Abstract
Laminated, microbially produced stromatolites within the rock record provide some of the earliest evidence for life on Earth. The chemical, physical, and biological factors that lead to the initiation of these organosedimentary structures and shape their morphology are unclear. Modern coniform structures with morphological features similar to stromatolites are found on the surface of cyanobacterial/microbial mats. They display a vertical element of growth, can have lamination, can be lithified, and observably grow with time. To begin to understand the microbial processes and interactions required for cone formation, we determined the phylogenetic composition of the microbial community of a coniform structure from a cyanobacterial mat at Octopus Spring, Yellowstone National Park, and reconstituted coniform structures in vitro. The 16S rRNA clone library from the coniform structure was dominated by Leptolyngbya sp. Other cyanobacteria and heterotrophic bacteria were present in much lower abundance. The same Leptolyngbya sp. identified in the clone library was also enriched in the laboratory and could produce cones in vitro. When coniform structures were cultivated in the laboratory, the initial incubation conditions were found to influence coniform morphology. In addition, both the angle of illumination and the orientation of the surface affected the angle of cone formation demonstrating how external factors can influence coniform, and likely, stromatolite morphology.
INTRODUCTION
Stromatolites, laminated organosedimentary structures, provide some of the earliest evidence for life on Earth (1, 2) and date back to ∼3.5 billion years ago (1, 3–5). Stromatolites found in the rock record can have a range of shapes from conical to domical to branching to columnar (6). Understanding how these different structures could form can provide a window into Earth's early history, including the physiological capabilities and diversity of its biota. Modern stromatolitic structures are one type of analog to help interpret how ancient stromatolites may have formed (7–9).
There has been much interest and study of modern stromatolites (7–15). Modern stromatolites are found in both carbonate- and silica-rich environments, have well-developed laminations, and harbor complex microbial communities that contain cyanobacteria, an abundance of Proteobacteria, Planctomycetes, Acidobacteria, and many other types of microbiota from all three domains (7, 13, 14, 16–20). The filamentous cyanobacteria are likely to be the main architects of modern stromatolites, performing such roles as trapping and binding sediment in an adhesive matrix of exopolymeric substances which aid in the precipitation of silica or carbonate (9, 14, 21, 22). The heterotrophs, which can be more abundant than the cyanobacteria (23), are thought to be important in the degradation of the exopolymeric substances, which in carbonate systems can promote calcium carbonate precipitation (24, 25).
Although the biogenicity of many stromatolites is uncertain, the conophyton cone-like stromatolites are less likely to form in the absence of biotic processes (26–28). Modern coniform structures can exist in various stages of lithification, likely caused by cooling and evaporation of silica-rich waters (15), can be laminated (15, 27), and can strongly resemble the conophyton found in the rock record (4). Modern coniform structures can be found in various siliceous springs in Yellowstone National Park (YNP) (14, 15, 27, 29, 30) at temperatures from 22 to 59°C (15, 29). These coniform structures are different from modern stromatolites, and yet understanding the microbial diversity in both stromatolites and cones may help to identify a core community, metabolisms, and physiological processes that can lead to vertical growth and lithification analogous to those in the rock record.
Lau et al. (30) examined the cyanobacterial diversity in microbial mat columns and cones from Black Sand Pool, YNP, by denaturing gradient gel electrophoresis (DGGE) of amplified 16S rRNA genes and subsequent DNA sequencing of extracted bands. These researchers found no difference between the cyanobacterial community in the cones and columns. Six different cyanobacteria were identified in the structures: one Leptolyngbya sp., one Synechococcus sp., and four sequences with no similarity to cultured cyanobacteria. Bosak et al. (29) also examined cyanobacterial diversity in cones and mats in four springs in YNP using similar techniques and found similar filamentous cyanobacteria (Leptolyngbya), as well as Synechocystis-like and Synechococcus-like species in the cones. Both studies (29, 30) found that filamentous cyanobacteria were more common in the cones and unicellular cyanobacteria were more common in the adjacent mats.
Cyanobacterial enrichment cultures have been shown to develop cone-like forms (herein referred to as cones or coniform structures) from a mat growing on an agar or sand surface in the laboratory (14, 27, 29, 31). Laboratory-grown cones have been used as a model to examine diffusion limitation controlling cone spacing (31), the effect of light intensity on morphology (27), and both the effect and the preservation of oxygen bubbles formed by oxygenic photosynthesis (27, 32). However, despite the relative simplicity of the physical coniform structure, the mechanism behind vertical movement out of a surface microbial mat to form a vertical cone remains uncertain. It may be driven by filament entanglement, gliding motility, phototaxis toward light (14, 15), bubble formation at ridges and mounds (8, 27, 32), diffusion limitation (31, 33), or more rapid accumulation of biomass in the upper part of the cone (26, 27, 33).
To understand the biogenesis of a coniform structure, we have investigated partially lithified cones from a living microbial mat at Octopus Spring, YNP. By characterizing the microbial diversity of a coniform community, this study builds on previous work to implicate the important role of Leptolyngbya in coniform formation and suggests that through its abundance, it is likely to be the primary architect of cones in Octopus Spring, YNP. In addition, to determine how environmental factors may shape coniform morphology, we quantitatively tested the hypothesis that phototaxis drives cone formation and found that phototaxis alone cannot explain the orientation of in vitro grown cones.
MATERIALS AND METHODS
Sampling site.
Coniform mat samples were obtained from Octopus Spring, YNP (Fig. 1). Octopus Spring has been well described (34–38). A couple of the orange, partially lithified, nonbranching coniform structures present on the mat surface (height = 2 to 3 cm) of the Octopus Spring outfall were removed under aseptic conditions. Temperatures in the cone-growing region were 17 to 35°C, and the laminar spring water flow was continuous at the time of sampling. The samples for culturing and microscopy were stored with spring water from the same site in a sterile 50-ml conical tube at 4°C. Samples were obtained in June 2008 and June 2010. Only the orange coniform structure was used for analysis. None of the analyses included the underlying green mat. The sample for 16S rRNA gene analysis was stored at −20°C prior to DNA extraction which occurred within 1 week of sample collection. This sample was obtained in June 2010.
Fig 1.
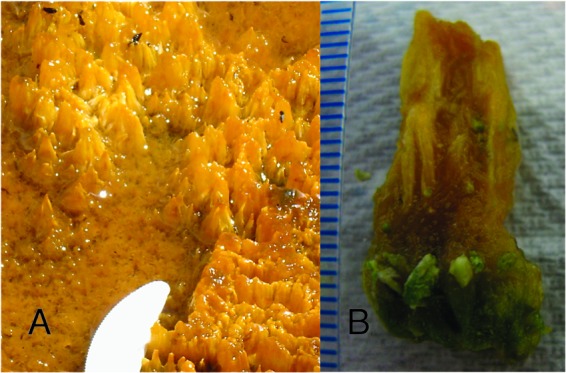
Coniform structures found in Octopus Spring, YNP. (A) Modern cones in their natural setting. Note the tip of plastic knife for scale. (B) Extracted 2.5-cm cone attached to underlying green mat material. The scale markers represent 1 mm.
16S rRNA gene clone library.
Environmental DNA was extracted by using a Mo Bio Powersoil DNA isolation kit, and 16S rRNA genes were amplified using the bacterial primers 8F and 1492R (39) and the archaeal primers 21Fa (40) and 1492R with standard thermocycling conditions of 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1.5 min, with a final 10-min extension at 72°C. Amplified product was cloned into TOPO PCR 2.1 (Invitrogen/Life Technologies) according to the manufacturer's protocol. Individual colonies were picked, and the clones were sequenced by rolling-circle amplification (Sequetech) using the M13 forward primer. Approximately 400 to 800 nucleotides (nt) of the sequence were obtained from 87 clones and used for analysis. Most sequencing reads were over 700 nt in length. Sequences were checked for possible chimeras with Pintail (41). Phylogenetic classification and sequence identity was based on the Ribosomal Database Project classifier (42) and by the basic local alignment search tool (BLAST) against the NCBI GenBank database. The Shannon-Wiener diversity index (43, 44) was calculated with operational taxonomic units defined as a >3% difference in 16S rRNA gene sequence. Representative sequences were deposited in GenBank with accession numbers KC236901 to KC236911.
Microscopy.
For confocal microscopy, a small, single coniform structure from Octopus Spring was extracted aseptically with a razor blade. Autofluorescence and staining with DAPI (4′,6′-diamidino-2-phenylindole) for DNA were used for the identification of chlorophyll a-containing cells and all cells, respectively. Mat sections were observed with a ×63 1.2 numerical aperture (NA) Plan Apochromat water-immersion objective lens, under an excitation beam from a Diode laser at 405 nm for chlorophyll a autofluorescence and at 543 nm for DAPI. Fluorescence emission was detected with band-pass filters at 412 to 590 nm and 604 to 691 nm, respectively.
For scanning electron microscopy (SEM), a 3- to 4-mm piece of the coniform structure from the tip was placed into a mix of 2% formaldehyde–2% glutaraldehyde in 50 mM sodium phosphate (pH 7.2) solution for 1.5 h. The samples were then washed twice with deionized water. The sample was dehydrated for 10 min each time with increasing percentages of ethanol (30, 40, 70, and 100%), covering the entire specimen with the solution. The sample was critically point dried in liquid carbon dioxide and sputter coated with 300 Å of gold palladium with a Pelco Model 3 sputter coater. Samples were examined on a Hitachi 2400 SEM.
Bacterial enrichment and culturing.
Cyanobacteria were enriched using both liquid and solid D media (45). Cyanobacteria enrichments were incubated under constant cool white fluorescent lamps or with a 12-h light/dark cycle at 32°C. Filaments grown on plates were transferred to fresh plates multiple times, followed by a dilution series in liquid medium. 16S rRNA genes were amplified with 8F and 1492R as described previously (39). The sequences were determined using the same primers. Gene sequences from cyanobacteria and heterotrophic isolates (see Table S1 in the supplemental material) were deposited in GenBank under accession numbers KC182742 to KC182752.
The coniform structures were grown as described by Walter et al. (14). Briefly, 50 ml of agar containing D medium was added to a 250-ml Erlenmeyer flask. The Leptolyngbya sp. strain C1 enrichment was streaked onto the agar surface. After 10 to 20 days at 28°C and constant light, 100 ml of liquid D medium was added to flood the well-developed mat. The culture was then incubated in a light/dark cycle or in constant light at 32°C. For structures incubated at an angle of >0° from the horizontal, 20 ml of solid medium was added to 50-ml tubes, the agar was solidified horizontally, and the initial culture was incubated horizontally to form a mat. After flooding of the culture with 30 ml of liquid D medium, the tubes were incubated at 34° from horizontal, and constant light was applied at 0° from horizontal (the side). A cardboard shade was placed horizontally on top of the tubes to decrease reflected light from above. Cultures were incubated for 130 days at 28°C, and then the cone orientation was determined. The number of cones at acute, normal, and obtuse angles from the agar surface was determined for more than 500 cones by five independent individuals, and the results were averaged. The approximate angle of the cones was measured from digital photographs.
Nucleotide sequence accession numbers.
Gene sequences were deposited in GenBank under accession numbers KC182742 to KC182752 and KC236901 to KC236911.
RESULTS AND DISCUSSION
Filamentous morphology abundant in coniform structures.
Confocal microscopy was performed along the entire length of the coniform structure from Octopus Spring to better understand the morphology of the cone on a microscopic level and to identify the relative qualitative abundance and location of cells with or without chlorophyll a. Representative images are shown in Fig. 2. Filamentous cyanobacteria, identified by autofluorescence from chlorophyll a, were found throughout the structure from the tip to the base with greatest abundance in the vertical midsection of the cone and a low abundance in the base of the cone. Surprisingly, the tip of the cone contains relatively low levels of chlorophyll a but relatively abundant filaments that exhibit a common directionality that may be indicative of phototaxis. The base of the cone also contains filamentous features but with lower amounts of chlorophyll a. These filamentous sheaths may be heterotrophs, bacteriochlorophyll containing phototrophs, low chlorophyll a-containing cyanobacteria, or empty sheaths of the primary cyanobacterial builders (9), which can be colonized by heterotrophs. Nonfilamentous chlorophyll a-containing cells were also observed. This pattern of a low density of autofluorescing cyanobacteria at the tip and a high density of autofluorescing cyanobacteria below the tip (middle) with decreasing density of cells with depth toward the mat is similar to the profile of carbon uptake and biomass in a cone (29). DAPI staining was used to observe non-chlorophyll a-containing microbes. DAPI staining identified microbial cells in all regions of the cone. Laboratory-grown cyanobacterial cones observed after several months of growth have abundant chlorophyll a autofluorescence in all regions of the cone (data not shown). The reason for the difference in chlorophyll a abundance in laboratory-grown cones and cones grown in situ has not been determined, but it may be related to the age of the cones, differences in nutrients between the growth media and the natural spring water, the loss of a microbial partner as a result of enrichment, or any other number of environmental conditions present in situ but absent in vitro.
Fig 2.
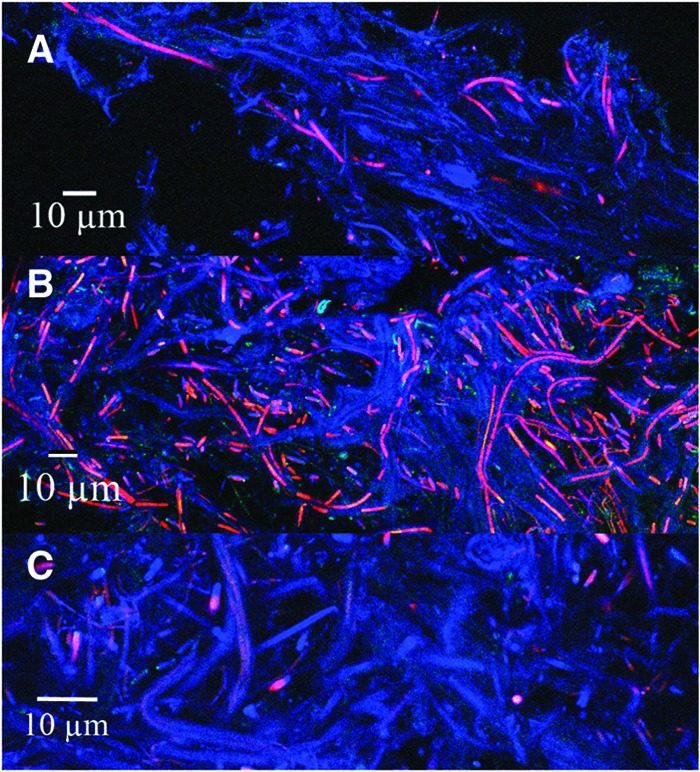
Confocal microscopy of coniform structures from Octopus Spring. (A) Tip of cone; (B) middle of cone; (C) bottom of cone. Chlorophyll a autofluorescence is shown in red; DAPI staining is shown in blue.
A sample from the tip of the cone was also observed by SEM (Fig. 3). The filaments characteristic of cyanobacteria dominate with what appears to be collapsed EPS (Fig. 3B). Some views (such as Fig. 3B) showed a common directionality of the filaments, but this was not found in all areas. Few nonfilamentous cells were observed in the sections examined. Both types of microscopy suggest the structures are dominated by filamentous cyanobacteria.
Fig 3.
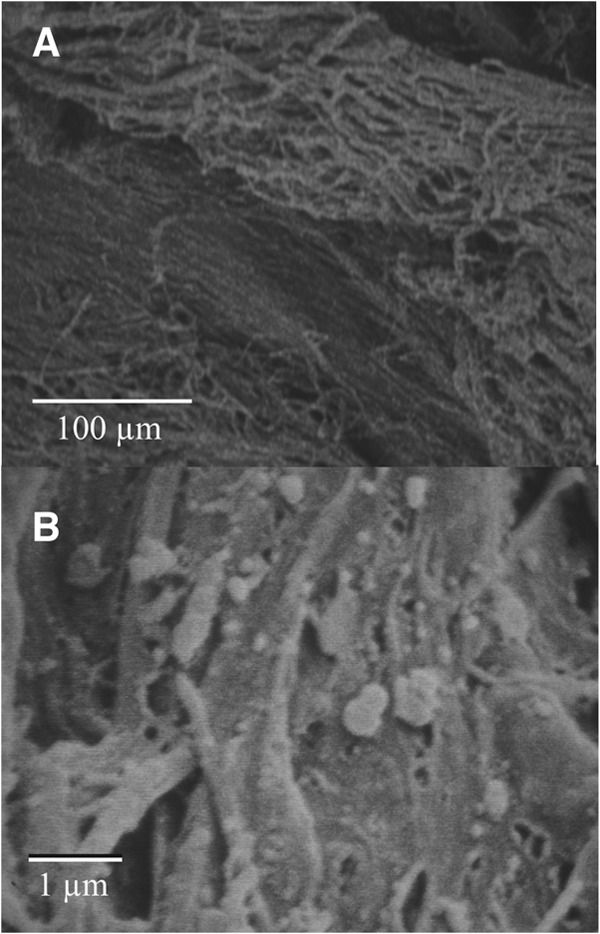
SEM images from the top portion of a cone. (A) Directional filaments; (B) filaments with potentially collapsed exopolymeric substances.
Leptolyngbya sp. dominates the 16S rRNA clone library.
To describe the microbial community present in the cones and potentially identify microbes playing a role in the formation and lithification of the coniform structures, the community members were identified by 16S rRNA gene analysis. Previous studies (29, 30) had looked closely at the cyanobacterial diversity in cones present in other YNP springs, but no studies had examined the community as a whole. 16S rRNA was amplified using bacterial primers and no amplification was achieved using the archaeal specific forward primer. Similarly, archaea were either not detected (13) or were present as a small percentage of clones in clone libraries from stromatolites (16, 46) and microbial mats in YNP (47, 48). The bacterial amplified product was cloned and 87 clones were sequenced. This number of clones was deemed sufficient because the dominant organism was identified, and it gave a snapshot of the low-abundance, yet diverse, community of noncyanobacteria that had not been previously examined in coniform structures. As shown in Fig. 4, the majority of the clones, 67, contained the 16S rRNA sequence of the cyanobacterium Leptolyngbya. This sequence is 99% identical to the sequenced cyanobacterial DGGE band R2A from cones from Black Sand Pool, YNP (30). This same sequence was also identified in several other coniform-forming environments in alkaline ponds and coniform structures in other YNP hot springs (29) and a cone-forming enrichment culture (27). Finding this same sequence in multiple studies of different cones in YNP suggests that this filamentous cyanobacterium may be one of the primary architects of these cone structures. The dominance of Leptolyngbya in a coniform structure clone library from Octopus Spring is also significant since Leptolyngbya was not abundant in the Synechococcus- and Chloroflexis-dominated Octopus Spring mats (36, 47–49), although the mats were not sampled at the same location or time. Leptolyngbya has also been found (i) in phototrophic layers of microbial mats in Fairy Falls, West Thumb, Obsidian Pool, and in the La Duke Hotspring areas of YNP (47, 50), (ii) in high abundance in the clone library from stromatolites from Ruidera Pools, Spain (13), (iii) the cyanobacterial fraction of “button” mats from Highborne Cay, Bahamas (51), and (iv) and in a microbial mat with tower features in Australia (52). Although not in high abundance, Leptolyngbya was also identified in Shark Bay and Bahamanian stromatolites (17–19). A siliceous stromatolite from Obsidian Pool Prime, YNP, was also found to be dominated by cyanobacteria (7, 9) but not by a Leptolyngbya sp. Another cyanobacterial clone in the library shows 98% identity to sequences retrieved from Fairy Springs water and glass rod biofilms (53) and Octopus Spring mat samples (49). The sequence was not found in other studies of coniform structures (29, 30).
Fig 4.
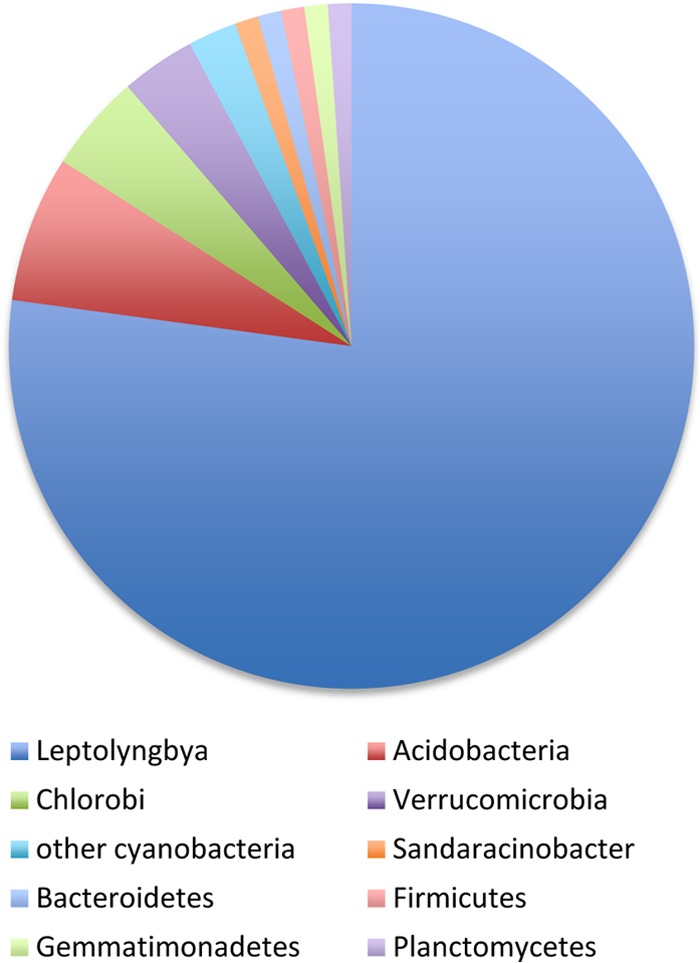
Abundance of phylotypes in the 16S rRNA gene clone library (n = 87 clones). Leptolyngbya sp. clones dominated the clone library.
The Acidobacteria were the next most abundant clone present in the library, comprising 7% of the sequences, with similarity to clones from Fairy Springs (53). The Acidobacteria phylum has recently been shown to contain photosynthetic organisms, and their presence in the cones is thus not surprising; however, the role they may play in the structure of the cones is uncertain and intriguing (35). Including Leptolyngbya, 92% of the clones had closest identity to sequences from YNP, including Fairy Springs (53), Octopus Spring (49), and Mammoth Spring (accession number AF445706). One of the clones was 99% identical (720 nt) to an aerobic bacteriochlorophyll a-containing alphaproteobacterium, Sandaracinobacter sibiricus (54), an orange-yellow microbe containing abundant carotenoids. This microbe may contribute to the orange pigment of the cones, although it was not identified in similarly pigmented phototrophic mats in YNP (47). Although most of the clones identified did show high similarity to other sequences from YNP, these similar sequences were not obtained from microbialites, suggesting that the lifestyle of these microorganisms may not be directly related to coniform formation.
The low abundance of non-Leptolyngbya sequences found in the coniform structure does bring into question the role of noncyanobacterial microbes in cone formation. A metagenomic study (55) of a freshwater microbialite identified an abundance of genes involved in regulation and cell signaling, suggesting that there may be significant coordination between microbialite community members such as cyanobacteria and heterotrophs. In the stromatolites of Highborne Cay, Bahamas, and Shark Bay, Australia, the microbial community was in fact dominated by alphaproteobacteria (7, 13, 14, 16–20). The role of heterotrophs in carbonate and silicate systems are likely different, but it would not be surprising to find that noncyanobacteria play a role in silica precipitation and cone morphogenesis in this system. However, if non-Leptolyngbya do play an important role in cone formation, it is not likely through their high abundance, which makes identifying their potential role more challenging. Such important but low-abundance contributors very well could have been missed in this library; however, our rarefaction analysis of this limited sequence data set indicates a reasonable plateau of community coverage. If stromatolites develop in a similar manner to the cones described here, it suggests initiation by a single builder species, followed by the development of a secondary community through an ecological succession pattern. If microbial species richness is related to lithification, as suggested by Baumgartner et al. (16), it may explain why cones can be found without any lithification at this same site. The Shannon-Wiener diversity index of the partially lithified cone was only 1.01, which is substantially less than the diversity indices determined for very well-lithified stromatolites (19, 51, 56, 57). It is possible that Leptolyngbya may be the initiating microorganism, but actual lithification may be driven by a secondary “inhabitant” community.
Enrichment of Leptolyngbya sp. strain C1 from Octopus Spring cones.
A cyanobacterial enrichment culture was obtained by multiple transfers of filaments from plates combined with liquid medium serial dilutions. The cyanobacterium in the enrichment (Leptolyngbya sp. strain C1) was filamentous, gliding, and capable of positive phototaxis. It also had the same 16S rRNA gene sequence of the Leptolyngbya sp. that dominated the clone library and is morphologically similar to the cyanobacteria identified by confocal microscopy (Fig. 1). The congruence of these techniques, as well as the presence of Leptolyngbya-like sequences in many cones and stromatolites (13, 18, 19, 29, 30, 51), suggests that the Leptolyngbya sp. strain C1 enrichment is relevant for mechanistic studies. The enrichment also contained Pseudomonas sp. strain OS4 (see Table S1 in the supplemental material). Based on microscopy, plate morphology, and 16S rRNA gene sequencing, the enrichment consists primarily of these two microbes. Several heterotrophic microbes with close relatives found in association with cyanobacteria were also isolated (see the supplemental material).
Factors affecting in vitro cone morphology.
The enrichment culture containing Leptolyngbya sp. strain C1 was tested for in vitro cone formation and found to form cones when grown on agar medium flooded with liquid medium as described previously (14) (Fig. 5). This cone forming enrichment culture was then used to identify growth conditions that affected cone morphology. Two different morphologies have been identified: cones and more-rounded nobs (Fig. 5). Cones form when a lawn of the enrichment culture is pregrown on agar medium prior to flooding. In this condition and prior to flooding, the lawn of the enrichment culture forms reticulated mats such as those described by Sumner and Shepard (58). When medium flooding occurs at the time of inoculation, aggregates of cells form in the liquid medium that eventually fall upon the agar surface. This may be one mechanism for the formation of the rounded nobs but may not explain all of the nobs that are formed. The shape (cone versus nob) may be an indicator of environmental conditions during formation.
Fig 5.
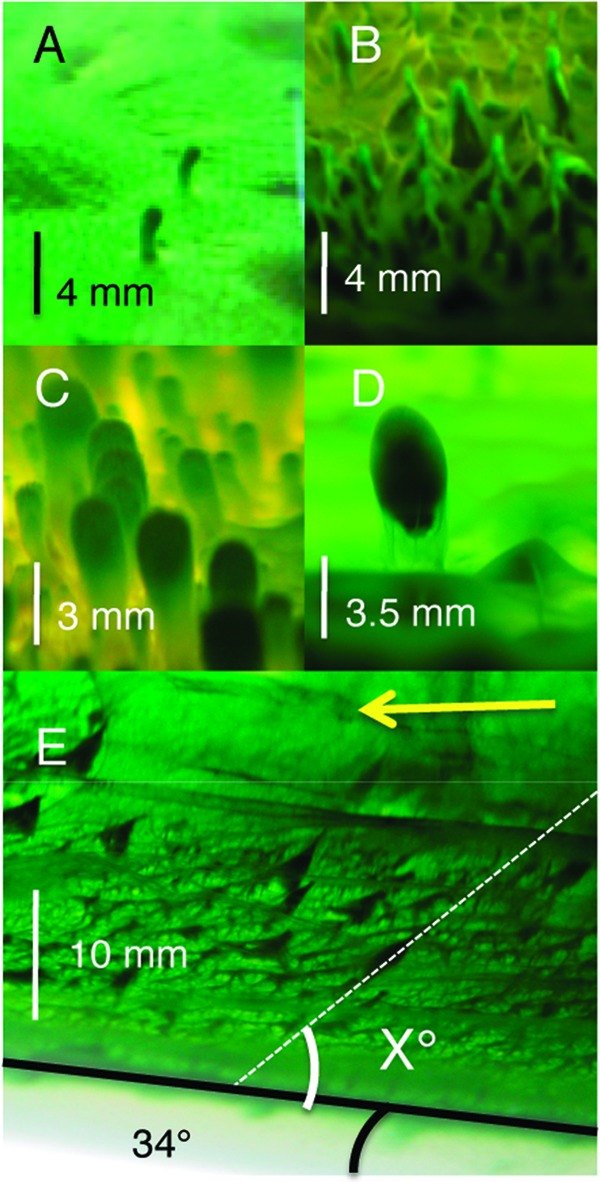
(A) Newly formed cyanobacterial enrichment cones 4 weeks after medium flooding. (B) Older cones 6.5 months after flooding. The matured structures become more similar to the structures found at Octopus Spring. (C) Cones grown in medium that was flooded upon inoculation instead of after formation of the mat on the agar surface, 3 months postinoculation. (D) An aggregate that has landed on the mat appears to be lifted up. (E) Cones formed on agar incubated at a 34° angle from horizontal. The arrow indicates the direction of light. “X” indicates the angle of cone inclination from the surface. Scale bars are approximate and are based on external flask markings. Potential magnification due to the liquid medium is not accounted for.
To test whether cone formation is driven by the biological process of phototaxis or the physically driven release of cyanobacterially formed gases (i.e., O2) as bubbles, we examined how the angle of illumination and surface orientation affected morphology. Cones were grown in tubes with the agar surface slanted at an angle of 34° with respect to horizontal and with the light source directed along the horizontal axis (Fig. 5). Under these growth conditions, cones took approximately twice as long to grow, and only very small peaks grew out of the mat surface farthest from the light. Cones grown under these conditions were classified as growing perpendicular to the agar surface, growing at an acute angle to the surface (toward the light), or growing at an obtuse angle to the agar surface (in the opposite direction of gravity). Of 500 cones examined, ca. 88% of the cones were oriented at angles roughly 58 to 75° from the agar surface toward the light source, and ca. 12% were oriented perpendicular to the surface. The cones did not lean directly toward the light source (34° from the agar surface) but were at an intermediate angle, suggesting that phototaxis toward the light may be playing a role, but another factor that would drive the cells to form cones oriented perpendicular to the agar surface is also at play. Cones leaning toward the light are consistent with previous observations (27), and individual cyanobacterial filaments have also been shown to orient normal to the surface (31). Additional experiments with angled agar surfaces have ruled out gravitational effects that could be pulling the cones down, leading to an acute angle of inclination (data not shown). No cones were oriented at obtuse angles. Awramik and Vanyo (59) have reported heliotropism in stromatolites from thermal springs in YNP and Hamelin Pool, Australia. In contrast, Petryshyn and Corsetti (60) and Mata et al. (8) examined stromatolites from Walker Lake and YNP, respectively, and, in both cases, surface normal growth dominated regardless of the angle of inclination which did not support a strong phototactic response toward light. In our cyanobacterial vertical structures, which are not stromatolites, we see a greater potential influence of phototaxis, and yet we still see an influence of factors that tend toward surface normal growth. If cyanobacterially produced oxygen bubbles are driving the initial formation of the cones from ridges or high points or driving the formation of tube-like structures (27), which should result in growth perpendicular to the true horizon and obtuse angles in our experimental setup, that orientation is not well preserved under these growth conditions and thus is less likely to be recorded from an environment in the rock record.
Laboratory-grown cones did not become lithified, and the addition of sodium silicate to the overlying medium at 0.1 g/liter and at saturation prevented cone formation. Cycling between wet and dry conditions is likely to be important for silica deposition (15), and such conditions were not replicated with these laboratory conditions. In addition, when the heterotrophic isolates (see Table S1 in the supplemental material) were added to the Leptolyngbya enrichment, there was no change in cone morphology, suggesting that under these conditions the heterotrophic isolates do not affect morphology.
Microscopy, molecular analysis, and enrichment cultures point to the filamentous cyanobacterium Leptolyngbya sp. strain C1 as the primary architect of the coniform structure from Octopus Spring, YNP. Laboratory-grown structures were used to identify factors that may affect the morphology of the cones including the presence of overlying liquid upon inoculation and the direction of light and surface orientation. To our knowledge, this is the first quantitative assessment using laboratory-manipulated conditions to determine the potential role of phototaxis, surface inclination, and bubble formation on cyanobacterial coniform orientation.
Future studies aimed at understanding the molecular mechanism that leads to surface normal growth should provide important insight into the biogenesis of coniform structures. These structures may share growth mechanisms similar to other vertical structures such as stromatolites and can provide important information about the complexity of life on early Earth.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steve Karl for microscopy assistance, Laura Beer for sample collection, and anonymous reviewers for improving the manuscript.
This research was supported by a CSUF incentive grant to H.A.J., CSUF associated student research grants to N.I.G., D.N., D.T.C., and F.O., and CSUF faculty-student creative activity grants to K.R. and N.G. Assistance was also provided by the International Geobiology Course funded by the National Science Foundation, NASA Exobiology, and the Agouron Institute.
A permit to conduct research in Yellowstone National Park is granted to J.R.S. by the Yellowstone Center for Resources.
Footnotes
Published ahead of print 14 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03008-12.
REFERENCES
- 1. Allwood AC, Grotzinger JP, Knoll AH, Burch IW, Anderson MS, Coleman ML, Kanik I. 2009. Controls on development and diversity of Early Archean stromatolites. Proc. Natl. Acad. Sci. U. S. A. 106:9548–9555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schopf JW. 1996. Are the oldest fossils cyanobacteria?, p 23–61 In Roberts DM, Sharp P, Alderson G, Collins MA. (ed), Evolution of microbial life. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 3. Allwood AC, Walter MR, Burch IW, Kamber BS. 2007. 3.43 billion-year-old stromatolite reef from the Pilbara Craton of western Australia: ecosystem-scale insights to early life on Earth. Precambrian Res. 158:198–227 [Google Scholar]
- 4. Allwood AC, Walter MR, Kamber BS, Marshall CP, Burch IW. 2006. Stromatolite reef from the Early Archaean era of Australia. Nature 441:714–718 [DOI] [PubMed] [Google Scholar]
- 5. Lowe DR. 1980. Stromatolites 3,400-Myr old from the Archean of Western Australia. Nature 284:441–443 [Google Scholar]
- 6. Grotzinger JP, Knoll AH. 1999. Stromatolites in Precambrian carbonates: evolutionary mileposts or environmental dipsticks? Annu. Rev. Earth Planetary Sci. 27:313–358 [DOI] [PubMed] [Google Scholar]
- 7. Berelson WM, Corsetti FA, Pepe-Ranney C, Hammond DE, Beaumont W, Spear JR. 2011. Hot spring siliceous stromatolites from Yellowstone National Park: assessing growth rate and laminae formation. Geobiology 9:411–424 [DOI] [PubMed] [Google Scholar]
- 8. Mata SA, Harwood CL, Corsetti FA, Stork NJ, Eilers K, Berelson WM, Spear JR. 2012. Influence of gas production and filament orientation on stromatolite microfabric. Palaios 27:206–219 [Google Scholar]
- 9. Pepe-Ranney C, Berelson WM, Corsetti FA, Treants M, Spear JR. 2012. Cyanobacterial construction of hot spring siliceous stromatolites in Yellowstone National Park. Environ. Microbiol. 14:1182–1197 [DOI] [PubMed] [Google Scholar]
- 10. Dill RF, Shinn EA, Jones AT, Kelly K, Steinen RP. 1986. Giant subtital stromatolites forming in normal salinity waters. Nature 324:55–58 [Google Scholar]
- 11. Hoffman P. 1976. Stromatolite morphogenesis in Shark Bay, Western Australia, p 261–272 In Walter MR. (ed), Stromatolites. Elsevier Scientific Publishing Company, Amsterdam, Netherlands [Google Scholar]
- 12. Playford PE, Cockbain AE. 1976. Modern algal stromatolites at Hamelin Pol, a hypersaline barred basin in Shark Bay, Western Australia, p 389–411. In Walter MR. (ed), Stromatolites. Elsevier Scientific Publishing Company, Amsterdam, Netherlands [Google Scholar]
- 13. Santos F, Pena A, Nogales B, Soria-Soria E, Del Cura MA, Gonzalez-Martin JA, Anton J. 2010. Bacterial diversity in dry modern freshwater stromatolites from Ruidera Pools Natural Park, Spain. Syst. Appl. Microbiol. 33:209–221 [DOI] [PubMed] [Google Scholar]
- 14. Walter MR, Bauld J, Brock TD. 1976. Microbiology and morphogenesis of columnar stromatolites (Conophyton, Vacerrilla) from hot springs in Yellowstone National Park, p 273–310 In Walter MR. (ed), Stromatolites. Elsevier Scientific Publishing Company, Amsterdam, Netherlands [Google Scholar]
- 15. Walter MR, Bauld J, Brock TD. 1972. Siliceous algal and bacterial stromatolites in hot spring and geyser effluents of Yellowstone National Park. Science 178:402–405 [DOI] [PubMed] [Google Scholar]
- 16. Baumgartner LK, Spear JR, Buckley DH, Pace NR, Reid RP, Dupraz C, Visscher PT. 2009. Microbial diversity in modern marine stromatolites, Highborne Cay, Bahamas. Environ. Microbiol. 11:2710–2719 [DOI] [PubMed] [Google Scholar]
- 17. Burns BP, Goh F, Allen M, Neilan BA. 2004. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ. Microbiol. 6:1096–1101 [DOI] [PubMed] [Google Scholar]
- 18. Foster JS, Green SJ, Ahrendt SR, Golubic S, Reid RP, Hetherington KL, Bebout L. 2009. Molecular and morphological characterization of cyanobacterial diversity in the stromatolites of Highborne Cay, Bahamas. ISME J. 3:573–587 [DOI] [PubMed] [Google Scholar]
- 19. Goh F, Allen MA, Leuko S, Kawaguchi T, Decho AW, Burns BP, Neilan BA. 2009. Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J. 3:383–396 [DOI] [PubMed] [Google Scholar]
- 20. Neilan BA, Burns BP, Relman DA, Lowe DR. 2002. Molecular identification of cyanobacteria associated with stromatolites from distinct geographical locations. Astrobiology 2:271–280 [DOI] [PubMed] [Google Scholar]
- 21. Arp G, Reimer A, Reitner J. 2001. Photosynthesis-induced biofilm calcification and calcium concentrations in phanerozoic oceans. Science 292:1701–1704 [DOI] [PubMed] [Google Scholar]
- 22. Reid RP, Visscher PT, Decho AW, Stolz JF, Bebout BM, Dupraz C, Macintyre LG, Paerl HW, Pinckney JL, Prufert-Bebout L, Steppe TF, DesMarais DJ. 2000. The role of microbes in accretion, lamination and early lithification of modern marine stromatolites. Nature 406:989–992 [DOI] [PubMed] [Google Scholar]
- 23. Walter MR, Burns BP, Anitori R, Butterworth P, Henneberger R, Goh F, Allen MA, Ibanez-Peral R, Bergquist PL, Neilan BA. 2009. Modern analogues and the early history of microbial life. Precambrian Res. 173:10–18 [Google Scholar]
- 24. Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT. 2009. Processes of carbonate precipitation in modern microbial mats. Earth-Sci. Rev. 96:141–162 [Google Scholar]
- 25. Dupraz C, Visscher PT. 2005. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 13:429–438 [DOI] [PubMed] [Google Scholar]
- 26. Batchelor MT, Burne RV, Henry BI, Jackson MJ. 2004. A case for biotic morphogenesis of coniform stromatolites. Physica A 337:319–326 [Google Scholar]
- 27. Bosak T, Liang B, Sim MS, Petroff AP. 2009. Morphological record of oxygenic photosynthesis in conical stromatolites. Proc. Natl. Acad. Sci. U. S. A. 106:10939–10943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grotzinger JP, Rothman DH. 1996. An abiotic model for stromatolite morphogenesis. Nature 383:423–425 [Google Scholar]
- 29. Bosak T, Liang B, Wu TD, Templer SP, Evans A, Vali H, Guerquin-Kern JL, Klepac-Ceraj V, Sim MS, Mui J. 2012. Cyanobacterial diversity and activity in modern conical microbialites. Geobiology 10:384–401 [DOI] [PubMed] [Google Scholar]
- 30. Lau E, Nash CZ, Vogler DR, Cullings KW. 2005. Molecular diversity of cyanobacteria inhabiting coniform structures and surrounding mat in a Yellowstone hot spring. Astrobiology 5:83–92 [DOI] [PubMed] [Google Scholar]
- 31. Petroff AP, Sim MS, Maslov A, Krupenin M, Rothman DH, Bosak T. 2010. Biophysical basis for the geometry of conical stromatolites. Proc. Natl. Acad. Sci. U. S. A. 107:9956–9961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bosak T, Bush JW, Flynn MR, Liang B, Ono S, Petroff AP, Sim MS. 2010. Formation and stability of oxygen-rich bubbles that shape photosynthetic mats. Geobiology 8:45–55 [DOI] [PubMed] [Google Scholar]
- 33. Petroff AP, Wu TD, Liang B, Mui J, Guerquin-Kern JL, Vali H, Rothman DH, Bosak T. 2011. Reaction-diffusion model of nutrient uptake in a biofilm: theory and experiment. J. Theor. Biol. 289:90–95 [DOI] [PubMed] [Google Scholar]
- 34. Brock TD. 1978. Thermophilic microorganisms and life at high temperature. Springer-Verlag, New York, NY [Google Scholar]
- 35. Bryant DA, Costas AM, Maresca JA, Chew AG, Klatt CG, Bateson MM, Tallon LJ, Hostetler J, Nelson WC, Heidelberg JF, Ward DM. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523–526 [DOI] [PubMed] [Google Scholar]
- 36. Ferris MJ, Ward DM. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klatt CG, Wood JM, Rusch DB, Bateson MM, Hamamura N, Heidelberg JF, Grossman AR, Bhaya D, Cohan FM, Kuhl M, Bryant DA, Ward DM. 2011. Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J. 5:1262–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Revsbech NP, Ward DM. 1984. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl. Environ. Microbiol. 48:270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Turner S, Pryer KM, Miao VP, Palmer JD. 1999. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46:327–338 [DOI] [PubMed] [Google Scholar]
- 40. DeLong EF. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Urbana, IL [Google Scholar]
- 44. Wiener N. 1948. Cybernetics: or control and communication in the animal and machine. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 45. Castenholz RW. 1988. Culturing methods for cyanobacteria. Methods Enzymol. 167:68–93 [Google Scholar]
- 46. Papineau D, Walker JJ, Mojzsis SJ, Pace NR. 2005. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl. Environ. Microbiol. 71:4822–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ross KA, Feazel LM, Robertson CE, Fathepure BZ, Wright KE, Turk-Macleod RM, Chan MM, Held NL, Spear JR, Pace NR. 2012. Phototrophic phylotypes dominate mesothermal microbial mats associated with hot springs in Yellowstone National Park. Microb. Ecol. 64:162–170 [DOI] [PubMed] [Google Scholar]
- 48. Ward DM, Ferris MJ, Nold SC, Bateson MM. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ferris MJ, Ruff-Roberts AL, Kopczynski ED, Bateson MM, Ward DM. 1996. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl. Environ. Microbiol. 62:1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brown II, Bryant DA, Casamatta D, Thomas-Keprta KL, Sarkisova SA, Shen G, Graham JE, Boyd ES, Peters JW, Garrison DH, McKay DS. 2010. Polyphasic characterization of a thermotolerant siderophilic filamentous cyanobacterium that produces intracellular iron deposits. Appl. Environ. Microbiol. 76:6664–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mobberley JM, Ortega MC, Foster JS. 2012. Comparative microbial diversity analyses of modern marine thrombolitic mats by barcoded pyrosequencing. Environ. Microbiol. 14:82–100 [DOI] [PubMed] [Google Scholar]
- 52. McGregor GB, Rasmussen JP. 2008. Cyanobacterial composition of microbial mats from an Australian thermal spring: a polyphasic evaluation. FEMS Microbiol. Ecol. 63:23–35 [DOI] [PubMed] [Google Scholar]
- 53. Boomer SM, Noll KL, Geesey GG, Dutton BE. 2009. Formation of multilayered photosynthetic biofilms in an alkaline thermal spring in Yellowstone National Park, Wyoming. Appl. Environ. Microbiol. 75:2464–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yurkov V, Stackebrandt E, Buss O, Vermeglio A, Gorlenko V, Beatty JT. 1997. Reorganization of the genus Erythromicrobium: description of “Erythromicrobium sibiricum” as Sandaracinobacter sibiricus gen. nov., sp. nov., and of “Erythromicrobium ursincola” as Erythromonas ursincola gen. nov., sp. nov. Int. J. Syst. Bacteriol. 47:1172–1178 [DOI] [PubMed] [Google Scholar]
- 55. Breitbart M, Hoare A, Nitti A, Siefert J, Haynes M, Dinsdale E, Edwards R, Souza V, Rohwer F, Hollander D. 2009. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Cienegas, Mexico. Environ. Microbiol. 11:16–34 [DOI] [PubMed] [Google Scholar]
- 56. Foster JS, Green SJ. 2011. Microbial diversity in modern stromatolites, p 385–405 In Seckbach J, Tewari V. (ed), Cellular origin, life in extreme habitats and astrobiology: interactions with sediments. Springer, Berlin, Germany [Google Scholar]
- 57. Myshrall KL, Mobberley JM, Green SJ, Visscher PT, Havemann SA, Reid RP, Foster JS. 2010. Biogeochemical cycling and microbial diversity in the thrombolitic microbialites of Highborne Cay, Bahamas. Geobiology 8:337–354 [DOI] [PubMed] [Google Scholar]
- 58. Sumner DY, Shepard RN. 2010. Undirected motility of filamentous cyanobacteria produces reticulate mats. Geobiology 8:179–190 [DOI] [PubMed] [Google Scholar]
- 59. Awramik SM, Vanyo JP. 1986. Heliotropism in modern stromatolites. Science 231:1279–1281 [DOI] [PubMed] [Google Scholar]
- 60. Petryshyn VA, Corsetti FA. 2011. Analysis of growth directions of columnar stromatolites from Walker Lake, western Nevada. Geobiology 9:425–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


