Abstract
The Toll-like receptor (TLR)/IL-1 receptor (IL-1R) superfamily plays fundamentally important roles in innate immune and inflammatory responses. Recent structural studies have begun to unveil the surprising concept that upon ligand stimulation TLR/IL-1Rs assemble large oligomeric intracellular signaling complexes, or signalosomes, to induce activation of ubiquitin ligases and kinases, leading eventually to activation of the transcription factors that are responsible for the expression of immune and inflammatory response genes. The different scaffolds of assembly identified from the structural studies provide a molecular foundation in understanding the formation of microscopically visible signaling clusters that have long been known to cell biologists. Here, we illustrate the potential mechanisms of assembly step by step from the membrane proximal interactions to the more downstream events. Formation of large oligomeric signalosomes may help to establish a digital, threshold response in TLR/IL-1R signaling.
Background
Toll-like receptors (TLRs) and interleukin-1 receptors (IL-1Rs) belong to a superfamily of evolutionary conserved transmembrane receptors involved in innate and inflammatory immune responses. To date, a total of 10 human TLRs (TLR 1-10) have been identified. They are characterized by an extracellular leucine-rich repeat (LRR) motif responsible for the identification of distinct microbial structures, referred to as pathogen-associated molecular patterns (PAMPs), enabling the innate immune system to recognize invading microorganisms and initiate protective immune responses (1, 2, 3, 4). Members of the IL-1R family are distinct from TLRs in their extracellular region, containing three immunoglobulin (Ig)-like domains. IL-1R family members interact with accessory proteins to form heterodimeric receptor signaling complexes (1).
The TLR/IL-1R superfamily share a common cytoplasmic domain, the Toll/IL-1R homology (TIR) domain, which is essential for initiating intracellular signaling. Recognition of cognate ligand triggers a signaling cascade by recruitment of TIR containing adaptor proteins through homotypic TIR/TIR interactions (1, 2, 3, 4). Myeloid differentiation protein 88 (MyD88), a TIR-containing adapter protein known to be essential for IL-1 and IL-18 signaling (5), was the first to be identified as involved in TLR signal transduction (6). Subsequently a number of other TIR-containing adapters were shown to participate in signaling from a subset of TLRs, including MyD88 adapter-like (Mal, also known as TIRAP) (7), TIR domain-containing adapter inducing IFNβ (TRIF, also known as TICAM1), and TRIF-related adapter molecule (TRAM, also known as TICAM2) (8, 9). TIR-containing receptors and TIR-containing adaptors interact to form membrane-proximal signaling complexes in the TLR/IL-1R pathway.
MyD88 has been shown to be a major adapter shared by almost all TLRs except TLR3 (10). In addition to its TIR domain, MyD88 possesses an N-terminal death domain (DD) (Fig. 1A). Upon stimulation, MyD88 dependent signaling propagates through the recruitment of IL-1R-associated kinase (IRAK) family members. IRAKs are multidomain proteins containing both a DD and a Ser/Thr kinase domain (KD). Targeted deletions in mice have identified IRAK1, IRAK2, and IRAK4 as positive regulators and IRAK-M as a negative regulator of signaling (11, 12, 13). Subsequent to MyD88 recruitment to receptor TIR domains, homotypic DD/DD interactions between MyD88 and IRAK family members lead to the formation of an oligomeric DD signaling scaffold. Formation of this signaling scaffold promotes IRAK activation, followed by recruitment and activation of TNF receptor-associated factor 6 (TRAF6).
Figure 1. Model of TLR/IL-1R:MyD88:IRAK signalosome formation.
A. Domain organization of TLR/IL-1R family members (top), MyD88 (middle), and IRAKs (bottom). B. Model of TLR/IL-1R signaling. Top left: IL-1RI (pink) bound to IL-1β (yellow) and co-receptor IL-1RAcP (light green). Top right: TLR1 (Cyan)/TLR2 (Green) dimer bound to lipopeptide (magenta). Cytosolic receptor TIR domains interact with MyD88 TIRs (light blue) to promote formation of the Myddosome DD complex. MyD88 DDs (blue) oligomerize with IRAK4 (red) and IRAK1/2 (orange) to promote phosphorylation and activation of IRAK kinase domains (bottom right).
TRAF6 is a RING-domain E3 ubiquitin (Ub) ligase (14). Together with a heterodimeric E2 Ub conjugating enzyme complex containing Ubc13 and Uev1A, it catalyzes the synthesis of Lys63 (K63)-linked polyubiquitin (polyUb) chains. Formation of K63-linked polyUb allows the recruitment of two downstream kinase complexes. The first is the TGFβ-activated kinase 1 (TAK1) complex, composed of TAK1 and the TAK1-binding proteins TAB1, TAB2, or TAB3 (15, 16, 17). The second is the IκB kinase (IKK) complex, consisting of the catalytic subunits IKKα and/or IKKβ and the regulatory subunit NF-κB essential modulator (NEMO; also known as IKKγ). Recruitment of these complexes to polyUb triggers phosphorylation and activation of their kinase activities. IKK phosphorylates NF-κB inhibitor (IκB) proteins, which are ubiquitinated by the E3 ligase complex SCFβ-TrCP and subsequently degraded by the proteasome. This allows NF-κB to enter the nucleus and, along with AP-1 and IRFs, induce target gene expression to elicit inflammatory and immune responses (18, 19).
Engagement of the TLR/IL-1R pathway in appropriate physiological environment initiates the development of protective immune responses. However, the complexity of this pathway renders itself susceptible to interruption and deregulation, leading to its association with many human diseases (20). For instance, inherited mutations or polymorphisms in TIR-containing adapters and IRAKs may cause either extreme sensitivity to or protection against infection (22). Other types of deregulation in the pathway contribute to both diseases in the immune system such as inflammatory disorders (22), autoimmune diseases (23) and allergy (24), as well as diseases beyond the immune system such as cancer (25), insulin resistance (26), atherosclerosis (27) and painful neuropathy (28). Given that the major components of the TLR/IL-1R pathway have been identified, obtaining the relevant structural information on the core signaling complexes would enhance the understanding of the molecular mechanism of signal transduction and assist the targeting of the pathway for therapeutics. In this review, we illustrate the structural features of the TLR/IL-1R signaling pathway and discuss recent advances in the field.
Receptor Clustering and TIR Domain Interactions
In humans, the 10-member TLR family recognizes a variety of pathogen associated molecular patterns (PAMPs) (29). Each TLR recognizes and binds its cognate PAMPs through an extracellular LRR domain. Agonistic ligand binding triggers signaling by promoting dimerization of TLRs (30) or altering the conformation of existing dimers (31). In addition, TLRs are known to partition into lipid raft microdomains upon ligand triggering and therefore higher order oligomers of TLRs may be critical for signal initiation (32, 33). Structural information is available on several dimeric TLR ectodomain-ligand complexes, including TLR4 bound to its co-receptor MD-2 and lipopolysaccharide (34), a TLR3 homodimer bound to dsRNA (35), heterodimers of TLR2/TLR1 (36) and TLR2/TLR6 bound to lipopeptides (37), and TLR5 bound to Salmonella flagellin (38). In each structure the TLRs adopt similar M-shaped dimers, bringing the C-terminal juxtamembrane ends in close proximity to each other (Fig. 1B, top right). Although the overall conformation of the TLR dimers is similar, each ligand is bound to a distinct surface of the LRR.
Cellular responses to the cytokine IL-1β require a receptor complex containing both IL-1RI and IL-1RAcP (29). Initial binding of IL-1β to IL-1RI requires subsequent association with IL-1RAcP to form an active signaling complex (39). The IL-1β binding extracellular regions of both receptors contain three Ig-like domains. Recently, the crystal structure of the ternary complex of IL-1β and the extracellular domains of IL-1RI and IL-1RAcP has been solved (Fig. 1B, top left) (40). This structure superimposes well with both the IL-1RI/IL-1β heterodimer structure (41) and the ternary complex structure containing the TIR-deficient decoy receptor IL-1RII bound to IL-1β and IL-1RAcP (42). In the IL-1β/IL-1RI/IL-1RAcP ternary complex, the juxtamembrane Ig-like domains of both receptors are brought in close proximity to each other.
Shared between all TLR and IL-1R family members is the cytoplasmic TIR domain. The TIR domain is small and globular, consisting of a parallel β-sheet surrounded by α-helices (43). Binding of an agonistic ligand to its receptor is thought to promote homotypic interactions of these domains, creating a nucleation platform to promote oligomerization with TIR containing adapter proteins. Several structures of TIR domains are available, including those of TLR1, TLR2 (43), TLR10 (44), the IL-1R family member IL-1RAPL (45), and the TIR containing adapters MyD88 (46) and Mal (47). Despite the abundance of TIR domain structural information, the nature of TIR domain oligomerization remains elusive. This is due in part to the difficulty of reconstituting stable TIR domain multimers in solution. Nevertheless, several models of TIR oligomerization have been proposed, based on crystal structures of TLR10 (44) and IL-1RAPL (45), mutagenesis, and molecular docking studies (46, 47).
Oligomeric DD Interactions and IRAK Activation
MyD88, the primary signaling adapter for TLR and IL-1R signaling, also contains an N-terminal DD. The DD is a member of the death domain (DD) superfamily, whose additional members include caspase recruitment domains, pyrin domains, and death effector domains. Superfamily members share a common fold that resembles a Greek key bundle of six antiparallel α-helices (48). DD containing proteins are critical components of apoptotic and inflammatory signaling. Through a conserved set of homotypic interactions, DDs form oligomeric molecular scaffolds responsible for activation of proapoptotic caspases (49, 50, 51), proinflammatory caspase (52), and kinases.
In the TLR/IL-1R pathways, the DDs of MyD88 associate with DDs of the IRAK family of Ser/Thr kinases (53). The DDs of MyD88 and IRAKs have been observed to assemble into oligomeric signaling platforms known as the Myddosomes (32) (Fig. 1B). The structure of a ternary Myddosome containing the DDs of MyD88, IRAK4, and IRAK2 has been solved (54). The DDs are arranged in a four layered tower, with six MyD88 forming the top two layers, followed by a layer of four IRAK4 DDs, and finally a layer of four IRAK2 DDs (Fig. 1B). The DDs form a single left-handed helical scaffold held together by three types of conserved DD interaction types (55). Shape and charge complementarity between the top and bottom surfaces of each layer confers a sequential assembly order to the Myddosome. This structure explains the observation that IRAK1 recruitment to the IL-1R complex is drastically reduced in IRAK4 deficient cells (56, 57). The DD of IRAK4 is monomeric in solution, while the DD of MyD88 is prone to oligomerization at high concentrations (54). This data suggest a strict hierarchical assembly mechanism that begins with ligand binding and receptor clustering. Juxtaposition of receptor TIR domains leads to MyD88 recruitment and DD oligomerization. The MyD88 DD oligomer then serves as a nucleation platform for the recruitment of IRAK4. Only after four IRAK4 molecules are present in the Myddosome can IRAK1/2 join the complex.
The DD interactions provide an oligomeric platform for IRAK kinase activation. In vitro kinase assays have shown that IRAK4 is able to auto-phosphorylate itself as well as phosphorylate the activation loop of IRAK1 (58). MyD88 has been shown to promote the phosphorylation of IRAK1 by IRAK4 (59). This data suggest a mechanism for signal propagation in which initial IRAK4 recruitment results in its auto-phosphorylation and activation. Activated IRAK4 is then able to phosphorylate and activate downstream IRAK1/2. Phosphorylated IRAK1/2 (60) then recruits TRAF6 to the membrane (61) through TRAF6 interaction motifs (62).
TRAF6 and PolyUb as Key Scaffolds
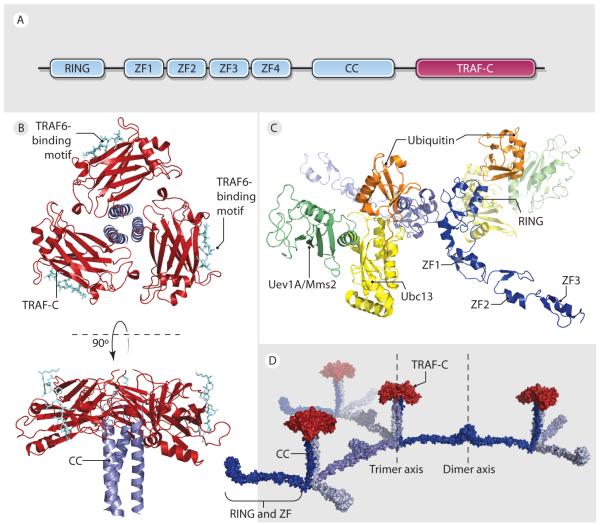
The ubiquitin ligase TRAF6 is essential for TLR/IL-1R mediated activation of NF-κB and AP-1 signaling pathways (63). Like other TRAFs, TRAF6 contains an N-terminal RING and zinc-finger (ZF) domains, followed by a C-terminal TRAF domain (Fig. 2A). Crystal structures of the C-terminal TRAF domain of TRAF6 and TRAF2 in complex with TRAF interaction motif peptides showed trimeric complexes (Fig. 2B) (62, 64), similar to the oligomerization state of TNFR1 bound to its ligand (65). In contrast, the N-terminal region of TRAF6 is unexpectedly dimeric, both in the crystal lattice and in solution, with the RING and ZF domains arranged linearly in a rigid golf club-like conformation (66, 67) (Fig. 2C). The RING and ZF domains mediate K63-linked polyubiquitination that is essential for NF-κB activation. Dimerization of TRAF6 is critical for E3 activity, as dimerization defective mutants are compromised in polyUb assembly and the ability to promote IκB phosphorylation (67).
Figure 2. Model of TRAF6 2-dimensional lattice.
A. Domain organization of TRAF6. B. Crystal structure of TRAF6 TRAF-C domain (red) trimerized with TRAF2 CC (light blue) based on TRAF2 crystal structure. C. Model of N-Terminal TRAF6 RING and ZF1-3 bound to Ubc13 (yellow)/Uev1A (green) E2 ubiquitin ligase complex and Ub (orange). D. Model of 2-dimensional TRAF6 lattice assembled through TRAF-C (red) trimerization and N-terminal domain (blue) dimerization.
The recruitment of TRAF6 to TLR/IL-1R signalosomes through IRAK1/2 promotes its oligomerization and E3 Ub ligase activity (68). TRAF6, in conjunction with the heterodimeric E2 enzyme Ubc13/Uev1A (Fig. 2C), catalyzes K63-linked polyubiquitination of target proteins including IRAK1, NEMO, and TRAF6 itself. Lys124 of TRAF6 is the major site of auto-polyubiquitination (69), and a point mutation here abolishes the ability of TRAF6 to activate TAK1 and IKK. The structure of a Ubc13/Mms2 (Uev1A homologue, hereafter referred to as Uev1A) complex with and without Ub has been reported (70, 71, 72) (Fig. 2C). The Ub is covalently linked to the active site residue of Ubc13. In addition, K63 of the Ub is bound the acceptor-binding site of an adjacent complex, revealing the mechanism by which Uev1A orients the acceptor Ub to promote formation of K63-linked polyUb chains (71). The structures of the TRAF6/Ubc13 complex and the Ubc13/Uev1A~Ub complex allow the generation of the model of the TRAF6/Ubc13/Uev1A~Ub quaternary complex (Fig. 2C).
Given the aggregation properties of TRAF6 in vitro and in cells, a 2-dimensional lattice model for infinite oligomerization of TRAF6 was proposed based on the symmetry mismatch between the dimeric N-terminal region and the trimeric C-terminal region (67) (Fig. 2D). Indeed, endogenous TRAF6 undergoes patch formation at the cell surface in response to TLR stimulation as shown by immunofluorescence and disruption of TRAF6 dimerization compromised the ability of TRAF6 to aggregate as shown by fluorescence energy transfer experiments (67). This proposed 2-dimensional lattice of TRAF6 facilitates polyUb synthesis on proteins in the complex such as IRAK1 and TRAF6 itself, which in turn promotes recruitment of downstream signaling proteins such as the TAK1 and the IKK complexes through interactions with polyUb chains (73). Interestingly, bacterial chemosensing receptors cluster at distinct regions of the cell and form stable ternary complexes with the histidine autokinase CheA and the adapter protein CheW as trimer-of-dimer arrays (74), in a manner that is reminiscent of the proposed 2-dimensional lattice of TRAF6. This structural parallel may translate into a mechanistic parallel in which both processes are highly cooperative due to the high order oligomerization of the participating proteins (75).
Recruitment and Activation of the TAK1 and IKK Kinase Complexes
TAK1 (also known as MAP3K7 and MEKK7) plays critical roles in signaling pathways stimulated by TGF-β, IL-1/TLR, TNF-α, LPS, and IL-8 (76, 77). The TAK1 kinase complex contains TAK1 and three kinase binding proteins (TAB1/2/3) (78). The activation of TAK1 requires TAB1, as TAK1 has no observable kinase activity when expressed alone but is active when co-expressed with TAB1 (79, 80). The TAK1/TAB1 complex is crucial for normal embryonic development and morphogenesis of the heart and lung (81). The structure of TAK1 and TAB1 reveals an extensive interface between a binding pocket on the C-terminal lobe of the TAK1 kinase domain and a TAB1 α-helix (82). This interaction promotes auto-phosphorylation of the TAK1 kinase activation loop (83, 84), likely through an allosteric mechanism (82).
The TAK1 binding proteins TAB2 and TAB3 facilitate TAK1 activation in the TLR/IL-1R signaling pathway by binding to K63-linked polyUb chains generated by TRAF6. TAB2 and TAB3 have similar domain structures, containing an N-terminal Ub-binding domain CUE, a coiled-coil (CC) region, a TAK1-binding domain, and a C-terminal NZF domain (85). The NZF domain binds to K63-linked polyUb chains much more strongly than it does to K48 chains or linear polyUb (86). The crystal structures of TAB2 and TAB3 NZF domains with K63-linked diubiquitin (diUb) have been reported (87, 88). TAB2 and TAB3 NZF domains share 79.3% sequence identity and are functionally redundant (89). Predictably, TAB2-NZF/diUb and TAB3-NZF/diUb structures are nearly identical. TAB2-NZF binds to K63-linked diUb via two distinct binding sites on adjacent Ub moieties and multiple NZF domains could recognize longer polyUb chains successively. The structures reveal that TAB2 preferentially binds K63-linked chains because it imposes a conformational constraint to the bound Ub chain that cannot be adopted by the linear linkage (88). Presumably, recruitment of the TAK1 kinase complex to the TRAF6/polyUb scaffold brings the kinases into proximity to promote auto-phosphorylation and activation. Active TAK1 can in turn phosphorylate and activate IKK.
The IKK complex was first identified in HeLa cells and is composed of the catalytic subunits, IKKα and/or IKKβ, and the regulatory subunit NEMO (also known as IKKγ) (90, 91). IKK phosphorylates IκBα, the inhibitor of NF-κB, tagging it for K48-linked ubiquitination and proteasomal degradation, allowing for NF-κB nuclear translocation and target gene activation (18). The IKKα and IKKβ kinases share 52% identity. The IKK complex can consist of either an IKKα/β heterodimer or IKKα and IKKβ homodimers (92) associated with NEMO. The C-terminal end of IKKα and IKKβ contains the NEMO-binding domain (93, 94), which interacts with the N-terminal kinase-binding domain of NEMO (18, 95). The crystal structure of this interacting complex showed that IKKβ and NEMO form a heterotetramer with two molecules from each protein (96). The four molecules pack into a parallel four-helical bundle (96).
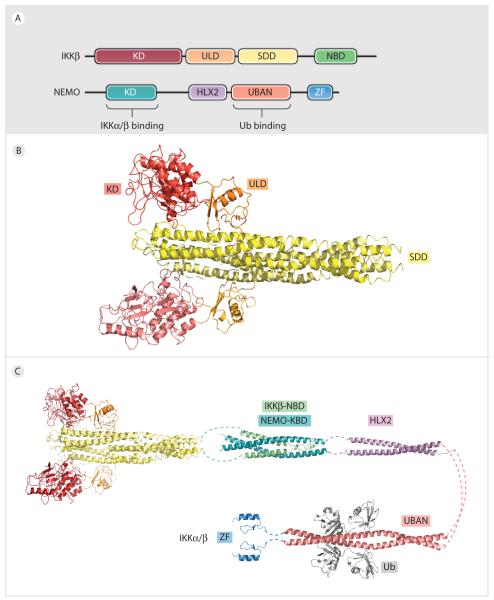
The recently reported crystal structure of almost full-length IKKβ showed that it contains three recognizable domains, the KD, the Ub-like domain (ULD), and an elongated α-helical scaffold/dimerization domain (SDD) (97) (Fig. 3A, 3B). The three domains interact mutually to form a tri-modular architecture. While the ULD appears to be important for IKKβ activity, the SDD mediates IKKβ dimerization and participates in substrate recognition (97). Full length IKKβ is dimeric in solution and mutations of the SDD disrupt this dimerization (97). Although SDD-mediated dimerization is critical for IKKβ activation but not for its kinase activity, the observed IKKβ dimer does not assume an arrangement that likely facilitates intradimer trans-auto-phosphorylation. This suggests a role for higher order oligomerization in IKKβ kinase activation. Indeed, IKKβ exists as dimers of dimers in the observed crystal forms, in which the activation loop is potentially within reach of the active site of a neighboring kinase (97). However, this IKKβ alone tetramer, possibly transient, is not observed in solution.
Figure 3. Model of IKK:NEMO:polyUb signalosome.
A. Domain organization of IKKβ and NEMO. B. Crystal structure of IKKβ dimer, showing IKKβ KD (red), ULD (orange), and SDD (yellow). C. Model of IKKβ:NEMO complex. Catalytic dimeric IKKβ KD:ULD:SDD domain interacts with NEMO-KBD (light blue) through a helical C-terminal NBD (green). The KBD of NEMO is followed by a the helical HLX2 domain (violet), the Ub (grey) binding UBAN domain (pink), and a C-terminal ZF (blue).
NEMO mediates the interaction with polyUb in the IKK complex using its ubiquitin binding in ABIN and NEMO (UBAN) domain and its C-terminal zinc-finger (ZF) domain (73, 98, 99, 100, 101). The UBAN domain of NEMO binds linear polyUb chains with higher affinity than K63 chains (102, 103). The crystal structures of the UBAN domain alone and in complex with both linear diUb and K63-linked diUb have been determined (102, 103, 104) (Fig. 3C). In both cases, the UBAN domain forms a parallel dimeric coiled coil extending about 130 Å in length. UBAN binds linear diUb through a conserved hydrophobic patch and C-terminal tail of the distal ubiquitin and an adjacent surface on the proximal ubiquitin. However, the UBAN domain does not make simultaneous contacts with both moieties of K63-linked diUb. This explains the much lower affinity between NEMO and K63-linked diUb (104), and provides evidence for linear polyUb mediated activation of IKK. In contrast to this is the result that the affinity of K63-linked polyUb for UBAN plus the C-terminal ZF is ~115 fold higher than that of the UBAN alone (105). Addition of the ZF has no effect on linear polyUb affinity. It is likely that both K63 and linear polyUb play important roles in different IKK pathways. A model for the full length IKK complex was proposed as an elongated, flexible dimeric coiled coil structure of NEMO bound to IKKβ (73) (Fig. 3C).
Given that higher order oligomerization such as tetramerization of NEMO has been observed and shown to be important for IKKβ activation (96, 106), it is reasonable to speculate that the IKK complex may be a tetramer or even higher order oligomer. A logical scenario might be that in the unstimulated state, the IKK complex is a transient high order oligomer, which leads to a basal level of IKK activation and associated NF-κB activity. During TLR/IL-1R signaling, the IKK oligomers may be stabilized through its interaction with K63-linked polyUb chains in the proposed 2-dimensional TRAF6 lattice, leading to robust IKK activation and much enhanced NF-κB transcriptional activity.
Conclusion
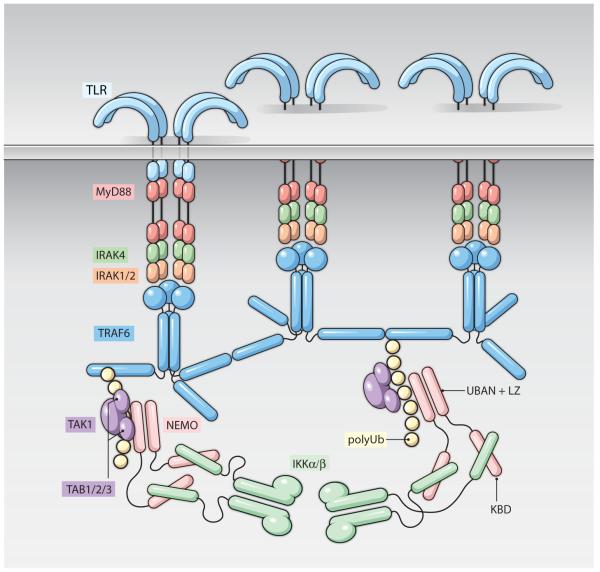
Signal transduction has traditionally been perceived as a linear string of recruitment and allosteric events that transfer and amplify the receptor activation signal to generate the proper cellular responses. Surprisingly, for TLR/IL1-R signaling, recent structural studies are challenging this conventional view. First, it has now been shown that large oligomers are involved in this signal transduction. Some of these oligomers are high order but defined oligomers while others are open-ended oligomers. Oligomerization appears to occur at all different levels of the signaling cascade, from receptors to adapters, from ubiquitin ligases to kinases, and eventually to activation of the transcriptional responses of the pathways. Second, instead of composed of successive signaling complexes, multiple signaling oligomers combine to form gigantic signalosomes (Fig. 4) in which multiple reactions can occur simultaneously including ubiquitination and phosphorylation.
Figure 4. Overview of TLR/IL-1R Signaling.
Ligand binding induces both dimerization and high order oligomerization of TLRs (blue, top). MyD88 (red) is recruited to receptors through homotypic TIR:TIR interactions and facilitates Myddosome assembly with IRAK4 (green) and IRAK1/2 (orange) through homotypic DD:DD interactions, leading to IRAK phosphorylation and activation. Activated IRAK1/2 interacts with TRAF6 and promotes the formation of an infinite 2D lattice, stimulating TRAF6 K63-linked Ub ligase activity. PolyUb (grey) is recognized by TAB2/3 (purple) and NEMO (pink) to allow for the recruitment and activation of TAK1 (purple) and IKK (teal) kinases. Activated IKK phosphorylates IκB, promoting its degradation and allowing for NFκB nuclear translocation.
At least three types of intracellular oligomeric scaffold may cooperate in the formation of the TLR/IL-1R signalosomes, the helical DD assembly scaffold, the infinite TRAF6 aggregation scaffold and the polyUb chain scaffold (Fig. 4). Coalescence of these intertwined interactions into gigantic signalosomes may provide at least two implications in signal transduction. First, the intrinsic cooperativity in the formation of these complexes would dictate a digital, all or none response, achieving a threshold control of innate immune signaling. Second, recruitment of all necessary components into one entity would allow most efficient enzymatic reactions, like in an assembly line of a factory. Because signal transduction of the TNF receptor superfamily shares many commonalities with TLR/IL1-R signaling, the concept of large oligomeric signalosomes may be transferred. In addition, it is tempting to predict that the same concept may be at play in a wide range of signaling systems as well as many other biological processes.
Gloss.
Innate immunity and inflammation make up the initial responses to injury or infection. Toll-like Receptors (TLRs) participate in recognition of invading viruses and microorganisms by recognizing molecules either not present or uncommon in the host cell, known as pathogen associated molecular patterns (PAMPs). The potent proinflammatory cytokine IL-1β is recognized by the receptor IL-1RI and the co-receptor IL-1RAcP. Upon recognition of their cognate ligand, these receptors initiate a shared cytosolic signaling cascade. Recent structural studies have begun to unveil the surprising concept that these signaling cascades coalesce into large oligomeric signaling complexes or signalosomes for signal propagation. Through phosphorylation and ubiquitination, this signaling results in the activation of IκB kinase (IKK). IKK then phosphorylates the NF-κB inhibitor IκB, promoting its degradation. Degradation of IκB allows for the transcription factor NF-κB to translocate into the nucleus and promote the expression of genes involved in innate and adaptive immunity, inflammation, cell survival, and proliferation.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nature reviews. Immunology. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B, Hoebe K, Du X, Ulevitch RJ. How we detect microbes and respond to them: the Toll-like receptors and their transducers. Journal of leukocyte biology. 2003;74:479–485. doi: 10.1189/jlb.0203082. [DOI] [PubMed] [Google Scholar]
- 3.Kopp E, Medzhitov R. Recognition of microbial infection by Toll-like receptors. Current opinion in immunology. 2003;15:396–401. doi: 10.1016/s0952-7915(03)00080-3. [DOI] [PubMed] [Google Scholar]
- 4.Brightbill HD, Modlin RL. Toll-like receptors: molecular mechanisms of the mammalian immune response. Immunology. 2000;101:1–10. doi: 10.1046/j.1365-2567.2000.00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science (New York, N.Y.) 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Molecular cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, Dunne A, Gray P, Harte MT, McMurray D, Smith DE, Sims JE, Bird TA, O’Neill LA. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 8.Kenny EF, O’Neill LAJ. Signalling adaptors used by Toll-like receptors: an update. Cytokine. 2008;43:342–349. doi: 10.1016/j.cyto.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Watters TM, Kenny EF, O’Neill LAJ. Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunology and cell biology. 85:411–419. doi: 10.1038/sj.icb.7100095. [DOI] [PubMed] [Google Scholar]
- 10.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi K, Hernandez LD, Galán JE, Janeway CA, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki N, Suzuki S, Duncan GS, Millar DG, Wada T, Mirtsos C, Takada H, Wakeham A, Itie A, Li S, Penninger JM, Wesche H, Ohashi PS, Mak TW, Yeh W-C. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature. 2002;416:750–756. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 13.Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, Saitoh T, Kawai T, Takeuchi O, Akira S. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nature immunology. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 14.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 15.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 16.Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, Ninomiya-Tsuji J, Matsumoto K. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Molecular cell. 2000;5:649–658. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Z, Ninomiya-Tsuji J, Qian Y, Matsumoto K, Li X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Molecular and cellular biology. 2002;22:7158–7167. doi: 10.1128/MCB.22.20.7158-7167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 19.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiological reviews. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 20.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nature immunology. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 21.Misch EA, Hawn TR. Toll-like receptor polymorphisms and susceptibility to human disease. Clinical science (London, England : 1979) 2008;114:347–360. doi: 10.1042/CS20070214. [DOI] [PubMed] [Google Scholar]
- 22.Fukata M, Vamadevan AS, Abreu MT. Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in inflammatory disorders. Seminars in immunology. 2009;21:242–253. doi: 10.1016/j.smim.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Fischer M, Ehlers M. Toll-like receptors in autoimmunity. Annals of the New York Academy of Sciences. 2008;1143:21–34. doi: 10.1196/annals.1443.012. [DOI] [PubMed] [Google Scholar]
- 24.Sabroe I, Parker LC, Wilson AG, Whyte MKB, Dower SK. Toll-like receptors: their role in allergy and non-allergic inflammatory disease. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2002;32:984–989. doi: 10.1046/j.1365-2745.2002.01451.x. [DOI] [PubMed] [Google Scholar]
- 25.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nature reviews. Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 26.Arkan MC, Hevener AL, Greten FR, Maeda S, Li Z-W, Long JM, Wynshaw-Boris A, Poli G, Olefsky J, Karin M. IKK-beta links inflammation to obesity-induced insulin resistance. Nature medicine. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 27.den Dekker WK, Cheng C, Pasterkamp G, Duckers HJ. Toll like receptor 4 in atherosclerosis and plaque destabilization. Atherosclerosis. 2010;209:314–320. doi: 10.1016/j.atherosclerosis.2009.09.075. [DOI] [PubMed] [Google Scholar]
- 28.Tanga FY, Nutile-McMenemy N, DeLeo JA. The CNS role of Toll-like receptor 4 in innate neuroimmunity and painful neuropathy. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5856–5861. doi: 10.1073/pnas.0501634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neill LAJ. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunological reviews. 2008;226:10–18. doi: 10.1111/j.1600-065X.2008.00701.x. [DOI] [PubMed] [Google Scholar]
- 30.Tsukamoto H, Fukudome K, Takao S, Tsuneyoshi N, Kimoto M. Lipopolysaccharide-binding protein-mediated Toll-like receptor 4 dimerization enables rapid signal transduction against lipopolysaccharide stimulation on membrane-associated CD14-expressing cells. International immunology. 2010;22:271–280. doi: 10.1093/intimm/dxq005. [DOI] [PubMed] [Google Scholar]
- 31.Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DCG, Monks BG, McKnight CJ, Lamphier MS, Duprex WP, Espevik T, Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nature immunology. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 32.Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. The Journal of biological chemistry. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fessler MB, Parks JS. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. Journal of immunology (Baltimore, Md. : 1950) 2011;187:1529–1535. doi: 10.4049/jimmunol.1100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science (New York, N.Y.) 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik S-G, Lee H, Lee J-O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Kang JY, Nan X, Jin MS, Youn S-J, Ryu YH, Mah S, Han SH, Lee H, Paik S-G, Lee J-O. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Yoon S.-i., Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Wilson IA. Structural basis of TLR5-flagellin recognition and signaling. Science (New York, N.Y.) 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korherr C, Hofmeister R, Wesche H, Falk W. A critical role for interleukin-1 receptor accessory protein in interleukin-1 signaling. European journal of immunology. 1997;27:262–267. doi: 10.1002/eji.1830270139. [DOI] [PubMed] [Google Scholar]
- 40.Thomas C, Bazan JF, Garcia KC. Structure of the activating IL-1 receptor signaling complex. Nature structural & molecular biology. 2012;19:455–457. doi: 10.1038/nsmb.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vigers GP, Anderson LJ, Caffes P, Brandhuber BJ. Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1beta. Nature. 1997;386:190–194. doi: 10.1038/386190a0. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Zhang S, Li L, Liu X, Mei K, Wang X. Structural insights into the assembly and activation of IL-1β with its receptors. Nature immunology. 2010;11:905–911. doi: 10.1038/ni.1925. [DOI] [PubMed] [Google Scholar]
- 43.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 44.Nyman T, Stenmark P, Flodin S, Johansson I, Hammarström M, Nordlund P. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. The Journal of biological chemistry. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 45.Khan JA, Brint EK, O’Neill LAJ, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. The Journal of biological chemistry. 2004;279:31664–31670. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- 46.Ohnishi H, Tochio H, Kato Z, Orii KE, Li A, Kimura T, Hiroaki H, Kondo N, Shirakawa M. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10260–10265. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valkov E, Stamp A, Dimaio F, Baker D, Verstak B, Roversi P, Kellie S, Sweet MJ, Mansell A, Gay NJ, Martin JL, Kobe B. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14879–14884. doi: 10.1073/pnas.1104780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park HH, Lo Y-C, Lin S-C, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annual review of immunology. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure (London, England : 1993) 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, Robinson CV, Siegel RM, Walz T, Wu H. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nature structural & molecular biology. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 53.Flannery S, Bowie AG. The interleukin-1 receptor-associated kinases: critical regulators of innate immune signalling. Biochemical pharmacology. 2010;80:1981–1991. doi: 10.1016/j.bcp.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 54.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Current opinion in structural biology. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lye E, Mirtsos C, Suzuki N, Suzuki S, Yeh W-C. The role of interleukin 1 receptor-associated kinase-4 (IRAK-4) kinase activity in IRAK-4-mediated signaling. The Journal of biological chemistry. 2004;279:40653–40658. doi: 10.1074/jbc.M402666200. [DOI] [PubMed] [Google Scholar]
- 57.Qin J, Jiang Z, Qian Y, Casanova J-L, Li X. IRAK4 kinase activity is redundant for interleukin-1 (IL-1) receptor-associated kinase phosphorylation and IL-1 responsiveness. The Journal of biological chemistry. 2004;279:26748–26753. doi: 10.1074/jbc.M400785200. [DOI] [PubMed] [Google Scholar]
- 58.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. The Journal of experimental medicine. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao Z, Henzel WJ, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science (New York, N.Y.) 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 61.Qian Y, Commane M, Ninomiya-Tsuji J, Matsumoto K, Li X. IRAK-mediated translocation of TRAF6 and TAB2 in the interleukin-1-induced activation of NFkappa B. The Journal of biological chemistry. 2001;276:41661–41667. doi: 10.1074/jbc.M102262200. [DOI] [PubMed] [Google Scholar]
- 62.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, Du K, Singh S, Pike JW, Darnay BG, Choi Y, Wu H. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 63.Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. BioEssays : news and reviews in molecular, cellular and developmental biology. 2003;25:1096–1105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 64.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 65.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 66.Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin Q, Lin S-C, Lamothe B, Lu M, Lo Y-C, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, Lenardo MJ, Darnay BG, Wu H. E2 interaction and dimerization in the crystal structure of TRAF6. Nature structural & molecular biology. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takatsuna H, Kato H, Gohda J, Akiyama T, Moriya A, Okamoto Y, Yamagata Y, Otsuka M, Umezawa K, Semba K, Inoue J-I. Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. The Journal of biological chemistry. 2003;278:12144–12150. doi: 10.1074/jbc.M300720200. [DOI] [PubMed] [Google Scholar]
- 69.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. The Journal of biological chemistry. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 71.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature structural & molecular biology. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 72.Moraes TF, Edwards RA, McKenna S, Pastushok L, Xiao W, Glover JN, Ellison MJ. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nature structural biology. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- 73.Zheng C, Yin Q, Wu H. Structural studies of NF-κB signaling. Cell research. 2011;21:183–195. doi: 10.1038/cr.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science (New York, N.Y.) 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 77.Landström M. The TAK1-TRAF6 signalling pathway. The international journal of biochemistry & cell biology. 2010;42:585–589. doi: 10.1016/j.biocel.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 78.Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. The EMBO journal. 2003;22:6277–6288. doi: 10.1093/emboj/cdg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kishimoto K, Matsumoto K, Ninomiya-Tsuji J. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. The Journal of biological chemistry. 2000;275:7359–7364. doi: 10.1074/jbc.275.10.7359. [DOI] [PubMed] [Google Scholar]
- 80.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. TAB1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science (New York, N.Y.) 1996;272:1179–1182. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 81.Komatsu Y, Shibuya H, Takeda N, Ninomiya-Tsuji J, Yasui T, Miyado K, Sekimoto T, Ueno N, Matsumoto K, Yamada G. Targeted disruption of the Tab1 gene causes embryonic lethality and defects in cardiovascular and lung morphogenesis. Mechanisms of development. 2002;119:239–249. doi: 10.1016/s0925-4773(02)00391-x. [DOI] [PubMed] [Google Scholar]
- 82.Brown K, Vial SCM, Dedi N, Long JM, Dunster NJ, Cheetham GMT. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. Journal of molecular biology. 2005;354:1013–1020. doi: 10.1016/j.jmb.2005.09.098. [DOI] [PubMed] [Google Scholar]
- 83.Sakurai H, Miyoshi H, Mizukami J, Sugita T. Phosphorylation-dependent activation of TAK1 mitogen-activated protein kinase kinase kinase by TAB1. FEBS letters. 2000;474:141–145. doi: 10.1016/s0014-5793(00)01588-x. [DOI] [PubMed] [Google Scholar]
- 84.Ono K, Ohtomo T, Sato S, Sugamata Y, Suzuki M, Hisamoto N, Ninomiya-Tsuji J, Tsuchiya M, Matsumoto K. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. The Journal of biological chemistry. 2001;276:24396–24400. doi: 10.1074/jbc.M102631200. [DOI] [PubMed] [Google Scholar]
- 85.Kanayama A, Seth RB, Sun L, Ea C-K, Hong M, Shaito A, Chiu Y-H, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Molecular cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 86.Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO reports. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. The EMBO journal. 2009;28:3903–3909. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nature structural & molecular biology. 2009;16:1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 89.Shim J-H, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee K-Y, Bussey C, Steckel M, Tanaka N, Yamada G, Akira S, Matsumoto K, Ghosh S. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes & development. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 91.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science (New York, N.Y.) 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 92.Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 93.May MJ, D’Acquisto F, Madge LA, Glöckner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science (New York, N.Y.) 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 94.Lo Y-C, Maddineni U, Chung JY, Rich RL, Myszka DG, Wu H. High-affinity interaction between IKKbeta and NEMO. Biochemistry. 2008;47:3109–3116. doi: 10.1021/bi702312c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marienfeld RB, Palkowitsch L, Ghosh S. Dimerization of the I kappa B kinase-binding domain of NEMO is required for tumor necrosis factor alpha-induced NF-kappa B activity. Molecular and cellular biology. 2006;26:9209–9219. doi: 10.1128/MCB.00478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rushe M, Silvian L, Bixler S, Chen LL, Cheung A, Bowes S, Cuervo H, Berkowitz S, Zheng T, Guckian K, Pellegrini M, Lugovskoy A. Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site. Structure (London, England : 1993) 2008;16:798–808. doi: 10.1016/j.str.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 97.Xu G, Lo Y-C, Li Q, Napolitano G, Wu X, Jiang X, Dreano M, Karin M, Wu H. Crystal structure of inhibitor of κB kinase β. Nature. 2011;472:325–330. doi: 10.1038/nature09853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 99.Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israël A. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 100.Mercurio F, Murray BW, Shevchenko A, Bennett BL, Young DB, Li JW, Pascual G, Motiwala A, Zhu H, Mann M, Manning AM. IkappaB kinase (IKK)-associated protein 1, a common component of the heterogeneous IKK complex. Molecular and cellular biology. 1999;19:1526–1538. doi: 10.1128/mcb.19.2.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bagnéris C, Ageichik AV, Cronin N, Wallace B, Collins M, Boshoff C, Waksman G, Barrett T. Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome. Molecular cell. 2008;30:620–631. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 102.Lo Y-C, Lin S-C, Rospigliosi CC, Conze DB, Wu C-J, Ashwell JD, Eliezer D, Wu H. Structural basis for recognition of diubiquitins by NEMO. Molecular cell. 2009;33:602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 104.Yoshikawa A, Sato Y, Yamashita M, Mimura H, Yamagata A, Fukai S. Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS letters. 2009;583:3317–3322. doi: 10.1016/j.febslet.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 105.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Véron M, Agou F, Israël A. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. The EMBO journal. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tegethoff S, Behlke J, Scheidereit C. Tetrameric oligomerization of IkappaB kinase gamma (IKKgamma) is obligatory for IKK complex activity and NF-kappaB activation. Molecular and cellular biology. 2003;23:2029–2041. doi: 10.1128/MCB.23.6.2029-2041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]