Abstract
A previous study suggested that delayed sample preparation can cause a false-positive plasma cytomegalovirus (CMV) PCR. We evaluated 78 samples submitted for CMV testing, and we report that a 24-h delay in plasma separation did not affect viral quantitation by real-time PCR. Laboratories should evaluate specimen handling requirements for their own PCR technology.
Cytomegalovirus (CMV) is an important cause of morbidity and mortality in immunocompromised individuals (2). Accordingly, monitoring of patients for CMV viremia has become an important laboratory tool for transplantation services. We recently developed a real-time PCR assay for detection of CMV. Studies comparing our CMV PCR assay with the older CMV antigenemia assay (fluorescent antibody stain for expression of pp65 on mononuclear cells) have demonstrated that PCR is superior for early detection of CMV-related disease (1). After discharge from our institution, posttransplant patients are monitored for CMV viremia for up to 1 year. Because this often requires shipping of samples from distant sites, it is critical to validate specimen handling strategies to ensure correct results.
For in-house patients, our PCR specimen handling protocols call for plasma to be separated immediately from EDTA-anticoagulated whole blood and stored frozen prior to testing. In contrast, the antigenemia test previously used for outpatients utilized EDTA-anticoagulated whole blood that was shipped and stored at room temperature for up to 48 h before processing. Although room temperature shipping of EDTA-anticoagulated whole blood is logistically simpler than immediate separation of plasma, it is unclear whether such handling allows accurate quantitation of CMV by real-time PCR. Shafer et al. reported that delayed separation of plasma increased the frequency of positive PCR results for CMV in latently infected patients (3). Those authors suggested that lysis of latently infected leukocytes released CMV into the plasma and thus increased the number of positive results. Shafer et al. used semiquantitative nested PCR followed by hybridization to radiolabeled probe, gel electrophoresis, and autoradiographic detection of amplified product. In contrast, our laboratory has used a rapid real-time TaqMan assay performed on the ABI 7700 (Boeckh et al., submitted). Schafer's assay amplified a region within the coding sequence of glycoprotein B (gB), while our amplicon spanned the junction between gB and UL123. Because of these differences, we felt it necessary to investigate whether delayed separation of plasma from EDTA-anticoagulated blood would increase the frequency of detection of CMV DNA by our assay.
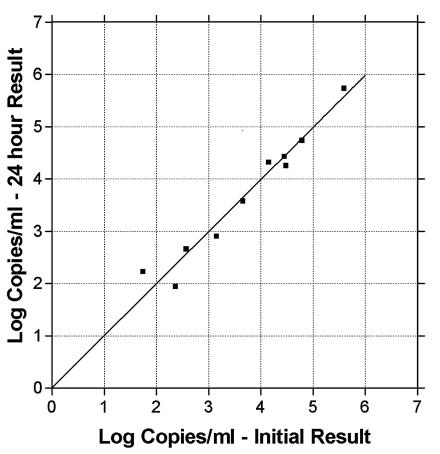
To evaluate the potential release of CMV DNA from cells in whole blood, it was necessary to establish the stability of CMV DNA in plasma after separation from cells. Ten previously PCR-positive plasma samples were chosen to encompass our clinical measuring range. These samples, stored at −20°C, were thawed, and a portion was separated for extraction and CMV DNA quantity determination. The remaining portion was stored for 24 h at room temperature and then extracted, and the CMV DNA quantity was determined (Fig. 1). No significant changes in quantities were observed between the initial and post-24-h results (slope = 0.965; R2 = 0.863).
FIG. 1.
CMV DNA in plasma is stable over 24 h at room temperature. Previously frozen plasma was thawed and split into aliquots. A portion was extracted immediately, and CMV DNA was quantitated by real-time PCR. The remaining portion was held at room temperature for 24 h before extraction and quantitation of CMV DNA by real-time PCR. Shown is the quantitation of CMV DNA for each plasma sample with immediate versus delayed extraction.
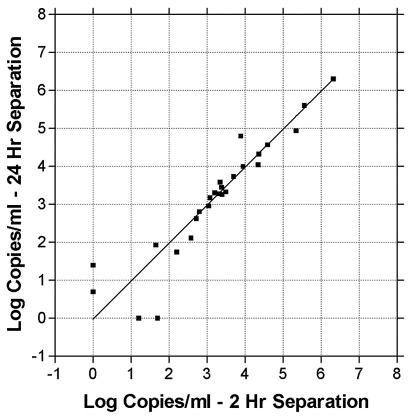
Next, we tested the effects of storing whole blood for 24 h before separating plasma. We obtained 78 consecutive EDTA-anticoagulated whole blood samples collected on-site that were submitted to the laboratory for CMV testing. Each sample was separated into two aliquots of 0.5 ml. From one aliquot, plasma was separated within 2 h. The other aliquot was held at room temperature for 21 to 27 h and then separated. The 2-h samples were either extracted and run on that day's CMV PCR clinical run if collected before noon or frozen at −20°C and run on the next day's CMV PCR run, as is our current clinical practice. Of the 78 samples, 74 showed qualitative agreement between the 2-h and 24-h time points (22 were CMV DNA positive at both time points, and 52 were negative) (Fig. 2). In agreement with Boeckh et al. (submitted), PCR was more sensitive than the antigenemia test, with only 12 of the 22 PCR-positive samples being positive by antigenemia. All of the antigenemia-positive samples were positive by PCR. For PCR-positive samples, no significant difference was seen between the CMV DNA quantities at 2 and 24 h (slope = 0.981; R2 = 0.957). Results from 14 of the 22 PCR-positive samples agreed within 0.2 log, 7 were within 0.5 log, and 1 differed by 0.9 log. Of the 78 total samples, 4 showed differing results between the 2-h and 24-hour time points. Two samples were negative at the 24-h time point but weakly positive at the 2-h time point (50 and 16 copies/ml). Similarly, two samples were negative at 2 h but weakly positive at 24 h (55 and 5 copies/ml). No samples were seen which were negative at 2 h and strongly positive at 24 h.
FIG. 2.
Storage of EDTA-anticoagulated whole blood at room temperature does not affect quantitation of CMV DNA by real-time PCR. A total of 78 consecutive EDTA-anticoagulated whole blood specimens submitted for CMV testing were split into aliquots. A portion was extracted within 2 h, and CMV DNA was quantitated by real-time PCR. The remaining portion was held at room temperature for 21 to 27 h before extraction and quantitation of CMV DNA by real-time PCR. Shown is the quantitation of CMV DNA for each plasma sample with immediate versus delayed extraction.
Our stability data differ from those obtained by Shafer et al. (3). In agreement with our results, Boom et al. evaluated this question in a single clinical specimen and found that quantitative CMV PCR results did not change even after 48 h of storage as EDTA-anticoagulated whole blood (1a). Those authors suggested that CMV DNA in plasma is present predominantly in fragments of 2,000 bp or less and that CMV assays utilizing small amplicons may detect higher levels of virus than those with larger amplicons. Boom's small amplicon was 138 bp, and the larger amplicon was 578 bp. Shafer et al. reported that the outer amplicon was 268 bp and the inner amplicon about 170 bp. Our amplicon was 150 bp. However, since Boom et al. reported similar stability of quantitative CMV PCR results using either the 138- or 578-bp amplicon (1a), it appears unlikely that amplicon size differences account for the CMV stability discrepancies between studies.
The ability to accurately measure CMV DNA in samples stored and transported as EDTA-anticoagulated whole blood offers several advantages to reference laboratories. Transport of whole blood at room temperature decreases shipping costs compared to those of shipping plasma on dry ice, and savings can be substantial for frequently monitored patients. Blood can be collected using standard EDTA tubes instead of more-costly plasma separation tubes. Quality control has been improved by performing all separations in our own laboratory. In our system, valid results can be obtained from EDTA-anticoagulated blood stored at room temperature for at least 24 h before separation of plasma. Individual laboratories should evaluate this according to their own technology for monitoring CMV.
REFERENCES
- 1.Boeckh, M., M. L. Huang, J. Ferrenberg, T. Stevens-Ayers, L. Stensland, W. G. Nichols, and L. Corey. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 42:1142-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Boom, R., C. J. Sol, T. Schuurman, A. Van Breda, J. F. Weel, M. Beld, I. J. Ten Berge, P. M. Wertheim-Van Dillen, and M. D. De Jong. 2002. Human cytomegalovirus DNA in plasma and serum specimens of renal transplant recipients is highly fragmented. J. Clin. Microbiol. 40:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nichols, W. G., and M. Boeckh. 2000. Recent advances in the therapy and prevention of CMV infections. J. Clin. Virol. 16:25-40. [DOI] [PubMed] [Google Scholar]
- 3.Schafer, P., W. Tenschert, M. Schroter, K. Gutensohn, and R. Laufs. 2000. False-positive results of plasma PCR for cytomegalovirus DNA due to delayed sample preparation. J. Clin. Microbiol. 38:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]