Abstract
In this study, the impacts of six potato (Solanum tuberosum) cultivars with different tuber starch allocations (including one genetically modified [GM] line) on the bacterial communities in field soil were investigated across two growth seasons interspersed with 1 year of barley cultivation, using quantitative PCR, clone library, and PCR-denaturing gradient gel electrophoresis (DGGE) analyses. It was hypothesized that the modifications in the tuber starch contents of these plants, yielding changed root growth rates and exudation patterns, might have elicited altered bacterial communities in the soil. The data showed that bacterial abundances in the bulk soil varied over about 2 orders of magnitude across the 3 years. As expected, across all cultivars, positive potato rhizosphere effects on bacterial abundances were noted in the two potato years. The bulk soil bacterial community structures revealed progressive shifts across time, and moving-window analysis revealed a 60% change over the total experiment. Consistent with previous findings, the community structures in the potato rhizosphere compartments were mainly affected by the growth stage of the plants and, to a lesser extent, by plant cultivar type. The data from the soil under the non-GM potato lines were then taken to define the normal operating range (NOR) of the microbiota under potatoes. Interestingly, the bacterial communities under the GM potato line remained within this NOR. In regard to the bacterial community compositions, particular bacterial species in the soil appeared to be specific to (i) the plant species under investigation (barley versus potato) or, with respect to potatoes, (ii) the plant growth stage. Members of the genera Arthrobacter, Streptomyces, Rhodanobacter, and Dokdonella were consistently found only at the flowering potato plants in both seasons, whereas Rhodoplanes and Sporosarcina were observed only in the soil planted to barley.
INTRODUCTION
Agricultural production is greatly assisted by a soil microbiota that exerts beneficial, i.e., growth-enhancing (1) and/or pathogen-suppressive, effects on crop plants (2, 3). Conversely, crop plants commonly affect the soil microbiota in the immediate environment of their roots via the so-called rhizosphere effect, i.e., the attraction of microbes to, and their stimulation by, the roots (2–5). The importance of the interaction between plant roots and local microbial communities for plant health cannot be overemphasized. Thus, a thorough understanding of how and to what extent crop plants affect the microbiota of soil, and vice versa, is key to our success in both intensive and extensive agricultural systems (2).
With the objective of continuous crop improvement, a plethora of potato lines with different agronomic or production characteristics have been developed over time. This currently includes both traditionally improved and genetically modified (GM) lines. However, a major, still unanswered question has been to what extent individual cultivars affect the microbiota surrounding their roots differently (6, 7). Such effects, leading to, for instance, highs and lows in the abundances and diversities of microbial groups at various levels, define a theoretical area of “normality” in the soil used for potato cropping. Such normality, which can be translated into a “normal operating range” (NOR), may then be used as a standard against which to weigh the effects of particular GM lines (7–9). The question of GM plant effects on the soil microbiota is particularly relevant for current GM plant admission procedures in different countries around the globe.
Several previous studies have provided partial answers with respect to the effects of, and normality under, potato crops across single and multiple growth seasons (7, 9–15). Thus, such studies have indicated that shifts in the soil microbiota occur primarily as responses to plant developmental stages, as well as being related to the plant line (7, 13, 14). However, the effects of cropping on the soil microbiota over multiple seasons, as well as those of the naturally occurring shifts in these communities over time, deserve more attention (10, 13).
The population dynamics of soil microorganisms are primarily driven by the proficiency of their members in sequestering the energy and carbon sources that emerge in soil (2, 16). This facet is especially relevant for the microbiota of the rhizosphere, where competition, in particular for the carbonaceous compounds that are exuded by the roots, is fierce (4). In fact, compounds in root exudates have been identified as major factors directing the abundance, diversity, and composition of the local (rhizosphere) soil microbiota (2, 16). Although often difficult to measure, it is plausible that the root-released products vary according to the plant cultivar type (genotype) and the plant developmental stage. Furthermore, the soil type, by its effect on both plant physiology and the soil microbiota, is likely to also play a role (7, 9). On the basis of these arguments, focus should be placed on the potential effects of all three parameters (plant genotype, plant developmental stage, and soil type) if a sensible NOR for potato cropping is to be established (8, 12, 14). In addition, other features of agricultural soil, i.e., the soil management regime and the natural fluctuations inflicted on the system due to weather or climatic conditions (temperature, pH, and moisture), are known to affect soil microbial communities (3). Thus, these factors also need to be considered in the definition of the NOR, as well as in appropriate GM impact studies (10, 12, 13).
In addition to the direct effects exerted by plant root exudates on the soil microbiota, it is important to assess the stability of the latter over the time following removal of the crop plants with respect to abundance, diversity, and community composition, particularly when GM plants have been used (6, 17–19). Moreover, an assessment of any putative effects of previous crops on the growth of subsequent ones is relevant (20). To achieve these aims, a selection of currently available advanced molecular methods (21–24) should be used.
In light of the above considerations, the objective of this study was to assess, using current advanced methods, the abundance, diversity, and structure of the bacterial communities in a field soil across three seasons. In the field, a potato-barley-potato rotation regimen was applied. In this cross-seasonal study, the bacterial communities in soil with six different potato cultivars, including a GM line, were analyzed, with an emphasis on three potato growth stages each in the two potato years and a crop rotation with barley in between. We hypothesized that the different tuber starch contents across the cultivars might have led to changed root growth and exudation patterns (7), resulting in altered bacterial communities at the roots. Such changes, however, were found to be minor in comparison to the changes in the whole (bulk) soil compartment.
(Part of the data used in this study for comparative purposes was published previously [7].)
MATERIALS AND METHODS
Field experiment: setup and soil sampling. (i) Field and potato cultivars.
The experiments were conducted over three successive years (2008 through 2010) in an experimental field located near the village of Buinen, The Netherlands. The soil in this field was characterized as a loamy sand. It contained 5% organic matter (OM) and was moderately acid (pH measured in KCl solution [pH-KCl], 5.0) (7). The field was under a strict agricultural rotation scheme used for potato production in The Netherlands. This scheme encompassed potatoes in the first year (2008), followed by barley the next year (2009), and potatoes in the last year (2010). For the two potato crops, six different cultivars, i.e., Aveka (A), Aventra (Av), Karnico (K), Modena (M) (genetically modified Karnico with lower amylose content in the tubers), Premiere (P), and Désirée (D), were used. These cultivars were genetically different from each other and differed in pedigree. For instance, cultivar A was related to D in the fifth generation and to K in the third generation (25). Also, the physiologies of the cultivars differed. Cultivars A, Av, K, and M produced tubers with high starch contents and had low and/or medium growth rates, whereas cultivars P and D yielded tubers with relatively low starch contents and had higher growth rates.
(ii) Experimental setup.
In the period preceding each growth season, the surface soil was treated by shoveling to remove any weed plants and improve soil aeration. The inter-potato crop barley was grown field-wide following sowing (2009). For the potato crops (2008 and 2010), randomized plot designs were applied. These included four replicate 3- by 3-m plots for each cultivar. The plots were separated from each other by 80-cm spacers, which were kept fallow. Twenty tubers were planted per replicate plot in four rows of five plants each. The fields were under routine agricultural regimes (including spraying with water in dry periods), common for potato and barley cultivation practices in The Netherlands. In 2008 and 2009, common NPK fertilizer mix was applied, whereas pig manure was used in 2010 (25 tons/ha).
(iii) Sampling.
Overall, the field was sampled at nine sampling moments. For the potato years, samples were taken at the young-plant (EC30), flowering (EC60), and senescence (EC99) stages (26). The times of sampling differed slightly between cultivars, depending on the time of flowering. Thus, the samplings were in April 2008 (before planting of potatoes), May 2008 (young plant), June 2008 (flowering), September 2008 (senescence), December 2008 (after removal of plants), July 2009 (soil under barley; the sampling date was chosen at the flowering of potato plants in an adjacent field), May 2010 (young potato plant), June 2010 (flowering), and September 2010 (senescence).
At sampling, four plants (potato years 2008 and 2010) were carefully removed from each plot and put together in one plastic bag (this yielded one composited rhizosphere soil sample per plot [see below]). Furthermore, composite bulk soil samples, each consisting of four (0- to 20-cm) cores sampled at each replicate of the six cultivars in areas at least 20 to 40 cm outside the plant rows, were prepared. All samples were taken to the laboratory and processed within 5 h. Bulk soil samples were thoroughly mixed and then used directly. To obtain rhizosphere soil samples, all soil loosely adhering to the plant roots was shaken off, after which the soil more tightly adhering to the root surface was brushed off. The latter soil constituted the rhizosphere soil, which was composited across the four plants sampled per plot.
Soil DNA extraction.
Subsamples from the bulk soil samples were used directly for DNA extraction, whereas for technical reasons, subsamples of three rhizosphere soil replicates were used. For the extractions, the Powersoil DNA extraction kit (Mo Bio Laboratories Inc., NY) was applied using a modified protocol (7). The amount, quality, and purity of the extracted soil DNA were checked in agarose gels, run for 1 h at 90 V and stained with ethidium bromide, upon exposure to UV.
qPCR.
Quantification of bacterial 16S rRNA genes was performed by quantitative PCR (qPCR) with primers 341F and 518R (7, 19). The PCR conditions were as follows: 95°C for 10 min, followed by 40 cycles of 15 s at 95°C, 20 s at 55°C, and 20 s at 72°C. The reaction mixture (total volume, 10 μl) contained 5 μl of DyNamo capillary SYBR green qPCR master mixture (Finnzymes, Helsinki, Finland), 0.3 pmol of forward and reverse primers, and 5 ng of template DNA. The qPCR standards (between 106 and 1010 target molecules per reaction) were prepared using PCR products made from Variovorax paradoxus DSM30034T (19). The 16S rRNA gene target numbers were taken as proxies for bacterial abundances across the samples (expressed as log numbers of 16S rRNA gene copies per g soil). Two-tailed Student t tests were performed to compare these numbers across samples.
Bacterial-community analyses. (i) PCR-DGGE fingerprints.
Bacterial PCR was performed on all DNA extracts, using primers GC-341F and GC-518R (35 cycles), as described previously (7). GC indicates the presence of a GC clamp. All denaturing gradient gel electrophoresis (DGGE) profiles were generated using the Ingeny Phor-U system (Ingeny International, Goes, The Netherlands). The amplicons obtained from the soil DNAs (200 ng), were loaded onto polyacrylamide gels (6% [wt/vol] acrylamide in 0.5× Tris-acetate-EDTA [TAE] buffer [2.42 g Tris base, 0.82 g sodium acetate, 0.185 g EDTA, 1 liter H2O]). Amplicons from different samples were cross-loaded over gels to control for gel-to-gel variability that might hamper the subsequent comparisons. Thus, gels that analyzed the relevant parameters—cultivar and time—were run. Gradients of 35 to 65% denaturant were used at 100 V for 16 h at 60°C. All gels were silver stained (27) and air dried, after which the profiles were digitized and stored as TIFF files for further analysis with GelCompar II (Applied Maths, Sint-Martens-Latem, Belgium). On the basis of normalized profiles, similarity matrices (consisting of defined band numbers within each profile) were generated using Pearson's correlation coefficient (r). Subsequently, the patterns were clustered using the unweighted pair group method with arithmetic averages (UPGMA) option of GelCompar II. In addition, data derived from the profiles (band analysis) were used for redundancy analysis (RDA) using Canoco (version 4.0 for Windows; PRI Wageningen, The Netherlands) (7). Profile similarities (based on relative band intensities and positions) were analyzed by performing canonical correspondence analyses with Monte Carlo permutation tests (CANOCO 4.0). Monte Carlo tests were based on 499 random permutations of the data to establish statistical significance.
Moving-window analyses (MWA) were used to calculate the rate-of-change parameter (Δt) for bulk soil over time during the season. First, the similarities of the densitometric curves of DGGE patterns were calculated based on the Pearson correlation coefficient. The percent change (percent change = 100 − percent similarity) was then calculated as described by Marzorati et al. (28). The percent change value matrix was used to perform moving-window analysis by plotting the values between consecutive sampling points. In addition, the change between the first and last samplings was calculated.
Rhizosphere soil samples from the 2008 and 2010 growth seasons were also compared using GelCompar. Principal-component analysis (PCA) and analysis of similarity (ANOSIM) were performed using Euclidean distance. ANOSIM was carried out using the program Primer v6 (Plymouth, United Kingdom), based on transformed data (fourth square). Samples were also grouped by growth stage and sampling year. Two-way cross-analyses with replicates were done with 5,000 permutations. The global R value, varying between −1 and 1, was used. An R value of 0 indicated completely random grouping, while an R value of 1 indicated that samples within a treatment group were more similar to each other than to any samples from the other treatment. A significant global R value indicated that there were differences between treatments somewhere in the analysis.
(ii) Cloning and sequencing of 16S bacterial rRNA gene amplicons generated from selected samples.
To allow monitoring of the changes in the bacterial-community makeup in bulk soil over 3 years, 12 clone libraries consisting of bacterial 16S rRNA gene fragments were generated (using primers 27F and 518R). The (replicate) bulk soil samples selected were those taken in April 2008 (before potato planting), June 2008 (at flowering), December 2008 (after removal of plants), June 2009 (barley field, at flowering of potato plants), July 2010 (potato flowering), and September 2010 (senescence). Following soil DNA extraction and PCR amplification, the amplicons were ligated into pGEM-T Easy vectors (pGEM-T Easy Vector System II, Promega, Madison, WI). The ligation mixtures were introduced into competent Escherichia coli JM109 cells by transformation, according to the manufacturer's instructions. Following plate incubation, white colonies (containing intact inserts) were randomly picked, taken up in 10 μl sterile water, and used as templates for PCR using the universal M13f/M13r vector primers. The resulting amplicons were sequenced (LGC, Berlin, Germany).
(iii) Analysis of clone libraries.
Sequences were checked, using BELLEROPHON v.3 (http://greengenes.lbl.gov) (29), for the occurrence of chimerae, which were removed from the subsequent analyses. The bacterial community structures, as evidenced by the cured clone libraries, were analyzed with fast UniFrac (30), using maximum-likelihood-based and Jack-knife trees. In addition, sequences were also subjected to library-shuffling analysis using LIBSHUFF to determine if the clone libraries were significantly different from each other (31).
In a second analysis, the sequences were compared with sequences in GenBank using BLAST-N searches to obtain the nearest phylogenetic neighbors (http://www.ncbi.nlm.nih.gov/BLAST). The sequences were then processed in MEGA (32). Finally, an RDP library comparison was done at a confidence threshold level of 80% (33). The program DOTUR (24) was used to create rarefaction curves, on the basis of a conservative operational taxonomic unit (OTU) cutoff of 97% similarity to determine the bias-corrected Chao1 estimator of richness. We subsequently aligned the sequences using the align.seqs command embedded in the program Mothur against Greengenes' core set (34). Use of Mothur (get.sharedseqs, listing and grouping sequences at the 0.03 similarity cutoff) further allowed us to find shared and unique OTUs. Sequence alignments and tree building of shared sequences were carried out using MEGA (32). The neighbor-joining algorithm was used, with bootstrapping using 1,000 replications. Pairwise sequence similarities were calculated with DNADIST (http://cmgm.stanford.edu/phylip/dnadist.html), using the Kimura 2-parameter algorithm (32).
Nucleotide sequence accession numbers.
The sequences generated in this study were deposited in GenBank under numbers JF910290 to JF911347.
RESULTS
Previous studies.
In previous work, we addressed the effects of the six potato cultivars on the soil bacterial communities in the Buinen field soil in a single-growth-season experiment (7) and later also applied deep pyrosequencing to the samples (9). Here, we extend these data to nine samplings done in the same field across 3 years (including the same first experimental year), using 16S rRNA gene-based qPCR, clone libraries, and PCR-DGGE. Thus, the majority of the data reported here are novel, whereas the previously published data (7) are used only for comparative purposes. This is indicated in the figures and tables wherever required.
Plant development over the two growth seasons.
In 2008, the young-plant stage (EC30) occurred for all cultivars around 30 days postplanting (p.p.), i.e., at the end of May. The flowering stages (EC60) occurred between 50 and 60 days p.p. for cultivars D and P and between 80 and 85 days p.p. for cultivars A, Av, K, and M (June). Finally, the senescence stages (EC99) were between 110 and 115 days p.p. for P and D, between 135 and 140 days p.p. for A, and between 145 and 150 days p.p. for Av, K, and M. The field was planted with barley in 2009, which developed mature plants from sowing in April up to harvest in August. The soil was sampled once, in June 2009, when potato plants in an adjacent field were flowering. In the third season (2010; the second potato season), the young-plant stage occurred again around 30 days p.p., flowering occurred between 50 and 60 days p.p., and the senescence stage was around 120 days p.p. for all cultivars. Since cultivar P showed a decline after July 2010, plants of this cultivar were not included in the senescence stage analyses.
Dynamics of bacterial abundances in the field (bulk and rhizosphere soils) as assessed by 16S rRNA gene-based qPCR. (i) Bulk soil.
The abundances of the bacterial populations, as determined in bulk soil across the nine samples taken over 3 years (three growth seasons) using the 16S rRNA gene copy number per g dry soil as a proxy, varied by roughly 2 orders of magnitude over time, i.e., from a minimum of about 1.4 × 107 to a maximum of 1.2 × 109 target genes per g dry soil. Barley cropping significantly enhanced the bacterial abundances (July 2009; see below) in comparison to those in the same period in 2008 and 2010 (i.e., flowering stage, first and second potato seasons). Besides the tendency of the target numbers to drop during the winter/spring periods, there were no other major trends in the bacterial dynamics with respect to effects of time or season or year. Specifically, from April to May 2008, the numbers went down, from 7.2 × 108 to 5.9 × 107, after which they remained roughly stable at 1.2 × 108 to 1.6 × 108 until September. Then, slight drops were noted, to about 3.6 × 107 in December 2008 (Fig. 1) (7). In the next year, when barley was grown in the field, the analyses (performed when potato plants in an adjacent field were flowering) revealed significantly raised target gene copy numbers, i.e., about 1.2 × 109 gene copies/g dry soil. In 2010, with the field again planted to potatoes, counts varied from about 1.4 × 107 (May) to 8.4 × 108 (September) per g dry soil. Interestingly, although the numbers fluctuated in the samples taken during the 3 years, those found in September 2010 were similar to those at the start of the experiment, i.e., April 2008. Hence, irrespective of the potato cultivar, the soil from Buinen field may be hypothesized to have a carrying capacity for bacterial cells corresponding to about 108 to 109 16S rRNA gene copies per g dry soil.
Fig 1.
Abundance of bacterial 16S rRNA gene copies by growth stage in Buinen soil. The error bars indicate standard errors of the mean.
(ii) Potato rhizosphere soil.
A first assessment addressed the putative effects on bacterial abundance of the rhizospheres of the six different potato cultivars in the two potato years. To assess such effects, the respective 16S rRNA gene copy numbers in the rhizosphere soils were compared to those of corresponding bulk soils. In both growth seasons, positive rhizosphere effects on the bacterial abundances were observed for all cultivars in all three plant growth stages, although they were not always significant. In 2008, the effects were significant (P < 0.05) for cultivars A, M, P, and D in the young-plant stage for all cultivars at flowering (Fig. 2) and for cultivars Av, K, M, and D at senescence. In the 2010 growth season, the effects were significant (P < 0.05) across cultivars at all growth stages, except for cultivars Av, K, and M at senescence.
Fig 2.
Abundance of bacterial 16S rRNA gene copies in rhizosphere and bulk soil from Buinen field per growth stage. The error bars indicate standard errors of the mean. Byp, young-plant stage; Bf, flowering stage; Bsn, senescence stage.
Considering the per-cultivar dynamics in the rhizosphere target numbers over time, in 2008, different patterns were found at the different cultivars, as follows. For cultivars A, Av, K, and M, the numbers increased from the young-plant to the flowering stages, remaining constant at the senescence stage (P < 0.05, except for cultivar A). In contrast, the numbers at cultivar D showed a significant increase from the flowering (1.4 × 109 targets/g dry soil) to the senescence (6.2 × 109) stages. In the 2010 growth season, the numbers showed progressively increasing trends from the young-plant to senescence stages for cultivars A (from 1.4 × 108 to 9.5 × 109 targets per g dry soil) and D (from 1.1 × 109 to 1.7 × 1010). In contrast, at cultivars K and M, these numbers increased from the young-plant to flowering stages and then decreased. Remarkably, an overall decreasing trend was noted for cultivar Av, i.e., from 1.7 × 109 (young plant) to 1.5 × 108 (senescence) targets per g dry soil.
Interestingly, some cultivars showed consistent seasonal patterns. Cultivars K and M behaved similarly, with the highest 16S rRNA gene copy numbers detected in the flowering stage and the lowest at the young-plant stage. Cultivar D showed the highest bacterial abundance at the senescence stage, whereas that at cultivar P remained generally constant over the seasons.
Dynamics of bacterial diversity and community composition in bulk and rhizosphere soils as assessed by PCR-DGGE.
The bacterial PCR-DGGE profiles in bulk and rhizosphere soils generated from all four replicate plots per cultivar and sampling moment (treatment) revealed very low within-treatment variation (data not shown). This suggested, in addition to technical consistency, generally low variability between the randomized plots within the individual treatments. To describe the sampling-to-sampling bacterial-community shifts, we first applied MWA across the bulk soil samples, as described below.
(i) Bulk soils.
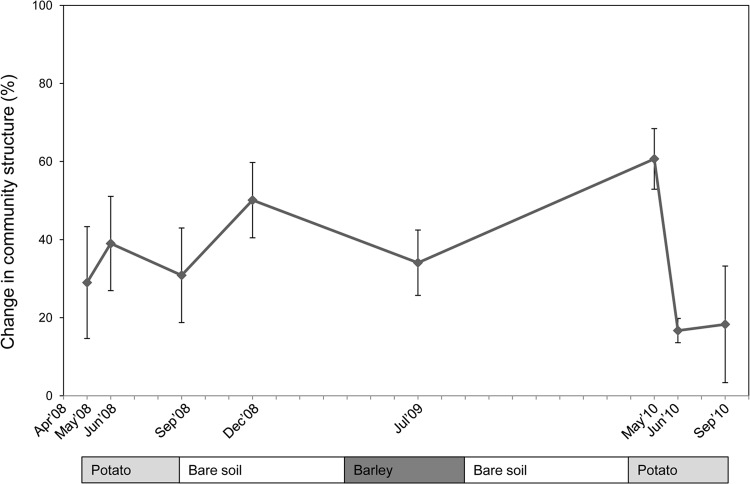
MWA applied to the PCR-DGGE profiles generated from the bulk soil samples revealed a 37% ± 9% change over the entire 2008 season. The profiles, in fact, showed progressively increasing differences in the communities from April through May, June, and September (Fig. 3) (7). Moreover, the patterns generated from the December 2008 bulk soils (after the removal of plants) were again different from the previous ones, with a high percentage of change (about 50%) observed in the period from September to December 2008 (Fig. 3). From December 2008 to July 2009 (barley), the percentage of variation was about 40%. Strikingly, in May 2010 (young-plant stage), the community had changed by 60% ± 7% in comparison to the previous sample from about 1 year before (June 2009). Following this, there were further changes in July (16% ± 3%) and September (18% ± 14%) compared to the respective previous samplings. A direct analysis of the total change in the community structure in bulk soil between April 2008 and September 2010 indicated that this amounted to 60% ± 15%. Conversely, it also indicated that about 40% of the dominant bacterial community detected by DGGE had remained present after about 2.5 years. We propose to designate such a persisting bulk soil community the core bacterial community (CBC) in soil under potatoes.
Fig 3.
Moving-window analyses (MWA) to evaluate the percent community change for bacterial communities in the bulk soil from Buinen field through different sampling times. A percent change value matrix was used to perform MWA by plotting the values between consecutive sampling points. The error bars indicate standard errors of the mean.
In a subsequent analysis, we also cross-compared the May 2008 and May 2010, the June 2008 and June 2010, and the September 2008 and September 2010 samples, as they represented the same phases in the development of potatoes in the field. The differences between the profiles of May 2008 and May 2010 amounted to only 25% ± 15%, indicating a modest “buffering” effect of the potato plants on these communities. The differences were greater between the profiles of June 2008 and June 2010, and September 2008 and September 2010, as in both cases, about 50% ± 10% changes were noted. Taken together, these observations indicated similarities of about 50 to 75% in the structures of the communities that were compared across the two potato seasons, which again suggested the existence of a core community resistant to change.
(ii) Statistical comparison of bulk and rhizosphere soil samples.
RDA applied to all PCR-DGGE profiles revealed that, at all potato growth stages in 2008 and 2010, the rhizosphere bacterial PCR-DGGE profiles grouped apart from the corresponding bulk soil patterns. This strongly indicated that the respective rhizospheres exerted concerted effects on the local soil bacterial-community structures (7). The effects were different with the individual cultivars. In 2008, the patterns from all cultivars except A grouped together in the young-plant stage, whereas at flowering, as well as senescence, cultivars P, D, and M grouped together, while A, Av, and K remained grouped as separate branches (P < 0.05) (7). In the 2010 season, at the young-plant stage, cultivars P, D, and M grouped together with K, while A and Av clustered apart. On the other hand, at flowering, cultivars P and D clustered together, whereas A, Av with K, and M formed a separate cluster. At senescence, cultivars D and M (P was not considered, as the plants had died out) grouped together, whereas A, Av, and K formed another group (data not shown).
Overall, and grossly consistent with these clusterings, two “core” dynamic groups of rhizosphere profiles, designated core groups I and II, could be distinguished across the 2 years, i.e., (i) core group I (associated with cultivars A, Av, and, mostly, K) and (ii) core group II (associated with cultivars P and D). Cultivar M appeared to accompany either core group I or II, in relation to its growth stage. Cultivar K (high starch) was found to mostly group with the high-starch-tuber cultivars, except for May 2010.
ANOSIM of all PCR-DGGE profiles revealed that both the season and the plant growth stage exerted significant effects on the bacterial-community structures at all cultivars (Table 1). UPGMA analysis also showed that cultivars clustered mainly based on the sampling year, indicating the importance of confounding factors, such as the weather or fertilization regime, on the microbial-community structures (data not shown).
Table 1.
ANOSIM of soil and cultivar effectsa
| Cultivar | Test for difference between: |
|||
|---|---|---|---|---|
| Growth stages (2008 and 2010) |
Yr (all growth stages) |
|||
| Global R | Significance | Global R | Significance | |
| A | 0.37 | 0.016 | 0.432 | 0.009 |
| Av | 0.383 | 0.003 | 0.457 | 0.006 |
| K | 0.653 | 0.002 | 0.841 | 0.002 |
| M | 0.494 | 0.003 | 0.802 | 0.002 |
| P | 0.381 | 0.029 | 0.466 | 0.01 |
| D | 0.679 | 0.001 | 0.79 | 0.001 |
PRIMER 6 software was used. The global R value is between −1 and 1, where an R value of 0 indicates completely random grouping, while an R value of 1 indicates that within-group variability is lower than differences from any samples from other groups. A significant global R value (boldface) indicates that there are differences between time points somewhere in the analysis.
Analysis of bacterial 16S rRNA gene clone libraries generated from bulk soil samples.
An important aim of this study was to assess the “normal” structure of the bacterial communities in soil used in potato cropping. We selected bulk soil samples for this comparative multiple-year study. The bacterial PCR-DGGE analyses had revealed differences between the communities in bulk soils collected at different time points. Of these, the samples from April 2008 (before planting), June 2008 (flowering), December 2008 (after removal of plants), July 2009 (barley), June 2010 (flowering potato), and September 2010 (senescent potato) were selected for the construction of 12 16S rRNA gene clone libraries (two replicate libraries for each sample).
(i) Statistical analysis of libraries.
After quality and chimera checks, LIBSHUFF analysis (31, 34) showed that the replicate libraries were similar to each other for each treatment (P > 0.05), whereas all of the different libraries, per treatment, were significantly different from any of the others (P < 0.05). Also, FastUnifrac analysis (30) also showed significant differences between the different libraries (data not shown). We therefore pooled the replicate libraries per sample type for further analysis. Thus, totals of 185, 176, 181, 183, 164, and 170 sequences were obtained from the respective samples. Rarefaction curves were generated for the six pooled libraries to assess the depth of sequence sampling and the library richness (97% cutoff for OTU separation). Unsurprisingly, none of the curves reached the plateau level (data not shown), and hence, the number of clones was insufficient to completely cover the diversity of bacterial OTUs in the samples. The rarefaction analysis did not show any significant effect of sampling time on the bacterial-community structures, although we could detect a slight increase in richness in 2010 compared to 2008. Based on the CHAO1 richness estimator, the estimated richness was highest in the December 2008 library (average value, 86/181 analyzed clones), whereas the April 2008 sample had the lowest richness (49/177 analyzed clones). The remaining libraries had a richness estimation of about 70 (Fig. 4).
Fig 4.

OTU richness estimation (CHAO1) at different sampling moments using a 97% cutoff for OTU determination. Bulk soils from April 2008 (before planting), June 2008 (flowering potato), December 2008 (after removal of plants), June 2009 (barley), July 2010 (flowering potato), and September 2010 (senescent potato) were used. bf, bulk soil, flowering stage; bsn, bulk soil, senescence stage (7).
(ii) Phylogenetic analysis of libraries.
Using the RDP database and comparison at a confidence threshold of 80%, the majority of the sequences (76 to 83%) were affiliated with recognized classes of bacteria, whereas the remaining minority (17 to 24%) had as-yet-unclassified bacteria as best hits (Fig. 5). There were significant differences in bacterial-community makeup, with respect to the prevalence of particular groups, between the bulk soil samples (Fig. 5 and 6A and B). For instance, sequences assigned to the Actinobacteria accounted for 15 to 22% of the clones from the bulk soils of April 2008 (before planting), June 2008 (flowering), December 2008 (after removal of plants), July 2009 (barley), and September 2010 (senescent potato), whereas this group made up 37% of the amplicons generated from the bulk soil of June 2010. In contrast, sequences affiliated with the Alphaproteobacteria were highly dominant in the aforementioned bulk soil samples (30 to 40%) (Fig. 6A), while they made up only 19% of the clone library from the June 2010 bulk soil (Fig. 6A). Curiously, it was found that Actinobacteria and Alphaproteobacteria were apparently selected differently at the different growth stages but reached very similar total relative abundances (i.e., around 50 to 52%) at each sampling moment (Fig. 6A). Furthermore, the prevalence of sequences affiliated with the Betaproteobacteria and Deltaproteobacteria was quite stable over the season, whereas that of Gammaproteobacteria peaked in June 2008 (Fig. 6B). Furthermore, neither Acidobacteria nor Gemmatimonadetes appeared to be highly selected in June and September 2010, and Firmicutes showed a rapid increase in the June 2010 sample (Fig. 6B). Moreover, the relative abundances of Gammaproteobacteria and Deltaproteobacteria were higher and those of Bacteriodetes lower in June 2008 than the general trend of the season. Also, the relative abundances of the Actinobacteria and Firmicutes were higher and those of Alphaproteobacteria lower than the overall trends in June 2010 (Fig. 6).
Fig 5.
Normal operating range (NOR) of different phyla and classes in bulk soil samples from the Buinen field, shown in three plots, as indicated. The boxes show the upper (75%) and the lower (25%) percentiles of the data. The whiskers indicate the highest and the lowest values (SPSS Statistics 16). Outliers are also indicated. bf08, bulk, flowering potato, June 2008; bf10, bulk, flowering potato, June 2010; bsn10, bulk, senescent potato, September 2010; after, after crop removal, December 2008 (7).
Fig 6.

Dynamics of dominating phyla and classes across time. (A) Actinobacteria, Alphaproteobacteria, and unclassified (Unc.) bacteria. (B) Betaproteobacteria, gammaproteobacteria, deltaproteobacteria, unclassified proteobacteria, acidobacteria, and firmicutes. B, bulk soil under potato or bare; Bbar, bulk soil under barley.
The relative abundance data from June 2008 and June 2010 were found to be outliers of the general trend in the overall results: some phyla/classes were detected that were obviously lower or higher in abundance than the remainder, e.g., increased relative abundances of Actinobacteria in June 2008 and Gammaproteobacteria in June 2010 (Fig. 5).
Altogether, 65 genera were found in the soils from the six sampling moments. Acidobacterium group 2 and Bradyrhizobium were the only two genus level taxa found at all sampling moments, indicating they may be integral parts of the CBC group. Also, Burkholderia and Gemmatimonas were found in five out of the six samples, whereas Blastococcus, Hyphomicrobium, Nitrospira, and Acidobacterium groups 1 and 16 were found in four out of the six samples analyzed. The April 2008 and June 2009 samples contained totals of 26 genera, whereas samples from December 2008, June 2008, and September 2010 encompassed only around 20. Also, samples collected in June 2010 had 29 genera in total. Interestingly, Arthrobacter, Streptomyces, Rhodanobacter, and Dokdonella were found only in the presence of potato plants (June 2008 and June 2010, flowering), whereas Ktedonobacter was specific to potato bulk soil, independent of the time of sampling. Rhodoplanes and Sporosarcina were observed only in the bulk soil from barley. Sequences of Aquicella, Acidobacterium group 7, Methylobacterium, Patulibacter, and Phenylobacterium were found to be specific to samples from December 2008.
We applied Mothur to the data set to find the level of unique versus shared sequences (get.sharedseqs command), listed in Table 2. Strikingly, 86 to 92% of OTUs defined at the 97% similarity cutoff level were unique to samples (“subjects”) regardless of the species. This finding indicates that time and environmental conditions have important effects on bacterial-community composition, and the soil harbored a considerable fraction of unique OTUs at each sampling point. For instance, only 9 OTUs were shared between the initial (April 2008) and the last (September 2010) sample. On the basis of database sequences, these shared OTUs were affiliated with sequences from Alphaproteobacteria, Gammaproteobacteria, Bradyrhizobium, and Methylibium sp., as shown in Fig. 7.
Table 2.
Number of shared OTUs for each pairwise comparison
| Samplea | No. of shared OTUsb |
|||||
|---|---|---|---|---|---|---|
| B-April08 | B-June08 | B-Dec08 | Bar-Jul09 | B-June10 | B-Sep10 | |
| B-April08 | 135 | 10 | 11 | 7 | 9 | 9 |
| B-June08 | 111 | 8 | 12 | 11 | 6 | |
| B-Dec08 | 139 | 8 | 5 | 4 | ||
| Bar-Jul09 | 137 | 13 | 9 | |||
| B-June10 | 121 | 9 | ||||
| B-Sep10 | 136 | |||||
Soil-month and year. B, bulk soil under potatoes (except Dec08, which was bare); Bar, bulk soil under barley.
The number of shared OTUs was derived from the unique OTUs0.03 (OTUs defined at the 97% similarity cutoff level) for each subject. The numbers were calculated using the get.sharedseqs command in the Mothur software (34).
Fig 7.
Neighbor-joining-based dendrogram of 16S rRNA gene sequences shared between April 2008 (B-April08) and September 2010 (B-Sep10). The tree was constructed using the Kimura two-parameter algorithm (complete deletion model). Nodal support in neighbor joining was evaluated by 1,000 bootstrap replications. The accession numbers of the closest BLAST-N matches are listed.
DISCUSSION
The results of this study showed that the bacterial communities in one field, Buinen, under a short potato rotation scheme (potato-barley-potato) are dynamic and undergo significant changes at temporal scales related to the seasonal measures. Specifically, our results provided evidence for the contention that (i) the bacterial abundances in the Buinen soil may fluctuate over time in a range of about 2 orders of magnitude, i.e., roughly between 107 and 109 cells (as indicated by the 16S rRNA gene proxy numbers) per g dry soil; (ii) the roots of all potato cultivars planted in the field exert positive rhizosphere effects on bacterial numbers; (iii) the diversities and community structures of the bacterial communities differ across time and are season dependent; (iv) the taxa Alphaproteobacteria and Actinobacteria are commonly prevalent in the Buinen soil, occasionally being inversely related to each other; and (v) the composition of the bacterial communities in soil is linked to particularities of different cultivars, which is potentially related to their different tuber starch contents.
Although the methods used in this study are well known and are known to work well on soil/rhizosphere DNA (13, 27, 35, 36), there are well-known caveats to them. Briefly, the extracted DNA may have been incomplete in coverage, and the PCR and subsequent DGGE/clone library analyses may have been subject to biases inherent in these methodologies, one of them being the coincidental colocation of bands on the gel. However, we performed the experiments in a comparative fashion and, hence, posit here that it is fair to compare our results, even in the face of the possible biases compared to absolute values.
There is still controversy with respect to the extent to which plants, and in particular different cultivars/genotypes within one plant species, drive the abundance and composition of bacterial communities in their vicinity (2, 4, 8, 20, 37–40). In this study, an overall population size-enhancing (positive) potato rhizosphere effect, albeit not always significant, on the root-associated bacterial communities was observed in virtually all cultivars over 2 years. This is consistent with findings by our group, as well as others (2, 12, 13, 15, 16). Here, we posit that it indicates the release of sufficient community-driving compounds from the potato plants to overwhelm any confounding effect of the year, e.g., via different temperature, moisture, or fertilization/tillage regimens. This is important, as it indicates that potato plants possess mechanisms that allow them to assemble their rhizosphere microbiota under the prevailing conditions in the Buinen soil. However, we obviously did not obtain data that would provide information on the physiologies of the respective responders to the conditions created in the rhizospheres under study, although one may speculate about rapid responses by r-strategist-type bacteria (fast-growing organisms or copiotrophs).
On the other hand, the effects of the crop versus the conditions associated with the year on the bacterial communities in the bulk soil were not evident. Thus, in order to understand such effects, replicate clone libraries prepared from bulk soil from six different time points, spanning 3 years, were compared. The 12 libraries, pooled into six per sampling time, varied in terms of bacterial diversities, indicating that the bulk soil bacterial consortia, within the confines of the experiment (given by the rather low numbers of representatives for particular bacterial taxa), were rather variable. Thus, some phyla became more dominant at particular sampling moments but dropped in prevalence at others. For instance, members of the Alphaproteobacteria and Actinobacteria became dominant or rare at opposed sampling moments. Whereas Actinobacteria were highly prevalent in July 2010, Alphaproteobacteria showed a significant decrease. A contrasting case occurred between the two groups in September 2010. We distill from these data that members of these two groups may compete for the same or similar niches, giving an advantage to one group in one sample but to the other in another sample, and never to the two groups simultaneously.
Interestingly, the relative abundance data from June 2008 and June 2010 constituted outliers of the general trend in the overall results. The June 2008 sample (taken during flowering of the potato) yielded the lowest number of different genera, whereas those from July 2009 (barley) and June 2010 (during flowering of the potato) revealed the highest numbers. These contrasting observations might have come about due to the combined effects of (i) the presence of a plant, (ii) the plant type, and (iii) environmental conditions, such as temperature and moisture.
Also, the presence of some bacterial types was plant dependent, as we found Arthrobacter, Streptomyces, Rhodanobacter, and Dokdonella only during the flowering stage of the potato in both seasons. In contrast, Rhodoplanes and Sporosarcina were observed only in the bulk soil under barley. Arthrobacter has been defined as a plant growth-promoting bacterium (PGPB), whereas Streptomyces species are key transformers of organic material in the soil, but particular species also cause (scab) disease in the potato (12). The drop in the prevalence of Streptomyces types in the other plant growth stages, provided they coincide with the pathogenic ones, might constitute a good argument for the application of crop rotation, reducing particular organisms (including clearly plant-pathogenic ones) in the soil bacterial community.
In addition, the PCR-DGGE analyses also revealed differences between the potato-planted bulk soils of the different years. This also provided evidence for the contention that environmental factors (temperature and rainfall) and/or management practices exert additional effects on the soil bacterial community.
Rhizosphere and cultivar effects were observed in different ways in the 2 years when potatoes were in the field (2008 and 2010). Overall, the same cultivars growing in the two different years did not show exactly the same trends, except for cultivar P. It was also seen that plants at the same growth stages in different years clustered apart. The differences in abundance and community makeup might have been caused by the different conditions with respect to either abiotic factors (temperature and rainfall) or management (levels and types of manure application). In 2008, mineral fertilizer was applied, whereas pig manure was applied in 2010. This change in the fertilization regime might also have affected the observed growth rate of the plants. It is an obvious possibility that different fertilizers, due to differences in nutrient contents and microbial loads, contribute to differentiation of rhizosphere microbial communities (37). These different levels of nutrients might also have caused the differences between the microbial communities of different cultivars that were in the young-plant stage in the 2010 field experiment. This was in contrast to the grouping of all cultivars in the young-plant stage in 2008. The difference in management may be a strong determinant of the NOR of the bulk soil communities; strikingly, the variability associated with the samples from 2008 was much smaller than that observed in 2010, in terms of both bacterial-community structure and abundance.
In this context, the diversity and community structure of the soil microbiota constitute important determinants of the stability of key soil functions, even though the relationship between functioning and microbial diversity in soil is not well defined (17, 41). In this study, although the number of clones was insufficient to completely inventory the different bacterial phylotypes, major shifts in the community compositions were observed. Moreover, MWA of the PCR-DGGE profiles showed that, irrespective of the shifts found, 40% of the community was actually still present in bulk soil after the ∼30-month experimental period. Since PCR-DGGE provides information on organisms down to about 0.1% of the total community (23, 28, 35), a core group of dominant bacterial-community members, denoted CBC, thus remained relatively stable. Organisms related to Acidobacterium group 2 and Bradyrhizobium may be key CBC members, as they were found in the clone libraries in all sampling periods. The role of such key CBC members may lie in the general transformation of carbonaceous compounds, although involvement of the latter group in nitrogen fixation is a clear possibility.
Generally, and across the two seasons, the high-starch- versus low-starch-tuber plants clustered in different groups with respect to their effects on the associated bacterial communities, especially in the flowering and senescence stages of plant growth. Thus, we surmised that the plant physiologies associated with the tuber starch phenotype were the drivers of these different communities in the rhizosphere. As has been suggested (2–4, 16), compounds excreted from roots constitute the link between plant roots and microbes in the rhizosphere. In light of the potential importance of the plant-associated microbiota for plant health, the rhizosphere microbiome may be thought of as representing the second genome of the plant (2). Some of the key factors affecting interactions in the rhizosphere, e.g., quorum sensing, have been identified (4). These findings support the idea that plant physiology affects its interaction with root-surrounding microbes (42).
At the bulk soil level, we found shifting communities across time and treatments. Thus, at the fine (rhizosphere) level, the NOR of the bacteriota under potatoes may have to be dichotomized, whereas we cannot affirm this for the gross (bulk soil) level (Fig. 5). In terms of its effect, the GM line M grouped differently, but generally with the low-starch-tuber cultivars, and it fell in the value range of the other cultivars for all measured variables in the two growth seasons in 2008 and 2010 (Fig. 5).
ACKNOWLEDGMENTS
We thank Eelco Hoogwout and Emilia Hannula for their help with soil sampling. The work was executed in fields laid out by Avebe (Foxhol, The Netherlands), and we gratefully acknowledge Paul Heeres and Peter Bruinenberg for their help in the field work.
This work was supported by the NWO-ERGO program (J.D.V.E.).
Footnotes
Published ahead of print 7 December 2012
REFERENCES
- 1. Rodríguez H, Fraga R. 1999. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17:319–339 [DOI] [PubMed] [Google Scholar]
- 2. Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends Plant Sci. 17:478–486 [DOI] [PubMed] [Google Scholar]
- 3. Garbeva P, van Veen JA, van Elsas JD. 2004. Microbial diversity in soil: selection of microbial population by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42:243–270 [DOI] [PubMed] [Google Scholar]
- 4. Badri DV, Weir TL, van der Lelie D, Vivanco JM. 2009. Rhizosphere chemical dialogues: plant-microbe interactions. Curr. Opin. Biotechnol. 20:642–650 [DOI] [PubMed] [Google Scholar]
- 5. Oger P, Mansouri H, Dessaux Y. 2000. Effect of crop rotation and soil cover on alteration of the soil microflora generated by the culture of transgenic plants producing opines. Mol. Ecol. 9:881–890 [DOI] [PubMed] [Google Scholar]
- 6. Bruinsma M, van Veen JA. 2003. Effect of genetically modified plants on microbial communities and processes in soil. Biol. Fertil. Soils 37:329–337 [Google Scholar]
- 7. Inceoglu Ö, Salles JF, van Overbeek LS, van Elsas JD. 2010. Effect of plant genotype and growth stage on the β-proteobacterial community associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 76:3675–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andreote F, Mendes R, Dini-Andreote F, Rossetto PB, Labate CA, Pizzirani-Kleiner AA, van Elsas JD, Azevedo JL, Araújo WL. 2008. Transgenic tobacco revealing altered bacterial diversity in the rhizosphere during early plant development. Antonie Van Leeuwenhoek 93:415–424 [DOI] [PubMed] [Google Scholar]
- 9. Inceoglu Ö, Al-Soud WA, Semenov AV, Salles JF, van Elsas JD. 2011. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS One 6:e23321 doi:10.1371/journal.pone.0023321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heuer H, Kroppenstedt R, Lottmann J, Berg G, Smalla K. 2002. Effects of T4 lysozyme release from transgenic potato roots on bacterial rhizosphere communities are negligible relative to natural factors. Appl. Environ. Microbiol. 68:1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lottmann J, Berg G. 2001. Phenotypic and genotypic characterization of antagonistic bacteria associated with roots of transgenic and nontransgenic potato plants. Microbiol. Res. 156:75–82 [DOI] [PubMed] [Google Scholar]
- 12. Rasche F, Hödl V, Poll C, Kandeler E, Gerzabek M, van Elsas JD, Sessitsch A. 2006. Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effect of soil, wild type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol. Ecol. 56:219–235 [DOI] [PubMed] [Google Scholar]
- 13. van Overbeek LS, van Elsas JD. 2008. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol. Ecol. 64:283–296 [DOI] [PubMed] [Google Scholar]
- 14. Weinert N, Meincke R, Gottwald C, Heuer H, Gomes NC, Schloter M, Berg G, Smalla K. 2009. Rhizosphere communities of genetically modified zeaxanthin-accumulating potato plants and their parent cultivar differ less than those of different potato cultivars. Appl. Environ. Microbiol. 75:3859–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weinert N, Piceno Y, Ding GC, Meincke RH, Heuer Berg G, Schloter M, Andersen G, Smalla K. 2011. PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol. Ecol. 75:497–506 [DOI] [PubMed] [Google Scholar]
- 16. Lynch J, Whipps J. 1990. Substrate flows in the rhizosphere. Plant Soil 129:1–10 [Google Scholar]
- 17. Harris J. 2009. Soil microbial communities and restoration ecology: facilitators or followers? Science 325:573–574 [DOI] [PubMed] [Google Scholar]
- 18. Lilley AK, Bailey MJ, Cartwright CL, Turner S, Hirsch P. 2006. Life in earth: the impact of GM plants on soil ecology? Trends Biotechnol. 24:9–14 [DOI] [PubMed] [Google Scholar]
- 19. Schmalenberger A, Kertesz M. 2007. Desulfurization of aromatic sulfonates by rhizosphere bacteria: high diversity of the asfA gene. Environ. Microbiol. 9:535–545 [DOI] [PubMed] [Google Scholar]
- 20. Bever JD, Westover KM, Antonovics J. 1997. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J. Ecol. 85:561–573 [Google Scholar]
- 21. Cavigelli M, Robertson G, Klug M. 1995. Fatty acid methyl ester (FAME) profiles as measures of soil microbial community structure. Plant Soil 170:99–113 [Google Scholar]
- 22. Hartmann M, Widmer F. 2006. Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices. Appl. Environ. Microbiol. 72:7804–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muyzer G, de Waal E, Uitterlinden A. 1993. Profiling of complex microbial populations by DGGE analysis of PCR-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schloss P, Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Vetten N, Wolters A, Raemakers K, van der Meer I, ter Stege R, Heeres E, Heeres P, Visser R. 2003. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nat. Biotechnol. 21:439–442 [DOI] [PubMed] [Google Scholar]
- 26. Hack H, Gall H, Klemke T, Klose R, Meier U, Stauss R, Witzenberger A. 1993. Phänologische entwicklungsstadien der Kartoffel (Solanum tuberosum L.). Nachrichtenbl. Deutsch. Pflanzenschutzd. 45:11–19 [Google Scholar]
- 27. Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturant gradients. Appl. Environ. Microbiol. 63:3233–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marzorati M, Wittebolle L, Boon B, Daffonchio D, Verstraete W. 2008. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ. Microbiol. 10:1571–1581 [DOI] [PubMed] [Google Scholar]
- 29. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence aligments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 30. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singleton D, Furlong M, Rathbun S, Whitman W. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 33. Cole J, Chai B, Farris R, Wang Q, Kulam-Syed-Mohideen A, McGarrell D, Bandela A, Tiedje JM. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169–D172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Elsas JD, Smalla K, Tebbe C. 2000. Extraction and analysis of microbial community nucleic acids from environmental matrices, p 29–51 In Jansson JK, van Elsas JD, Bailey MJ. (ed), Tracking genetically engineered microorganisms. Biotechnology Intelligence Unit. Landes Bioscience, Austin, TX [Google Scholar]
- 36. van Elsas JD, Duarte GF, Keijzer-Wolters AC, Smit E. 2000. Analysis of the dynamics of fungal communities in soil via fungal-specific PCR of soil DNA followed by denaturing gradient gel electrophoresis. J. Microbiol. Methods 43:133–151 [DOI] [PubMed] [Google Scholar]
- 37. Aira M, Gómez-Brandón M, Lazcano C, Bååth E, Domínguez E. 2010. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 42:2276–2281 [Google Scholar]
- 38. Duineveld B, Rosado AS, van Elsas JD, van Veen JA. 1998. Analysis of the dynamics of bacterial communities in the rhizosphere of chrysanthemum via DGGE and substrate utilization patterns. Appl. Environ. Microbiol. 64:4950–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Felske A, Wolterink A, van Lis R, Akkermans ADL. 1998. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands). Appl. Environ. Microbiol. 64:871–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ibekwe A, Grieve C. 2004. Changes in developing plant microbial community structure as affected by contaminated water. FEMS Microbiol. Ecol. 48:239–248 [DOI] [PubMed] [Google Scholar]
- 41. Lupwayi N, Rice W, Clayton G. 1998. Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol. Biochem. 30:1733–1741 [Google Scholar]
- 42. Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD. 2012. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7:e30438 doi:10.1371/journal.pone.0030438 [DOI] [PMC free article] [PubMed] [Google Scholar]