Abstract
Lactic acid bacteria, especially lactobacilli, are common inhabitants of the gastrointestinal tract of mammals, for which they have received considerable attention due to their putative health-promoting properties. In this study, we describe the development and application of luciferase-expressing Lactobacillus plantarum and Lactococcus lactis strains for noninvasive in vivo monitoring in the digestive tract of mice. We report for the first time the functional in vitro expression in Lactobacillus plantarum NCIMB8826 and in Lactococcus lactis MG1363 of the click beetle luciferase (CBluc), as well as Gaussia and bacterial luciferases, using a combination of vectors, promoters, and codon-optimized genes. We demonstrate that a CBluc construction is the best-performing luciferase system for the noninvasive in vivo detection of lactic acid bacteria after oral administration. The persistence and viability of both strains was studied by bioluminescence imaging in anesthetized mice and in mouse feces. In vivo bioluminescence imaging confirmed that after a single or multiple oral administrations, L. lactis has shorter survival times in the mouse gastrointestinal tract than L. plantarum, and it also revealed the precise gut compartments where both strains persisted. The application of luciferase-labeled bacteria has significant potential to allow the in vivo and ex vivo study of the interactions of lactic acid bacteria with their mammalian host.
INTRODUCTION
Lactococci and lactobacilli are lactic acid bacteria (LAB) which comprise a large variety of microorganisms that are applied to industrial and artisanal dairy, meat, or plant fermentation. Some selected strains are believed to be beneficial to human and animal health and are marketed as probiotics. Their major field of activity is believed to be the gastrointestinal tract (GIT), although many secondary effects outside the gut have been described. Therefore, it is important to understand the interactions between the bacteria administered and the host intestinal system. Unfortunately, the survival and metabolic activities of these bacteria in the GIT often remain uncertain, impairing our understanding of all the beneficial health effects of these organisms. LAB strains may adapt to the conditions in the intestinal tract, as shown by genetic screening and complementary studies that identified genes in Lactobacillus plantarum and Lactobacillus reuteri, that are specifically induced in the GIT of mice or by whole-genome transcriptome profiling of L. plantarum and Lactobacillus johnsonii in mice or humans (for a review, see reference 1). A better understanding of the survival and metabolic activities of LAB would be facilitated by direct in vivo monitoring of these processes in terms of both spatial and temporal evolution.
Bioluminescence, the production of light by luciferase-catalyzed reactions, is a versatile reporter technology with multiple applications both in vitro and in vivo (2). In vivo bioluminescence imaging (BLI) represents one of the most outstanding uses of the technology by allowing the noninvasive localization of luciferase-expressing cells in real time within a small animal (2). Moreover, using luciferase as a reporter of gene expression, it is possible to establish when and where a gene function is needed.
Luciferases are a large family of enzymes that catalyze the oxidation of a substrate, generically called luciferin, to yield oxyluciferin with the concomitant production of light. Three main luciferin-luciferase systems have been utilized for BLI.
The first system is represented by the firefly luciferase from Photinus pyralis (FFluc) and the click beetle luciferase from Pyrophorus plagiophthalamus (CBluc), which use d-luciferin as the substrate, and they depend on ATP and result in the production of a yellow-green light and green-orange light, respectively. Red click beetle and firefly luciferase variants with different emission wavelengths have also been developed, but these have not been fully investigated for in vivo applications (2).
The second system includes the luciferases from the marine organisms Renilla reniformis (a cnidarian species) and Gaussia princeps (a copepod species) and the substrate coenlenterazine. The signal produced by G. princeps (Gluc) has been reported to be stronger than that of FFluc, even though the light emitted is in the blue range and therefore is more susceptible to tissue absorption and scattering. The facts that Gluc is strongly resistant to heat and extreme pH and is secreted by eukaryotic cells also make this system very attractive.
Bacterial luciferases, found in the terrestrial bacterium Photorhabdus luminescens and in marine bacteria from the genera Vibrio and Photobacterium, constitute the third luciferin-luciferase system. These luciferases are heterodimeric enzymes that use FMNH2 and a long-chain aldehyde as the substrates. Bacterial luciferases are encoded by the genes luxAB that form an operon (luxCDABE) together with three additional genes (luxCDE) whose products synthesize the long-chain aldehyde. The main advantage of this system is that it does not need exogenously added substrate, but again the light produced is in the blue range.
A genetic approach using transcriptional fusions of luciferase genes (luxAB) and selected promoters has been developed to study responses of lactococci and lactobacilli to the GIT environment (3–5). However, determining luciferase activity in these studies necessitates the sacrifice of the animals at different time points, dissection of the GIT, and addition of the luciferase substrate. More recently, the whole luxABCDE operon was placed under the control of the nisin-inducible nisA promoter and expressed in L. lactis for rapid detection of nisin in food and milk (6). However, these constructs have never been applied to the imaging of bacteria in vivo.
Our laboratory selected the human isolate L. plantarum NCIMB8826. This strain has become one of the model strains in LAB research, especially since its early genome publication in 2003 (7). This bacterium is also a promising probiotic strain with good technological properties and has the ability to survive and persist in the human GIT (8). We also selected the dairy starter derivative L. lactis MG1363, which is used in complex food fermentations but is also one of the best-characterized, low-GC, Gram-positive bacteria, and it has been extensively studied for its genetic properties. L. lactis MG1363 is known to have a shorter survival in the human GIT than, e.g., L. plantarum (8). Both strains are also extensively used as mucosal vectors for new molecules with targeted activity in different eukaryotic hosts (9).
We report here on the functional expression in vitro of both Gluc and CBluc (red-emission variant, CBRluc) in L. lactis MG1363 and L. plantarum NCIMB8826 using a combination of vectors, promoters, and codon-optimized genes. We demonstrate that CBRluc is the best-performing luciferase system for the noninvasive detection of both bacteria in vivo in the digestive tract of mice after oral administration. We were able to demonstrate differences in gut persistence between the strains and could follow the precise gut localization of the strains over time.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. The L. plantarum codon-optimized gluc gene under the control of Pldh, the L. lactis codon-optimized gluc gene under the control of Pusp45, the L. plantarum codon-optimized cbrluc gene under the control of Pldh, and the L. lactis codon-optimized cbrluc gene under the control of Pusp45 (the four DNA fragments were synthesized by Eurogentec, Belgium) were cloned into pNZ8148 as BglII-XbaI fragments. The four resulting constructs were subsequently introduced into L. lactis MG1363 and L. plantarum NCIMB8826 by electrotransformation, as described elsewhere (10) and named L. plantarum-Gluc, L. lactis-Gluc, L. plantarum-CBRluc, and L. lactis-CBRluc, respectively. The pNZ8048luxABCDE vector was also introduced into L. lactis NZ9800 and L. plantarum Int-1. The strains were named L. lactis-lux and L. plantarum-lux, respectively. Induction of luxABCDE production in recombinant L. lactis and L. plantarum was performed using nisin as previously described (6, 11). L. lactis MG1363 and L. plantarum NCIMB8826 containing the empty vector pNZ8148 (named L. lactis-pNZ8148 and L. plantarum-pNZ8148, respectively) served as controls in all of the in vitro and in vivo experiments. Strain stability was tested by standard methodology in our laboratory (11).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Lactococcus lactis subsp. cremoris MG1363 | Plasmid free | 10 |
| L. lactis NZ9800 | Strain derived from L. lactis MG1363, ΔnisA, non-nisin producer, pepN::nisRK | 24 |
| Lactobacillus plantarum NCIMB8826 | Originally isolated from human saliva | NCIMB |
| L. plantarum NCIMB8826 Int-1 | NCIMB8826 containing nisRK genes stably integrated to the tRNASer locus | 11 |
| Escherichia coli MC1061 | araD139 Δ(ara-leu)7696 lacX74 galV galK hsr-hsm rpsL | Invitrogen |
| Plasmids | ||
| pNZ8148 | Cmr, L. lactis pSH71 replicon | MoBiTech |
| pMEC252 | pNZ8148 carrying Gluc cDNA optimized for L. plantarum codon fused to the L. plantarum Pldh promoter (lactate dehydrogenase) | This study |
| pMEC253 | pNZ8148 carrying Gluc cDNA optimized for L. lactis codon fused to the L. lactis Pusp45 promoter | This study |
| pMEC256 | pNZ8148 carrying CBRluc cDNA optimized for L. plantarum codon fused to Pldh | This study |
| pMEC257 | pNZ8148 carrying CBRluc cDNA optimized for L. lactis codon fused to Pusp45 | This study |
| pNZ8048luxABCDE | pNZ8048 vector carrying the insert luxABCDE fused to the PnisA promoter | 6 |
Cmr, resistance to chloramphenicol.
Escherichia coli strains were cultured in Luria-Bertani broth at optimal growth temperatures. L. lactis was grown at 30°C in M17 medium (Difco, Becton, Dickinson, Sparks, MD) supplemented with 0.5% glucose. L. plantarum was grown at 37°C in MRS medium (Difco, Becton, Dickinson). Chloramphenicol (Sigma-Aldrich, St. Quentin Fallavier, France) was added to culture media for bacterial selection when necessary at a final concentration of 20 μg/ml for E. coli and 10 μg/ml for lactic acid bacterial strains.
In vitro bioluminescence quantification.
The level of each recombinant luciferase was evaluated in triplicate. Recombinant bacteria were grown in M17 or MRS medium overnight (stationary phase). When the optical density (OD) reached 2, 50 μl of each bacterial culture was distributed in black microplates (Nunc, Thermo Fisher, NY) and imaged after addition of either 50 μl of the Biolux Gluc substrate (New England BioLabs, France), which is the substrate for Gluc, or 50 μl of the Bright-Glo Luciferase (Promega, Madison, WI), which is the substrate for CBRluc. Luminescence was measured at room temperature on the in vivo imaging system (IVIS) Lumina XR (Caliper Corp., Alameda, CA) using Living Image software (Caliper, PerkinElmer) and acquiring the signal for 1 to 30 s. Each individual well which contains bacterial culture was determined manually as a region of interest (ROI). The luminescence of LuxABCDE transformants was measured in a similar way but without adding substrate. Strains were compared on the basis of photons per second (p/s) measured per ml of culture. L. lactis MG1363 and L. plantarum NCIMB8826 containing the empty vector pNZ8148 were used to measure the background luminescence.
Preparation of bacterial strains and administration to mice.
Bacterial strains were grown overnight (stationary phase), harvested by centrifugation, and washed with phosphate-buffered saline (PBS). Mice received 5 × 1010 CFU in 200 μl gavage buffer (0.2 M NaHCO3 buffer containing 1% glucose, pH 8). Eight-week-old female BALB/c mice were purchased from Charles River (St. Germain sur l'Arbresle, France). Experiments were performed in an accredited establishment (no. A59107; Institut Pasteur de Lille) according to European guidelines (number 86/609/CEE), and animal protocols were approved by the local ethics committee.
In vivo persistence of LAB in the GIT of mice.
Groups of mice received a daily dose of 5 × 1010 CFU of live L. plantarum-CBRluc, L. lactis-CBRluc, L. lactis-lux, or L. lactis-Gluc intragastrically for one or four consecutive days. Control mice received L. plantarum-pNZ8148 and L. lactis-pNZ8148 control strains in the different experiments. Fecal samples were collected individually at different time points and mechanically homogenized in MRS medium at 100 mg of feces/ml. Dilutions were plated onto the selective media described above and incubated before enumeration. No chloramphenicol-resistant bacterium was detected in noninoculated mice. Mice were sacrificed by cervical dislocation, and mouse digestive tracts (from stomach to rectum) were immediately excised for ex vivo bioluminescence imaging. According to Foucault et al. (12) and Rhee et al. (13), the intestines were injected with air to enhance the bioluminescent signal and immediately imaged with IVIS.
In vivo bioluminescence imaging.
Bioluminescence imaging was performed using a multimodal IVIS Lumina XR (Caliper, PerkinElmer), which consists of a cooled charge-coupled-device camera mounted on a light-tight specimen chamber. Prior to bioluminescent imaging, mice were anesthetized with 2% isoflurane. d-Luciferin potassium salt (Caliper, PerkinElmer) at 30 mg/ml was then administered to animals inoculated with CBRluc-expressing strains by intragastric inoculation (200 μl/mouse). To image mice administered strains expressing GLuc, XenoLight Rediject Coelenterazine (Caliper, PerkinElmer) at 150 μg/ml was administered via the intraperitoneal route (100 μl/mouse). Mice were placed into the camera chamber of the IVIS, where a controlled flow of 1.5% isoflurane in air was administered through a nose cone via a gas anesthesia system (Temsega, Lormont, France). A grayscale reference image under low illumination was taken as an overlay prior to quantification of emitted photons over 1 s to 5 min, depending on signal intensity and using the software program Living Image (Caliper, PerkinElmer). For anatomical localization, a pseudocolor image representing light intensity (blue, least intense, to red, most intense) was generated using the Living Image software and superimposed over the grayscale reference image. For each individual mouse, there was only one ROI corresponding to the mouse digestive tract, and this ROI was determined manually. Bioluminescence was quantified using the Living Image software (given as p/s). Seventy-five mg of barium sulfate was given twice by the oral route in 200 μl gavage buffer 3 h and 10 min prior to imaging as an X-ray contrast agent. The mouse was then imaged in both bioluminescence and X-ray modes using the multimodal IVIS Lumina system.
RESULTS
Characterization of bioluminescent L. lactis and L. plantarum.
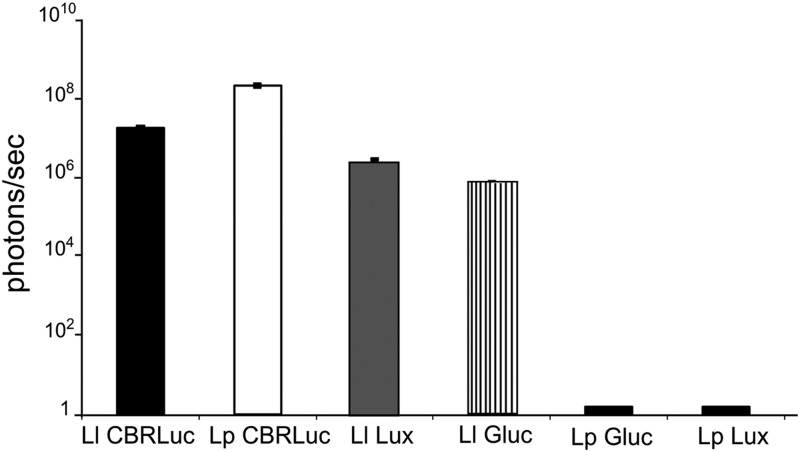
We have observed that the production of different luciferases in L. lactis and L. plantarum did not affect the growth of the strain (data not shown). Luciferase production by the different recombinant L. lactis and L. plantarum strains was evaluated in vitro directly on bacterial cultures (Fig. 1) and in culture supernatants after centrifugation. Results, expressed in p/s/ml of culture, show that the maximum bioluminescence signal was obtained with L. plantarum-CBRluc with mean values of 2 × 108 p/s. A high luminescent signal was produced by L. lactis-CBRluc and L. lactis-lux with mean values of 1.7 × 107 and 2.7 × 106 p/s, respectively. The lowest bioluminescence signal was obtained with L. lactis-Gluc with mean values of 7 × 105 p/s. We did not detect any bioluminescent signal in culture supernatants of these different recombinant strains, showing that the luciferase production was strictly intracellular. No bioluminescent signal was detected in cultures of L. plantarum-Gluc or L. plantarum-lux.
Fig 1.
Bioluminescence measured in cultures of L. lactis-CBRluc, L. plantarum-CBRluc, L. lactis-lux, and L. lactis-Gluc. Strains were cultured overnight (stationary phase). All strains were adjusted to an OD of 2 and distributed in black microplates. The values represented correspond to the means from three independent cultures measured in triplicate. Results are given as p/s/ml of culture, and the background of L. plantarum-pNZ8148 and L. lactis-pNZ8148 has been subtracted from each respective value. The error bars indicate standard deviations. Ll, L. lactis; Lp, L. plantarum. Overall differences between the groups were assessed using the Kruskal-Wallis nonparametric test. Each strain was statistically different from the others, and we decided not to show those results for clarity purposes.
The stability of the different plasmids in the recombinant strains was tested in vitro by subculture in M17 or MRS medium over a 10-day period with replica plating of an aliquot on medium containing chloramphenicol; the bioluminescent signal was also monitored in parallel. In L. lactis, plasmids pMEC253, pMEC257, and pNZ8048luxABCDE were remarkably stable, with 100% of bioluminescent colonies after 10 days of daily subculture. In L. plantarum, pMEC256 was also stable, with 100% of bioluminescent colonies after 10 days of daily subculture.
The excellent correlation (R2 > 0.98) between CFU counts and bioluminescent signals obtained after serial dilutions of total cultures from L. plantarum-CBRluc, L. lactis-CBRluc, and L. lactis-lux indicated that photon emission levels accurately reflect bacterial numbers in total cultures (data not shown). The bioluminescence system allowed the detection of bacterial quantities as low as 5 × 104 CFU/ml for L. plantarum-CBRluc, 5 × 105 CFU/ml for L. lactis-CBRluc, and 5 × 106 CFU/ml for L. lactis-lux. We did not find a good correlation between the CFU counts and the bioluminescent signal for L. lactis-Gluc, because the bioluminescence signal was too low already to be detected after the first 10-fold dilution.
Transit of bioluminescent L. lactis and L. plantarum in mice after one oral administration.
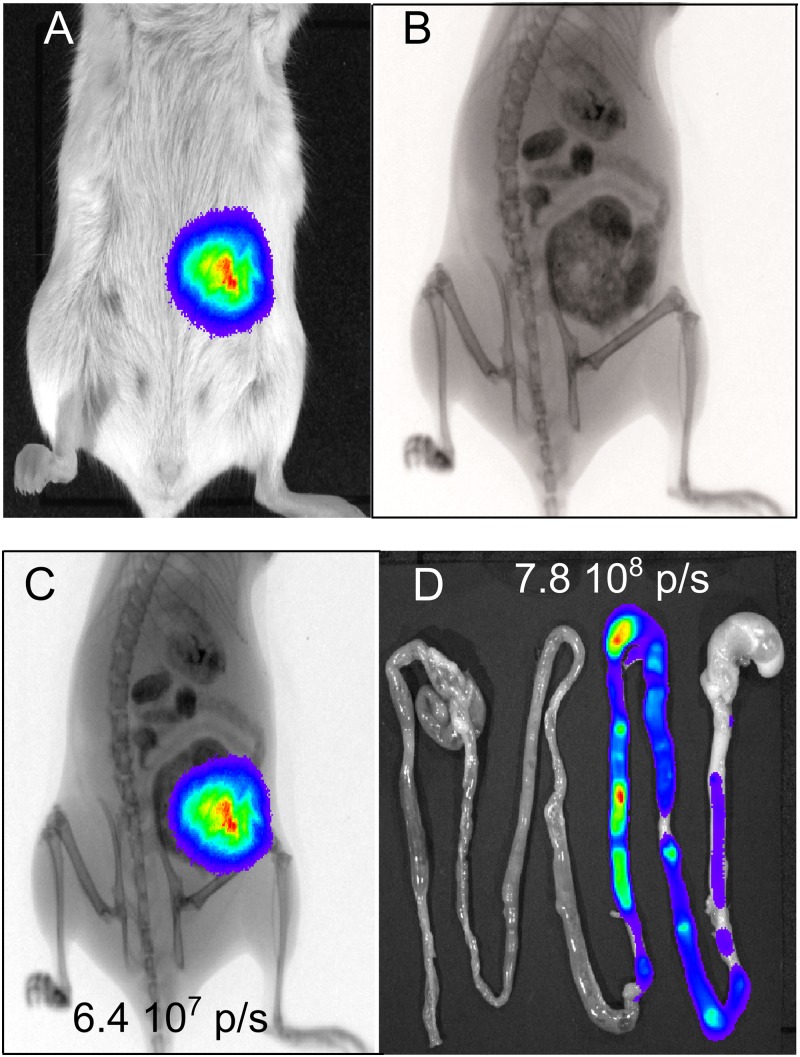
To determine the spatial and temporal transit of L. plantarum-CBRluc, L. lactis-CBRluc, and L. lactis-lux in the GIT of mice after a single oral administration, the signal produced by the recombinant strains was measured in vivo by imaging. Mice were also imaged in both bioluminescence and X-ray modes for anatomical coregistration of bioluminescent signals from the GIT (Fig. 2). In addition, the intestines of some mice were removed at different time points and imaged ex vivo. Bacteria expressing the CBR luciferase do not produce luciferin, and the substrate has to be added exogenously, whereas the ATP is available endogenously with the L. lactis-lux strain. d-luciferin is usually injected via the intraperitoneal route and distributes rapidly throughout the mice (14). After intraperitoneal injection of 200 μl/mouse of the substrate at 30 mg/ml in PBS to mice administered L. lactis-CBRluc or L. plantarum-CBRluc, the bioluminescent signal peaked 5 min postinoculation and decreased rapidly (data not shown). After oral administration of 200 μl/mouse of the substrate at 30 mg/ml to mice 1 h before the administration of L. plantarum-CBRluc or L. lactis-CBRluc, the transcutaneous bioluminescent signal was detectable immediately after substrate introduction, and the maximal signal in the digestive tracts was obtained after 1 h. The signal reached a plateau that lasted approximately 5 h and then started to decline slowly 7 h postinoculation (data not shown). Similar kinetic results have been shown by Foucault et al. with luciferin given by the oral route to mice colonized in the digestive tract with nonpathogenic E. coli strains expressing FFluc (12). In all subsequent experiments, d-luciferin was given intragastrically 1 h before the administration of L. plantarum-CBRluc and L. lactis-CBRluc.
Fig 2.
Monitoring of intestinal transit of L. lactis and L. plantarum by bioluminescence imaging in whole animals. L. lactis-CBRluc, L. plantarum-CBRluc, and L. lactis-lux (5 × 1010 CFU) was inoculated intragastrically into mice, and the bioluminescent signal was measured transcutaneously in whole animals at different time points postfeeding. (A) The intensity of the transcutaneous photon emission is represented as a pseudocolor image. (B) The same mouse was imaged in X-ray mode after barium sulfate administration. (C) The mouse was imaged in both bioluminescence and X-ray modes, and the bioluminescent signal was quantified in the whole animal. (D) The digestive tract of the mouse was then dissected after sacrifice, and the bioluminescent signal was quantified on intact organs. A representative mouse is shown.
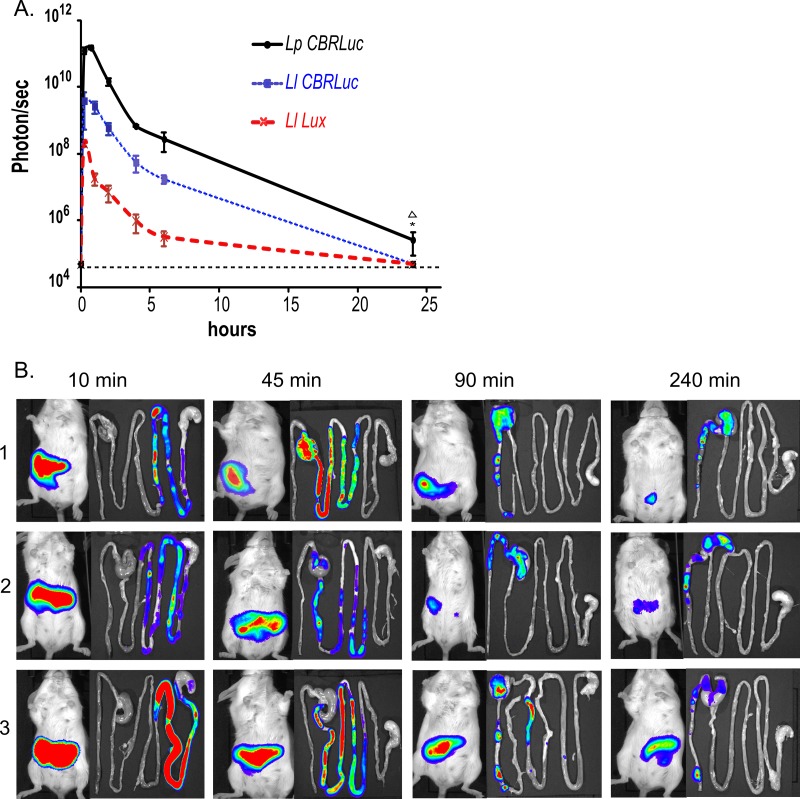
At 0 and 30 min and 1, 1.5, 2, 4, 6, and 24 h after the administration of L. plantarum-CBRluc, L. lactis-CBRluc, and L. lactis-lux, respectively, to mice, the bioluminescent signal was quantified by imaging directly on three anesthetized mice (Fig. 3A). The same three mice were used during the whole experiment. Before the administration of strains to mice (time zero), we did not detect a bioluminescent signal (2 × 104 p/s being the background signal obtained with the IVIS). The signal became very intense 5 min after administration: L. plantarum-CBRluc emitted a bioluminescent signal (mean value of 3 × 1011 p/s) approximately 100 and 1,000-fold superior to that of L. lactis-CBRluc (mean value of 3 × 109 p/s) and L. lactis-lux (mean value of 1 × 108 p/s), respectively. The bioluminescent signals of L. plantarum-CBRluc and L. lactis-CBRluc remained at a plateau until almost 1 h, whereas the signal of L. lactis-lux declined very rapidly after 30 min. After 24 h the signals of both L. lactis strains decreased to the background level, whereas the signal of L. plantarum-CBRluc declined to 2 × 105 p/s. While the three kinetic curves of the bioluminescent signals of L. plantarum-CBRluc, L. lactis-CBRluc, and L. lactis-lux show a similar overall decline, the signals of L. plantarum-CBRluc are always higher than those of L. lactis-CBRluc, and the signals of L. lactis-CBRluc are always higher than those of L. lactis-lux, as observed in the in vitro experiments.
Fig 3.
Transit of L. lactis and L. plantarum in the digestive tract of mice. Groups of mice were fed once with 5 × 1010 CFU of L. lactis-CBRluc, L. plantarum-CBRluc, or L. lactis-lux. At each time point, the bioluminescent signals in p/s in whole animals (A) are plotted for each set of three mice, with standard deviations. Two mice were sacrificed at each time point, and (B) one representative image of one mouse and its digestive tract are shown after 10, 45, 90, and 240 min in mice fed with L. lactis-CBRluc (1), mice fed with L. lactis-lux (2), and mice fed with L. plantarum-CBRluc (3). d-Luciferin was given intragastrically 1 h before administration of bacteria, and the bioluminescence signal was measured 3 h after inoculation of the substrate. For the bioluminescent signals, overall differences between the groups were assessed using the Kruskal-Wallis nonparametric test, and those found to be significant (P < 0.05) are indicated with an asterisk for comparison between L. plantarum-CBRluc and L. lactis-CBRluc and a triangle between L. plantarum-CBRluc and L. lactis-lux. The background level for the bioluminescent signal is represented by a dashed line.
We also determined the localization of the bioluminescent bacteria in the GITs of mice ex vivo (Fig. 3B). We found that it took approximately 90 min for the three strains to reach the cecum/colon. By 4 and 6 h after intragastric administration, bacteria were localized throughout the cecum and colon (data not shown). After 24 h, no bioluminescent L. lactis-CBRluc or L. lactis-lux bacteria could be detected anymore in the intestines of mice, while L. plantarum-CBRluc bacteria were still localized in the cecum and colon (data not shown). Moreover, quantification of the bioluminescent signal (in p/s) was done ex vivo in the intestines of mice (Fig. 3B). Results showed that the values of the signals obtained ex vivo were systematically higher than the ones obtained from the anesthetized animal (data not shown). No doubt the signal gets weakened by the passage through the mouse tissue and skin compared to a signal obtained directly from the intestine.
Enumeration of bacteria and quantification of their bioluminescent signal in mouse feces after a single oral administration.
We monitored the number of L. plantarum-CBRluc, L. lactis-CBRluc, and L. lactis-lux bacteria as well as their respective bioluminescent signals in mouse feces at different time points after the oral administration of bacteria (Fig. 4A and B). The number of viable bacteria increased with time in feces, reaching its maximum level after 2 h with approximately 109 CFU/100 mg feces for each strain. This peak correlated perfectly with the maximum level of bioluminescent signal of 8 × 109 p/s/100 mg of feces and 2 × 109 p/s/100 mg feces after 2 h for L. plantarum-CBRluc and L. lactis-CBRluc, respectively. For L. lactis-lux, the maximum level of bioluminescent signal was significantly lower, reaching only 107 p/s/100 mg of feces. The number of both L. lactis-CBRluc and L. lactis-lux organisms remained at a plateau for about 4 h and then declined. After 24 h, this number reached approximately 105 CFU/100 mg feces with a bioluminescent signal corresponding to the background level for both L. lactis strains. After 24 h, the number of L. plantarum-CBRluc organisms reached 2 × 106 CFU/100 mg feces, with a bioluminescent signal of 2 × 106 p/s/100 mg feces. After 72 h, no L. lactis-CBRluc could be found in the feces, while the number of L. plantarum-CBRluc organisms was still approximately 104 CFU/100 mg feces, although the bioluminescent signal corresponded to the background level.
Fig 4.
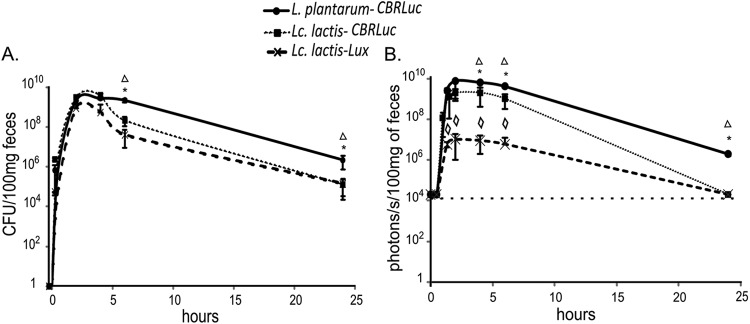
Transit of L. lactis and L. plantarum in feces of mice. Groups of mice were fed once with 5 × 1010 CFU of L. lactis-CBRluc, L. plantarum-CBRluc, and L. lactis-lux. At every time point, averages of the CFU counts per 100 mg of feces (A) and p/s per 100 mg of feces (B) are plotted for each set of three mice, with standard deviations. Overall differences between groups were assessed using the Kruskal-Wallis nonparametric test, and those found to be significant (P < 0.05) are indicated with an asterisk for comparison between L. plantarum-CBRluc and L. lactis-CBRluc, a triangle between L. plantarum-CBRluc and L. lactis-lux, and a diamond between L. lactis-CBRluc and L. lactis-lux. The background level for the bioluminescent signal is represented by a dashed line.
Persistence of bioluminescent L. lactis and L. plantarum in mice after 4 oral administrations.
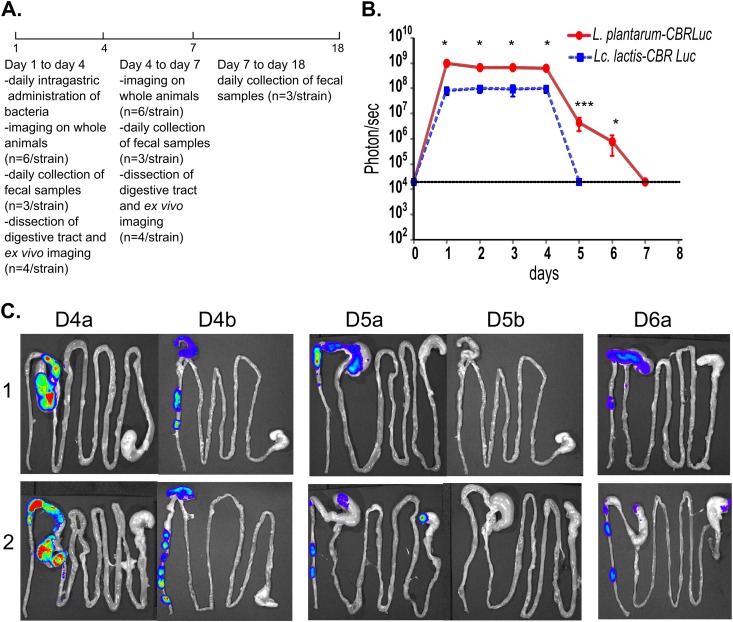
To study more thoroughly the persistence of bioluminescent strains in mice after multiple daily oral administrations, we chose the two most bioluminescent strains, L. plantarum-CBRluc and L. lactis-CBRluc. Groups of mice were given a daily dose of 5 × 1010 CFU of live L. plantarum-CBRluc and L. lactis-CBRluc intragastrically for four consecutive days. The experimental design is described in Fig. 5A. The bioluminescent signal was quantified every day by bioluminescence imaging directly on 6 anesthetized mice (Fig. 5B). The same 6 mice were used during the whole experiment. The signal was very strong for both strains on day 1 and remained at a plateau until day 4. L. plantarum-CBRluc emitted a bioluminescent signal (mean value of 7 × 108 p/s) that was 8-fold superior to the L. lactis-CBRluc signal (mean value of 9 × 107 p/s). The respective signal measured for both strains on days 1 to 4 was similar to the bioluminescence corresponding to the 3-h time point represented in Fig. 3A, which was obtained after a single oral administration of the strains. The signal of L. lactis-CBRluc then declined to the background level at day 5, whereas the signal of L. plantarum-CBRluc declined to the background level only at day 7.
Fig 5.
Persistence of L. lactis and L. plantarum in the digestive tract of mice after four oral daily administrations. (A) The experimental design. Groups of mice were fed once daily with 5 × 1010 CFU of L. lactis-CBRluc and L. plantarum-CBRluc for four consecutive days (days 1 to 4). (B) From day 1 to 8, p/s in whole animals for each set of six mice are plotted with standard deviations. (C) Four mice were sacrificed from day 4 to day 7, and representative images of the digestive tracts of two mice are shown (1 and 2) at day 4 (D4), 5 (D5), and 6 (D6) in mice fed with L. plantarum-CBRluc (a) and mice fed with L. lactis-CBRluc (b). For the bioluminescent signals, differences between groups were assessed using the Mann-Whitney nonparametric test, and those found to be significant are indicated with one (P < 0.05) or three (P < 0.001) asterisks. The background level for the bioluminescent signal is represented by a dashed line.
From day 1 to day 4, both strains were localized predominantly in the cecum and colon as of 3 h after the intragastric inoculation of bacteria (data not shown for day 1 to day 3). At day 5, L. plantarum-CBRluc was also localized predominantly in the cecum and colon (Fig. 5C, D5a1), but remarkably, a strong bioluminescent signal was also found in the stomach of 1 mouse out of 4 (Fig. 5C, D5a2). This could be explained by the fact that mice are coprophagic, hence their fecal material likely served as a secondary source of L. plantarum. On day 5, no signal was measured in the intestines of mice orally administered L. lactis-CBRluc. On day 6, L. plantarum-CBRluc was localized predominantly in the cecum and colon, but a bioluminescent signal was also found in the stomach and ileum of 1 mouse out of 4. Once again, this result could be explained by the fact that mice are coprophagic.
The persistence of bioluminescent bacteria and their respective bioluminescent signals were also investigated in mouse feces (Fig. 6A and B). The L. plantarum-CBRluc strain persisted for 12 days after the last inoculation (day 4) and maintained itself at levels ranging from 107 to 102 CFU/100 mg of feces from days 5 to 14. L. lactis-CBRluc was detected in feces at lower counts after day 4 and for fewer days than L. plantarum.
Fig 6.
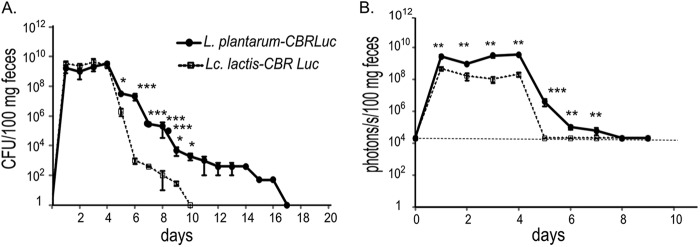
Persistence of bioluminescent bacteria in feces of mice after four oral administrations. Groups of mice were fed once daily with 5 × 1010 CFU of L. lactis-CBRluc and L. plantarum-CBRluc for 4 days (days 1 to 4). Feces were collected daily from days 1 to 14. Means of the CFU per 100 mg of feces (A) and p/s per 100 mg of feces (B) are plotted for each set of six mice, with standard deviations. Differences between groups were assessed using the Mann-Whitney nonparametric test, and those found to be significant are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001. The background level for the bioluminescent signal is represented by a dashed line.
The L. plantarum-CBRluc bioluminescent signal was detected in feces for up to 3 days after the last dose (day 4), while the L. lactis-CBRluc signal was detected at lower levels and was undetectable at day 5. The bioluminescent system allowed the detection of bacterial quantities in feces as low as 106 CFU/100 mg of feces for L. lactis-CBRluc and 105 CFU/100 mg feces for L. plantarum-CBRluc.
DISCUSSION
Mouse models are essential to study the persistence and localization of LAB. However, the conventional approaches are frequently limited by the need to sacrifice large numbers of animals to establish the precise localization of these bacteria. We used BLI for real-time monitoring of Lactococcus lactis MG1363 and Lactobacillus plantarum NCIMB8826 by using several recombinants that express the click beetle luciferase as well as the bacterial Lux operon.
We first optimized the CBRluc and Gluc luciferases for use in these LAB. Our results demonstrate that L. plantarum-CBRluc produced the highest luminescence with a signal 10 times brighter than the one with L. lactis-CBRluc. We made other L. lactis CBRluc constructs with two different L. lactis-specific strong promoters, but we could not obtain a higher bioluminescent signal (data not shown). Moreover, the signal obtained with L. lactis-CBRluc was approximately 30 and 10 times brighter than the one obtained with L. lactis-Gluc and L. lactis-lux, respectively. The comparison could not be made for the recombinant L. plantarum strains, as no bioluminescent signal was found with L. plantarum-lux or L. plantarum-Gluc. These results could be explained by the fact that both lux and Gluc constructs were structurally very unstable in L. plantarum.
Very few studies compared the use of different bioluminescent reporters in the same host: Andreu et al. optimized the use of firefly, Gaussia, and bacterial luciferases in mycobacteria and found that FFluc produced the highest luminescence in vitro, 10 times brighter than that obtained with Lux and 100 times that of Gluc (15). These conclusions are similar to ours for L. lactis, except that we used CBRluc instead of FFluc, but these two luciferases belong to the same luciferase system, and both require d-luciferin and ATP (2). Andreu et al. also showed that FFluc emitted a bioluminescent signal 10 times superior to that of the Lux system in mycobacteria in vivo (15).
We proceeded to explore if the bioluminescence signal obtained with the recombinant strains was strong enough for the imaging of bacteria in vivo. The signal from Gluc-producing L. lactis in mice could not be distinguished from the background produced by the substrate alone (data not shown). This result was predictable, since the signal obtained was already low in vitro. Andreu et al. also found that the signal from Gluc-producing Mycobacterium smegmatis in mice could not be distinguished from the strong background signal produced by coelenterazine alone (15). However, published work with eukaryotic cells states that the Gluc signal is 1,000 times stronger than that of FFluc in cell culture and is as bright as the FFluc signal in vivo with no background detected in vivo, even using a 20-fold higher concentration of coelenterazine (16).
Importantly, LAB bioluminescence could be detected in mice after a single oral administration of either CBRluc- or Lux-producing L. lactis or CBRluc-producing L. plantarum. Bioluminescence was also detected ex vivo in the digestive tract and feces of mice. The highest bioluminescence was obtained with L. plantarum-CBRluc and L. lactis-CBRluc. The L. plantarum-CBRluc signal was approximately 10- and 1,000-fold superior to those of L. lactis-CBRluc and L. lactis-lux, respectively.
The substrate might be a limiting factor in the operon lux reporter system. Luminescence depends on the intracellular FMNH2 concentration, which is directly correlated with the metabolic activity of the cells (17). This dependency is well documented for Gram-positive bacteria and is illustrated by a rapid decline in luminescence upon entry into the stationary growth phase. Coexpression of the lux operon together with a gene encoding a protein that would supply reduced flavin mononucleotide could increase bacterial luminescence (18).
In the past, it has been possible to successfully lux label a range of intestinal pathogens, such as E. coli, Citrobacter rodentium, and Yersinia enterocolitica, and bioluminescence signals could readily be detected from the GIT (12–14, 19, 20). Intestinal commensal bacteria such as Bifidobacterium breve UCC2003 and E. coli K-12, which colonize the GIT, have also been lux labeled and detected in the GIT of mice (12, 21). However, externally administered LAB generally persist but do not replicate actively or colonize permanently the GIT. The CBRluc system, which requires local addition of exogenous substrate, seems more suited to detect such bacteria in vivo. Moreover, it is known that the longer wavelength in the red light spectrum exhibits better penetration through living tissues (2). However, luciferin accessibility to bacteria which are associated with the intestinal mucosa and luminal bacteria might be different and could affect the signal obtained. This clearly needs further investigation.
We found that bioluminescent L. lactis and L. plantarum had similar GIT transit dynamics in the early phase after administration of the strains to mice, even though the bioluminescent signal always remained higher for L. plantarum. Differences between the two strains were observed 24 h postfeeding, with a higher bioluminescent signal in whole animals for L. plantarum associated with a significantly higher number of bacteria and bioluminescent signal in feces of mice. Bacteria in feces were also enumerated after 48 and 72 h, and L. lactis was eliminated more rapidly than L. plantarum. Marco et al. also showed similar transit dynamics of viable L. plantarum WCFS1, a clone of L. plantarum NCIMB8826, by enumeration of bacteria in feces and in gut compartments after a single oral administration (22). However, L. plantarum was not selectively monitored on MRS medium, and it was not possible in their study to conclude whether this organism was still present once the numbers of Lactobacillus cells in mouse feces returned to the initial level 24 h after administration of bacteria (22). We extended these observations by demonstrating that while the majority of L. plantarum bacteria transited the GIT, a small but persistent population of this organism was retained in mice. We confirmed these observations after 4 oral daily administrations, as L. plantarum cells were able to persist in mice for up to 6 days after inoculation. In contrast, L. lactis could not be observed 24 h after the last inoculation in whole animals by bioluminescence imaging. The differences in transit dynamics between both strains were already shown by Grangette et al. in mice after three oral daily administrations (23) and in humans by Marteau et al. after one oral administration in fermented milk (8). We have shown that these differences are clearly more emphasized after multiple daily oral administrations of the bacteria compared to a single administration.
Oozeer et al. observed similar transit dynamics with a Lactobacillus casei strain expressing luxAB (4) and Bacillus subtilis spores fed once to human flora-associated mice. They did not find luciferase activity in the stomach or the duodenum-jejunum compartments after sacrifice of the animals and addition of the substrate (5). We did not find bioluminescent L. lactis or L. plantarum in the stomach either. These results reflect the adverse conditions exposed to the ingested microorganisms and the absence of protein synthesis in that compartment of the gastrointestinal tract. However, in contrast to the study of Oozeer et al., we did find luciferase activity for both strains in the duodenum-jejunum compartment. Cell concentrations in that compartment might have been too low in their study to elicit a measureable luciferase activity, whereas bioluminescence imaging might be more sensitive ex vivo on dissected organs. We could not detect luciferase activity in the duodenum-jejunum 90 min after oral administration of the respective bacteria, reflecting the rapid transit in that gut compartment. Luciferase activity could still be detected, however, in the cecum and colon up to 6 h after the administration of bacteria, reflecting the active physiological state of bacteria in these compartments.
Cecum and colon were found to be the predominant sites for persistent L. plantarum in mice. Interestingly, enteric pathogens such as E. coli, C. rodentium, and Y. enterocolitica exhibit murine colon and cecal colonization (12, 14, 20, 21). B. breve was also shown to colonize the cecum (21). The cecum may be the site which allows certain pathogens and nonpathogens to adapt to the intestinal environment and where genes required for adaptation of the colon are activated. The cecum may also act as a reservoir shedding bacteria into the colon.
The application of luciferase-labeled bacteria has significant potential to allow further study of the interactions of LAB with a mammalian host. This system may be used to analyze gene expression during transit and persistence in the digestive tract, for in situ real-time investigation of promoter activities both in vitro and in vivo, or for the study of the impact of gene mutations on the course of the transit and persistence of lactobacilli in vivo.
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support of the CNIEL and Syndifrais.
We heartily thank Meliza Sendid for her participation in the construction of the recombinant strains and Lucie Caerou for her participation in animal experiments and careful analysis. We thank the BioImaging Center of Lille (Frank Lafont) for the use of the Lumina IVIS. We also thank Kevin Francis and Béatrice David from Caliper for their great help in setting up the project and for the use of the Living Image software.
Footnotes
Published ahead of print 30 November 2012
REFERENCES
- 1. Bron PA, van Baarlen P, Kleerebezem M. 2012. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 10:66–78 [DOI] [PubMed] [Google Scholar]
- 2. Andreu N, Zelmer A, Wiles S. 2011. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol. Rev. 35:360–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corthier G, Delorme C, Ehrlich SD, Renault P. 1998. Use of luciferase genes as biosensors to study bacterial physiology in the digestive tract. Appl. Environ. Microbiol. 64:2721–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oozeer R, Goupil-Feuillerat N, Alpert CA, van de Guchte M, Anba J, Mengaud J, Corthier G. 2002. Lactobacillus casei is able to survive and initiate protein synthesis during its transit in the digestive tract of human flora-associated mice. Appl. Environ. Microbiol. 68:3570–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oozeer R, Mater DD, Goupil-Feuillerat N, Corthier G. 2004. Initiation of protein synthesis by a labeled derivative of the Lactobacillus casei DN-114 001 strain during transit from the stomach to the cecum in mice harboring human microbiota. Appl. Environ. Microbiol. 70:6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Immonen N, Karp M. 2007. Bioluminescence-based bioassays for rapid detection of nisin in food. Biosens. Bioelectron. 22:1982–1987 [DOI] [PubMed] [Google Scholar]
- 7. Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vesa T, Pochart P, Marteau P. 2000. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14:823–828 [DOI] [PubMed] [Google Scholar]
- 9. Daniel C, Roussel Y, Kleerebezem M, Pot B. 2011. Recombinant lactic acid bacteria as mucosal biotherapeutic agents. Trends Biotechnol. 29:499–508 [DOI] [PubMed] [Google Scholar]
- 10. Wells JM, Wilson PW, Le Page RW. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629–636 [DOI] [PubMed] [Google Scholar]
- 11. Pavan S, Hols P, Delcour J, Geoffroy MC, Grangette C, Kleerebezem M, Mercenier A. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foucault ML, Thomas L, Goussard S, Branchini BR, Grillot-Courvalin C. 2010. In vivo bioluminescence imaging for the study of intestinal colonization by Escherichia coli in mice. Appl. Environ. Microbiol. 76:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rhee KJ, Cheng H, Harris A, Morin C, Kaper JB, Hecht G. 2011. Determination of spatial and temporal colonization of enteropathogenic E. coli and enterohemorrhagic E. coli in mice using bioluminescent in vivo imaging. Gut Microbes 2:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wiles S, Pickard KM, Peng K, MacDonald TT, Frankel G. 2006. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect. Immun. 74:5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andreu N, Zelmer A, Fletcher T, Elkington PT, Ward TH, Ripoll J, Parish T, Bancroft GJ, Schaible U, Robertson BD, Wiles S. 2010. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One 5:e10777 doi:10.1371/journal.pone.0010777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11:435–443 [DOI] [PubMed] [Google Scholar]
- 17. Duncan S, Glover LA, Killham K, Prosser JI. 1994. Luminescence-based detection of activity of starved and viable but nonculturable bacteria. Appl. Environ. Microbiol. 60:1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szittner R, Jansen G, Thomas DY, Meighen E. 2003. Bright stable luminescent yeast using bacterial luciferase as a sensor. Biochem. Biophys. Res. Commun. 309:66–70 [DOI] [PubMed] [Google Scholar]
- 19. Gahan CG. 2012. The bacterial lux reporter system: applications in bacterial localisation studies. Curr. Gene Ther. 12:12–19 [DOI] [PubMed] [Google Scholar]
- 20. Trcek J, Fuchs TM, Trulzsch K. 2010. Analysis of Yersinia enterocolitica invasin expression in vitro and in vivo using a novel luxCDABE reporter system. Microbiology 156:2734–2745 [DOI] [PubMed] [Google Scholar]
- 21. Cronin M, Sleator RD, Hill C, Fitzgerald GF, van Sinderen D. 2008. Development of a luciferase-based reporter system to monitor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol. 8:161 doi:10.1186/1471-2180-8-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marco ML, Bongers RS, de Vos WM, Kleerebezem M. 2007. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl. Environ. Microbiol. 73:124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grangette C, Muller-Alouf H, Geoffroy M, Goudercourt D, Turneer M, Mercenier A. 2002. Protection against tetanus toxin after intragastric administration of two recombinant lactic acid bacteria: impact of strain viability and in vivo persistence. Vaccine 20:3304–3309 [DOI] [PubMed] [Google Scholar]
- 24. Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291 [DOI] [PubMed] [Google Scholar]