011. DNA-based detection of the fungal pathogen Geomyces destructans in soil from bat hibernacula. Mycologia 103:241–246 [DOI] [PubMed] [Google Scholar]
- 16. Lorch JM, Gargas A, Meteyer CU, Berlowski-Zier BM, Green DE, Shearn-Bochsler V, Thomas NJ, Blehert DS. 2010. Rapid polymerase chain reaction diagnosis of white-nose syndrome in bats. J. Vet. Diagn. Invest. 22:224–230 [DOI] [PubMed] [Google Scholar]
- 17. Muller LK, Lorch JM, Lindner DL, O'Connor M, Gargas A, Blehert DS. 6 September 2012. Bat white-nose syndrome: a real-time TaqMan polymerase chain reaction test targeting the intergenic spacer region of Geomyces destructans. Mycologia. [Epub ahead of print.] doi:10.3852/12-242 [DOI] [PubMed] [Google Scholar]
- 18. Casadevall A. 2005. Fungal virulence, vertebrate endothermy, and dinosaur extinction: is there a connection? Fungal Genet. Biol. 42:98–106 [DOI] [PubMed] [Google Scholar]
- 19. Fisher MC, Henk DA, Briggs CJ, Brownstein LC, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verant ML, Boyles JG, Waldrep W, Jr, Wibbelt G, Blehert DS. 2012. Temperature-dependent growth of Geomyces destructans, the fungus that causes bat white-nose syndrome. PLoS One 7:e46280 doi:10.1371/journal.pone.0046280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lorch JM, Lindner DL, Gargas A, Muller LK, Minnis AM, Blehert DS. 16 October 2012. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia. [Epub ahead of print.] doi:10.3852/12-207 [DOI] [PubMed] [Google Scholar]
- 22. King JL, Guidry C. 2004. Müller cell production of insulin-like growth factor-binding proteins in vitro: modulation with phenotype and growth factor stimulation. Invest. Ophthalmol. Vis. Sci. 45:4535–4542 [DOI] [PubMed] [Google Scholar]
- 23. White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY [Google Scholar]
- 24. Lindner DL, Banik MT. 2009. Effects of cloning and root-tip size on observations of fungal ITS sequences from Picea glauca roots. Mycologia 101:157–165 [DOI] [PubMed] [Google Scholar]
- 25. Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118 [DOI] [PubMed] [Google Scholar]
- 26. R Development Core Team 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- 27. Sturtz S, Ligges U, Gelman A. 2005. R2WinBUGS: a package for running WinBUGS from R. J. Stat. Softw. 12:1–16 [Google Scholar]
- 28. Kéry M. 2010. Introduction to WinBUGS for ecologists. Elsevier, New York, NY [Google Scholar]
- 29. Plummer M, Best N, Cowles K, Vines K. 2006. CODA: convergence diagnosis and output analysis for MCMC. R News 6:7–11 [Google Scholar]
- 30. Chaturvedi S, Rudd RJ, Davis A, Victor TR, Li X, Appler KA, Rajkumar SS, Chaturvedi V. 2011. Rapid real-time PCR assay for culture and tissue identification of Geomyces destructans: the etiologic agent of bat geomycosis (white-nose syndrome). Mycopathologia 172:247–256 [DOI] [PubMed] [Google Scholar]