Abstract
Maternal immune activation during prenatal development, including treatment with the viral RNA mimic, polyriboinosinic-polyribocytidilic acid (poly IC), serves as a widely used animal model to induce behavioral deficits reminiscent of schizophrenia and related disease. Here, we report that massive cytokine activation after a single dose of poly IC in the prenatal period is associated with lasting working memory deficits in adult offspring. To explore whether dysregulated gene expression in cerebral cortex, contributes to cognitive dysfunction, we profiled the cortical transcriptome, and in addition, mapped the genome-wide distribution of trimethylated histone H3-lysine 4 (H3K4me3), an epigenetic mark sharply regulated at the 5′end of transcriptional units. However, deep sequencing-based H3K4me3 mapping and and mRNA profiling by microarray did not reveal significant alterations in mature cerebral cortex after poly IC exposure at embryonic days E17.5 or E12.5. At a small set of loci, H3K4me3 was sensitive to activation of cytokine signaling in primary cultures from fetal forebrain but adult cortex of saline- and poly IC-exposed mice did not show significant differences. A small subset of transcription start sites (TSS), including Disrupted-in-Schizophrenia 1 (Disc1), a schizophrenia risk gene often implicated in gene-environment interaction models, showed altered H3K4me3 after prenatal poly IC but none of these differences survived after correcting for multiple comparisons. We conclude that prenatal poly IC is associated with cognitive deficits later in life, but without robust alterations in epigenetic regulation of gene expression in the cerebral cortex.
Maternal infection during pregnancy with a variety of infectious agents—including virus such as influenza, rubella, herpes simplex virus and others, as well as bacteria and parasite such as toxoplasma have been shown to increase risk of neuropsychiatric disease in offspring, including schizophrenia and autism(Brown and Derkits, 2010). The magnitude of the problem is illustrated by the potentially large number of affected cases—up to one third according to some estimates (Brown and Patterson, 2011)—, highlighting the need for an in depth analyses of the underlying pathophysiology of disease and the prevention thereof. Consequently, several model systems have been developed in order to explore the consequences of immune activation in the pregnant rodent. These include infection with certain types of influenza virus, protozoa or bacterial lipopolysaccarides(Kneeland and Fatemi, 2012). While each of these models show important differences in pathophysiology and translability to the human condition, one of the most popular models involves the viral RNA mimic polyriboinosinic–polyribocytidilic acid (poly IC) which is thought, via actication of various proinflammatory cytokines, to negatively affect brain function and behavior in adult offspring(Boksa, 2010; Kneeland and Fatemi, 2012; Meyer et al., 2009). In particular, poly IC administered as a single dose during mid- or late gestation is widely used to induce behavioral alterations reminiscent of schizophrenia and related disease. These include deficits in cognition, social behaviors, sensorimotor gating and filtering (acoustic startle inhibition) and heightened sensitivity to psychotogenic drugs including dopaminergic agonists and NMDA receptor antagonists(Meyer et al., 2008). However, while there is broad consensus that the prenatal poly IC model is associated with abnormal behaviors later in life, surprisingly little is known about the underlying cellular mechanisms. For example, long-range synchronization of prefrontal-hippocampal assemblies (Dickerson et al., 2010) and plasticity of hippocampal synapses is abnormal, together with decreased expression of presynaptic proteins (Oh-Nishi et al., 2010). There is also conflicting evidence for loss of neurons (or lack thereof) after prenatal poly IC exposure(Oh-Nishi et al., 2010; Zuckerman et al., 2003). However, a molecular signature in the adult brain, indicative of poly IC exposure at a much earlier developmental stage, has yet to be determined. Remarkably, recent microarray studies on cerebral cortex and other brain regions prenatally exposed to polyIC or influenza virus identified subtle changes in only a small set of 40–60 genes (Fatemi et al., 2005; Smith et al., 2007). This would suggest that the behavioral defects after prenatal poly IC are not accompanied by a robust change in the gene expression program of the cerebral cortex. Given the important role of the prenatal poly IC paradigm in the context of maternal immune activation and offspring neuropsychiatric disease, the goal of the present study was to further explore gene expression functions in the cerebral cortex of mice exposed to maternal poly IC or saline injections during prenatal development, using microarrays for genome-wide transcript profiling. Because changes in chromatin structure and function are tightly linked to transcriptional regulation, the present study also employed chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq) to chart on a genome-wide scale the distribution of trimethylated histone H3-lysine 4 (H3K4me3). This chromatin mark is involved in epigenetic control of gene expression and sharply regulated at transcription start sites (TSS) of many active genes, and while it shows on a genome-wide scale a loose correlation with gene expression levels, it is associated at specific promoters not only with activation but also repression of transcription(Guenther et al., 2007; Kanhere et al., 2010; Shi et al., 2006). Because H3K4me3 is highly regulated during cortical development and involved in the neurobiology of schizophrenia, we speculated that epigenetic dysregulation after fetal poly IC exposure will include TSS-associated changes in cortical histone H3K4me3 methylation a multiple genes.
We report that exposure to poly IC at late gestation (embryonic day 17) leads to deficits in working memory of the adult animal. However, gene expression profiling in adult cerebral cortex revealed only minimal changes and furthermore, a very small subset of the annotated transcription start sites (TSS) from altogether less than 30 genes, including Disrupted-in-Schizophrenia 1 (Disc1), showed altered histone H4K4me3 methylation after prenatal poly IC (compared to saline exposed controls) but none of these differences survived correction for multiple comparisons. Taken together, the findings reported here suggest that the molecular pathology after prenatal poly IC exposure could be very subtle, and may include epigenetic dysregulation of Disc1 and other illness-relevant genes without changes in steady state levels of the corresponding RNAs.
Materials and Methods
Animals
C57BL/6J mice 10–12 weeks of age were obtained from Jackson Laboratories (Bar Harbor, ME) and housed in a temperature and humidity-controlled environment and maintained on a 12 hour light-dark cycle, with food and water provided ad libitum. Mice were allowed to acclimate for at least one week prior to breeding. Mice were mated overnight and the presence of a vaginal plug constituted embryonic day 0.5 (E0.5).
Poly(I:C) injections
On E12.5 or E17.5, pregnant females received an injection of 5 mg/kg poly(I:C) (potassium salt; Sigma, St. Louis, MO) freshly reconstituted in sterile 0.9% sodium chloride via the tail vein route under mild physical constraint. The injection volume was 5 ml/kg. Control females received an injection of saline solution.
Maternal Serum Cytokine ELISA
A subset of females (n = 3 per group) was sacrificed three hours following poly(I:C) or saline injection and blood was collected via cardiac puncture. Blood was allowed to clot at room temperature for 1 hour, centrifuged at 12,000 rpm at 4°C, and the serum aliquoted and stored at −20°C until the cytokine analysis was performed.
Levels of four cytokines (interleukin-6, IL-6; tumor necrosis factor alpha, TNFα; interleukin 1 beta, IL-1β; and interferon gamma, IFNγ) were measured in maternal serum using a SearchLight® Proteome Array (Aushon BioSystems; Billerica, MA), a quantitative multiplexed sandwich ELISA containing capture antibodies spotted on the bottom of a 96-well polystyrene microtiter plate. The bound proteins were detected by the subsequent addition of a biotinylated detection antibody, streptavidin-horseradish peroxidase (HRP), and chemiluminescent substrate; chemiluminescent signal was measured with the SearchLight Imaging System’s cooled charge-coupled device (CCD) camera.
Fetal Brain Cytokine ELISA
Another group of pregnant females were injected with poly(I:C) at E12.5 or E17.5, sacrificed 3 or 6 hours later, the fetal pups removed, and the brains dissected and frozen at −80°C. Fetal brain tissue was prepared for cytokine ELISA by dounce-homogenizing 3 whole brains per litter in 400 μl 0.1 M phosphate-buffered saline (PBS), pH 7.4, containing 24 μl/ml Complete Protease Inhibitor Cocktail (Roche Diagnostics; Indianapolis, IN) for 1 min on ice. Tissue homogenate was transferred to a 1.5 ml tube, centrifuged for 5 min at 13,000 rpm at 4°C, and the supernatant transferred to a new tube. IL-6 protein was measured as described above.
Cell culture work
Cells were prepared from forebrain of embryonic day 14.5 (E14.5) SASCO SD rat embryos (Charles River Laboratories; Wilmington, MA). Live cells were plated at 1.2 – 1.4 × 106 cells per 100 mm polystyrene Petri dish (VWR; West Chester, PA) coated with poly-L-lysine (Sigma; St. Louis, MO), 15 μg/ml poly-L-ornithine (Sigma), and 1 μg/ml fibronectin (R&D Systems; Minneapolis, MN), and treated daily with 1 μg/ml fibroblast growth factor 2 (FGF2) (R&D Systems). At 6 days in vitro (DIV), cells were passaged and stored in dimethyl sulfoxide (DMSO) in liquid nitrogen. For experiments intended for RNA and protein extraction, cells were removed from liquid nitrogen, re-suspended in media with D-MEM/F-12 (Invitrogen; San Diego, CA) with 2 μg/mL FGF2, plated on pre-coated polystyrene 6-well plates (BD Biosciences; Franklin Lakes, NJ) at 0.27 – 0.28 × 106 cells per well, and grown until approximately 95% confluent (for experiments with undifferentiated cultures). Alternatively, for experiments with differentiated cultures, comprised primarily neurons and a smaller proportion of astrocytes and undifferentiated cells(Huang and Akbarian, 2007; Huang et al., 2007), more dense plating was performed (0.69 – 1.0 × 106 cells per well) and after expansion for 1–3 days FGF2 was withdrawn and the cells allowed to differentiate for 3 days before treatment with IL-6 and IL-6R. For dose-response experiments, the six wells of each 6-well plate were treated with 0, 5, 10, 50, 100, or 150 ng/ml of recombinant human interleukin-6 (IL-6; Peprotech; Rocky Hill, NJ) and recombinant human soluble interleukin-6 receptor (IL-6R; Peprotech) for 12 hours. As an additional control, some plates were co-treated with the above doses of IL-6 and IL-6R plus 1.5 μg of IL-6 neutralizing antibody (#CBL2117; Millipore). Cells intended for RNA extraction were harvested as follows. Following IL-6 / IL-6R treatment, cells were rinsed with PBS, incubated in 1× Hank’s Balanced Salt Solution (HBSS; Invitrogen) for 5 min at 37°C, and detached by forcefully pipetting the HBSS over the cells with a fine-tipped pipette. The cell suspension was transferred to a 1.5 ml tube, centrifuged for 10 min at 8000 rpm, and the cell pellet frozen at −80°C. Plates containing cells for protein extraction were rinsed with PBS and stored at −80°C.
Behavior
Working memory was assessed in 10-week old offspring employing the T-maze. Mice tend to alternate between the two arms of the maze because they prefer to explore the more novel environment, a behavior known as spontaneous alternation, and this behavior is dependent upon an intact working memory [131]. The apparatus was constructed from white plexiglass and consisted of a home arm and a perpendicular arm. The mouse was placed in the home arm and allowed to choose between the left and right arms, at which point the opposite arm was blocked off, forcing the mouse to return to the beginning; the blocked arm was then opened and the mouse was then allowed to make another choice. Mice were given 15 choices for a total of 14 possible alternations; the same test was administered on two consecutive days. A total of 78 E12.5 offspring were tested (saline n = 51 [9 litters]; poly(I:C) n = 27 [8 litters]) and 62 E17.5 offspring were tested (saline n = 28 [10 litters]; poly(I:C) n = 34 [12 litters]).
Microarray
Cortex was dissected from 10 fresh frozen E17.5 brains (poly(I:C) = 5, saline = 5) and six E12.5 brains (poly(I:C) = 3, saline = 3) and the RNA extracted and processed for Affymetrix GeneChIP Mouse Gene 1.0 ST arrays. Total RNA was extracted employing the RNeasy Lipid Tissue Mini Kit (Qiagen; Valencia, CA). Following dissection, right cortex from each mouse was frozen at −80°C dounced in 1 mL QIAzol Lysis Reagent and RNA isolated according to the manufacturer’s instructions. RNA integrity (RIN) assessed with a 2100 Bioanalyzer (Agilent; Santa Clara, CA). Labeled, fragmented single-stranded cDNA was prepared and hybridized to Affymetrix GeneChip Mouse Gene ST arrays, then washed, and scanned with the Affymetrix Gene Chip Saccner 3000 7G according to the manufacturer’s instructions.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cortex as described above and, from different animals, selectively from the prefrontal cortex (Van De Werd et al., 2010). qRT-PCR was performed using the Quantifast SYBR Green RT-PCR kit according to manufacturer protocol (Qiagen; Valencia, CA). Primer sequences for 18S and hprt have been previously described (Jiang et al., 2010; Schleicher et al., 2005). Other primer sequences used are as follows: disc1 (5′-GGATGGCATACATTGCTCAGGGAA -3′ and 5′-TATCGTAATCGCCATCCTCGACCA-3′) nostrin1 (5′-TTAGCTACGCCAAAGGGCTTCAGA-3′). All reactions were carried out on an Applied Biosciences 7500 and analyzed using the Pfaffl method(Pfaffl, 2001).
Microarray data analysis
Quality of microarray data was assessed employing the Bioconductor package, arrayQualityMetrics(Kauffmann et al., 2009). Microarray data was then uploaded to MicroArray Computational Environment 2.0 (MACE), which employs Robust Multiarray Average (RMA) to preprocess raw oligonucleotide microarray data. The preprocessed data were stored as base 2 log transformed real signal numbers and used for fold-change calculations and statistical tests. Mean signal values and standard deviations were first computed for each gene across samples and the fold-change of expression of a gene between treatment groups was calculated by taking the ratio of these mean signal values. To determine differential expression of genes MACE internally conducts a Student t-test with the expression signal values of the two hybridizations for all genes in the set. GO enrichment analysis was performed by using topGO package from Bioconductor.
Chromatin immunoprecipitation (ChIP)
Nuclei were extracted from cortex of 6 twelve-week old E17.5 poly(I:C) offspring and 6 saline offspring; mice were derived from 6 independent litters with n = 3 males and 3 females per group. Whole cortex was dissected, frozen at −80°C, and nuclei extracted in hypotonic lysis buffer and purified under a sucrose gradient by ultracentrifugation. Nuclei were resuspended in 200 microliter buffer (10 mM Tris-base, 4 mM MgCl2, and 1 mM CaCl2; pH 7.5). Chromatin was digested with micrococcal nuclease (4 U/ml) for 5 min at 37°C and the reaction was stopped by the addition of 0.5 M EDTA, pH 8 (1:10). Subsequently, 350 microliter of Hypotonic Solution (0.2 mM EDTA, PMSF 1:1000, DTT 1:3000, and benzamidine 1:2000; pH 8.0) was added to each sample, and samples were pre-cleared by rotating for 30 minutes with 45 microliter of protein G agarose beads (Millipore; Billerica, MA) re-suspended in 1′ Frozen Storage Buffer (FSB; 20–29 mM Tris-base; 5 mM EDTA, and 50 mM NaCl; pH 7.5). Samples were then centrifuged for 5 min at 3000 rpm at 4°C and the supernatant transferred to a new tube; 500 ml of H3K4me3 antibody solution (4 microgram rabbit anti-H3K4me3 antibody [#07-473; Millipore] and 15 microgram H3K4me2 peptide [Abcam; Cambridge, MA] pre-incubated for 1 hour in 1×FSB with 0.2 mM EDTA, pH 8) and 50 ml of 10′ FSB were added and samples were rotated overnight at 4°C. The next day, samples were incubated with 90 ml protein G agarose beads re-suspended in 1×FSB for 1 hour at 4 °C and then centrifuged for 1 min at 3000 rpm and the supernatant discarded. Beads were rotated for 3 min with 1 ml Low Salt Buffer (20 mM Tris-base, 150 mM NaCl, 2 mM EDTA, 1% Triton-X100, and 0.1% SDS; pH 8.0), centrifuged for 1 min at 3000 rpm, and the supernatant discarded; beads were then sequentially washed with 1 ml each High Salt Buffer (20 mM Tris-base, 500 mM NaCl, 2 mM EDTA, 1% Triton-X100, and 0.1% SDS; pH 8.0), Lithium Chloride Buffer (250 mM LiCl, 10 mM Tris-base, 1 mM EDTA, 1% Igepal CA-630, and 1% deoxycholic acid; pH 8.0), and TE Buffer (10 mM Tris-base and 1 mM EDTA; pH 8.0). Chromatin was eluted from beads by rotating for 15 min in 250 ml Elution Buffer (100 mM NaHCO3 and 1% SDS) and then centrifuging for 1 min at 3000 rpm; the supernatant was transferred to a separate tube and set aside. An additional 250 microliter Elution Buffer was added to the beads and vortexed for 15 min; samples were centrifuged for 5 min at 13,000 rpm and the supernatant combined with the first 250 microliter. Proteins were digested by incubating samples with 2.5 microliter proteinase K (10 mg/ml), 10 microliter of 0.5 M EDTA (pH 8.0), and 25 microliter Tris-HCl (pH 6.5) for 3 hours at 52°C, and DNA extracted, precipitated and washed using standard procedures. DNA was re-suspended in 36 microliter 4 mM Tris-HCl (pH 8.0) and 2 microliters used for measuring DNA concentration; the remaining 34 microliter were used for preparation of libraries for deep sequencing.
Deep sequencing of chromatin immunoprecipitates
Immunoprecipitated DNA was processed for deep sequencing as follows. The ends of the DNA fragments were blunted using the End-it DNA Repair kit (Epicentre; Madison, WI), the reaction cleaned with the QIAquick Gel Extraction kit (Qiagen; Valencia, CA), and the fragments A-tailed by incubating with 0.2 mM dATP and 0.8 U/μl Exo-minus Klenow DNA polymerase in 1′ Klenow Buffer for 1 hour at room temperature. Subsequently, the reaction was again cleaned with the QIAquick Gel Extraction kit and the Genomic Adaptor Oligo Mix (lllumina; San Diego, CA) was ligated to fragments overnight at 16°C employing the Fast Link kit (Epicentre). The next day, fragments were PCR-amplified with Illumina single-read primers, the reaction cleaned with the QIAquick kit, and fragments around 250 base pairs gel-purified. The H3K4me3 ChIP libraries were deep sequenced by an Illumina Genome Analyzer (GA II).
ChIP-Seq Data analysis
Raw sequence reads were aligned to the rat reference genome (version rn4) using Bowtie (Langmead et al., 2009) to determine the total number of reads per library, as well as the number of uniquely-mappable, multi-mappable, and non-mappable reads. Only uniquely aligned reads with up to 1 mismatch were considered in this analysis and reads mapping to forward and reverse strands were pooled. The aligned sequence data was converted to ELAND format and, concurrently, normalized for sequencing depth by randomly sampling the same number of reads from each library (the lowest number of uniquely-mappable reads). Genomic regions significantly enriched for reads—called peaks—were detected with MACS (Zhang et al., 2008) and the percentage of proximal (< 2 kb from a transcription start site, or TSS), medial (2–10 kb from TSS), and distal (> 10 kb from TSS) peaks were determined. Data from normalized libraries was uploaded to the University of Southern California (UCSC) Genome Browser, which allows visualization of H3K4me3 levels in each sample genome-wide.
After confirming that each library was of good quality (i.e., possessing both a high percent of uniquely-mappable reads and proximal peaks), group differences in H3K4me3 levels within TSS (0 bp) and +500 bp were measured employing the DESeq Bioconductor package (Anders and Huber, 2010; Gentleman et al., 2004). Read counts per gene region were calculated using the Genominator package(Bullard et al., 2010) from Bioconductor 2.7, the count tables annotated, and differential H3K4me3 was inferred using the DESeq. Comparing treatment groups with DESeq offers an advantage over MACS, because it measures all reads within defined genomic intervals. Thus, it is possible to detect regions with H3K4me3 levels significantly altered by treatment that do not necessarily contain a statistically significant peak as called by MACS.
To evalulate significance of differences between the poly IC and saline group, the list of transcripts was filtered according read counts and genes were kept in the list if the read count for a gene was at least 10 in any replicate for either poly IC or saline. This left 8308 genes. The p values of differential expression for the filtered genes was adjusted with the multtest package (KS Pollard, HN Gilbert, Y Ge, S Taylor, S Dudoit (2011). multtest: Resampling-based multiple hypothesis testing. R package version 2.7.1. http://CRAN.R-project.org/package=multtest).
Results
Cytokine levels after poly IC injections
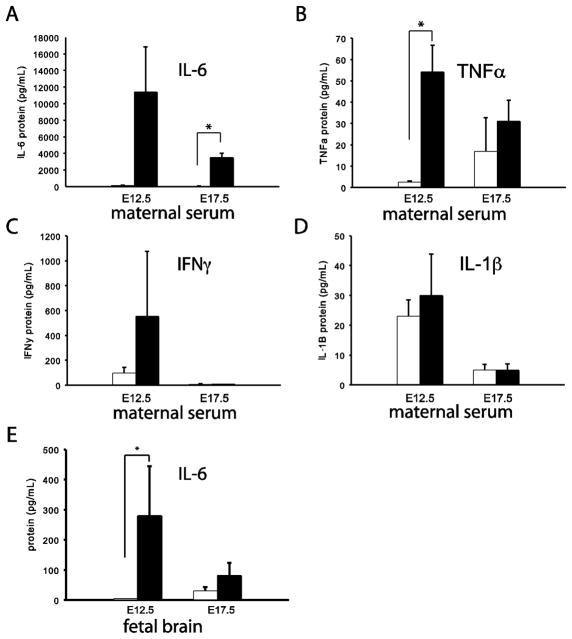
Because interleukin 6 (IL-6) from maternal tissues is now thought to be a key mediator for the effects of prenatal poly IC on the developing brain, including the behavioral abnormalities later in life(Hsiao and Patterson, 2011; Samuelsson et al., 2006; Smith et al., 2007), we measured IL-6 levels in maternal serum three hours after a tail vein injection of 5mg/kg poly IC. Indeed, there was a significant, 100–1000 fold increase in IL-6 levels in serum of poly IC treated E17.5 dams, compared to saline controls (Fig. 1A. This poly IC-mediated upregulation of IL-6 levels at E17.5 was specific, because other cytokines showed either a less robust (tumor necrosis factor alpha (TNFa) or no increase (interleukin 1 beta, IL-1b, interferon gamma, IFNg) in the maternal serum (Fig. 1B–D). Similar poly IC-mediated changes were observed in serum of E12.5 dams (Fig. 1A–D). There was also an increase in IL-6 levels in the poly IC-exposed fetus when sampled 3 hours post injection, albeit at least in brain these changes showed considerable variability from litter to litter (Fig. 1E). Following 6 hours post poly IC injection, fetal brain IL-6 injection were indistinguishable from baseline (data not shown).
Figure 1.
(A) IL-6, (B) TNF-a, (C) IFN-g and (D) IL-1b protein levels in maternal serum 3 hours following poly(I:C) (black bars) or saline (white bars) on embryonic day 12.5 (E12.5) and E17.5 (n = 3 mice/group) group). (E) IL-6 in fetal brain 3 hours after poly(I:C) injection (n=3–4 litters/group) * = P <0.05 (two tailed t-test).
Working memory in adult mice is altered after E17.5 poly IC
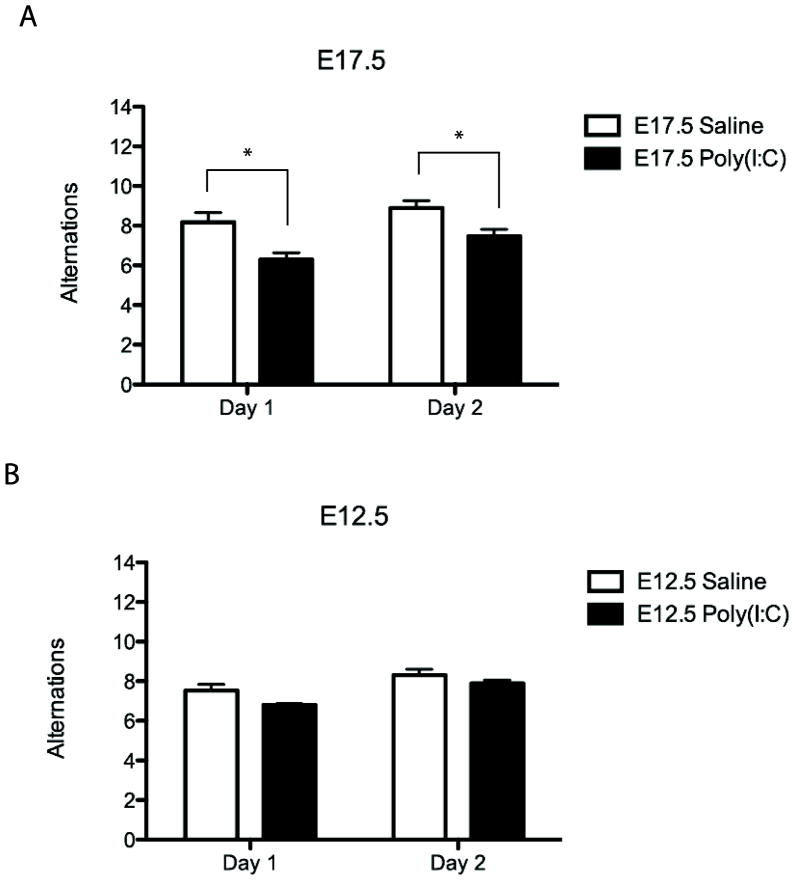
The profile of poly IC-induced behavioral changes in adult offspring differs depending on the timing of the insult, with prominent deficits in sensorimotor gating when administered during the first half of gestation while robust changes in working memory and cognition occur after exposure during at later prenatal stages(Meyer et al., 2008). Because in the present series, poly IC induced a robust and consistent increase in IL-6 serum levels of E17.5 dams, we predicted that their adult offspring will show deficits in alternating choices in the T-maze, a working memory paradigm. Indeed, this is what we observed. The E17.5 poly IC mice showed a significant decrease in the number of alternating entries into maze’s two arms, in comparison to E17.5 saline animals (Fig. 2A). These behavioral sequelae of prenatal poly IC were specific for the E17.5 timepoint, because E12.5 poly IC did not negatively impact working memory function (Fig. 2B), which is consistent other studies (Meyer et al., 2008).
Figure 2. T-maze performance and working memory is impaired in adult E17.5 poly IC mice.
(A) Adult mice from mothers injected with poly(I:C) on E17.5 demonstrate significantly fewer numbers of T-maze alternations compared to saline on both days 1 and 2 of testing (saline n = 28 [10 litters]; poly(I:C) n = 34 [12 litters]). (B) Adult mice from mothers injected with poly(I:C) on E12.5 do not demonstrate a significantly different number of alternation on day 1 or day 2 of T-maze (saline n = 51 [9 litters]; poly(I:C) n = 27 [8 litters]). * = P < 0.05 (two-tailed t-test).
No significant changes in the cortical transcriptome of poly IC exposed mice
A recent microarray study on adult cerebral cortex of animals exposed to poly IC at E12.5 revealed only a small number of transcripts (N=61) showing altered expression in comparison to saline controls(Smith et al., 2007). This is surprising given that the prenatal poly IC paradigm elicits robust behavioral changes in the adult. Therefore, we wanted to re-examine this issue by microarray-based gene expression profiling of adult cerebral cortex from E12.5 and E17.5 poly IC exposed animals and their saline-treated controls (N=5 animals/group). Remarkably, and in good agreement with the aforementioned earlier work (Smith et al., 2007), our studies revealed for both the E12.5 and E17.5 poly IC groups only subtle changes in expression. Most transcripts showed less than 2-fold changes from control and none of these differences were significant when corrected with the False Discovery Rate (FDR); Supplemental Figures S1A and S1B highlight for each poly IC group the top 50 differentially expressed genes compared to each of their control group, respectively. We conclude that poly IC – exposure during prenatal development has relatively mild, and mostly non-significant effects on the cortical transcriptome in adulthood.
Transcription-start site (TSS)-associated histone methylation markings (H3K4me3) are maintained in cerebral cortex of poly IC exposed mice
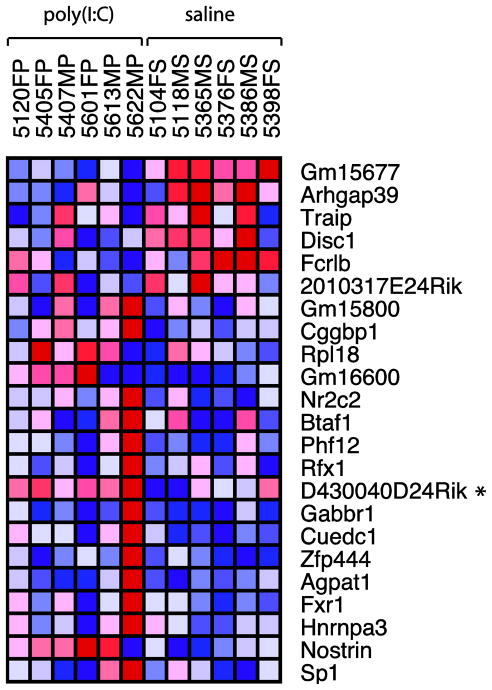
The overall non-significant differences in steady state RNA levels of saline and poly IC exposed mice would suggest that the cortical transcriptome is only minimally affected after prenatal poly IC. However, regulation of gene expression could be a highly dynamic process not fully captured by measurement of baseline RNA levels. Therefore, in order to further examine transcriptional regulation after poly IC and maternal immune activation, we conducted genome-wide mappings of tri-methyl-lysine 4 (H3K4me3). This epigenetic mark is linked to the transcriptional initiation complex and activated RNA polymerase II, thereby providing a docking site at the 5′end of genes for chromatin remodeling complexes that either facilitate or repress transcription(Shilatifard, 2008). To determine whether prenatal exposure to inflammation was capable of inducing long-lasting H3K4me3 landscapes in cortical chromatin, ChIP-Seq was performed on whole cerebral cortex of 12 week old E17.5 poly(I:C) and saline mice. We chose to focus on the E17.5 offspring, because these mice displayed significantly impaired working memory as assessed by T-maze. Alignment of raw reads to mouse genome revealed total library sizes ranging from 2.4 × 106 to 23.0 × 106 total reads (Fig. 3). A high percentage of total reads were uniquely mappable to mouse genome (87% for both saline and poly(I:C) mice), while smaller percentages were multi-mappable or non-mappable (8.5% and 4.0%, respectively) (Fig. 3A). Following normalization to the lowest number of uniquely-mappable reads (1.94 × 106), analysis with MACS determined similar numbers of total peaks in poly(I:C) and saline mice (14,907 and 14,847, respectively), a high percentage of which were proximal (within 2000bp) to annotated TSS (79% in both treatment groups), demonstrating the H3K4me3 immunoprecipitation worked efficiently across all experimental conditions(Fig. 3B). Comparison of H3K4me3 levels from nucleosomes positioned within 500 bp from the 36,902 annotated TSS in the mouse genome (mm9, ENSEMBL) in poly(I:C) cortex to saline revealed less than 25 loci that showed subtle fold-changes (Fig. 4). In total, 18 genes demonstrated increased H3K4me3 levels between 1.25 and 1.61-fold, while 5 genes were decreased between -1.26 and -2.47-fold. Interestingly, among these was the susceptibility gene disrupted in schizophrenia 1 (Disc1), which was decreased -1.34-fold in the poly(I:C) mice (Fig. 4,5), the GABA-B receptor gene Gabbr1, and fragile X gene Fxr1 as a negative regulator of protein synthesis and various molecules involved in gene expression control (Fig. 4 and Supplemental Table T1). However, none of the p-values for the H3K4me3 changes at the 24 genes survived correction for multiple comparisons using the False Discovery Rate (FDR). Furthermore, consistent with the negative findings from the aforementioned microarray studies, follow up qRT-PCR studies revealed that the H3K4me3 deficits at the TSS of Disc1 in the poly IC exposed cortex were not associated with a robust alteration in levels the corresponding gene transcripts (Disc1/Hprt ‘housekeeping’ gene transcript, mean ± S.D; poly IC group, 0.94 ± 0.22; saline 0.89 ± 0.12, N=4/group). Similarly, other genes showing subtle H3K4me3 differences between the poly IC and saline groups, including Nostrin (nitric oxide synthase traffic inducer) did not show significant alterations on the level of gene transcript (Nostrin/Hprt ‘housekeeping’ gene transcript, mean ± S.D; poly IC, 1.13 ± 0.18; saline 1.12 ± 0.26, N=4/group). Lack of significant differences between poly IC and saline groups were further confirmed when instead of Hprt the18S rRNA was used for normalization (data not shown). Similarly, when RNA quantification was done specifically on prefrontal cortex tissue, differences in Nostrin or Disc1 RNA levels between poly IC- and saline-exposed animals were subtle (<20%) and in part dependent on the choice of housekeeping gene (Supplemental Figure S2).
Figure 3. H3K4me3 ChIP-Seq of adult mouse cortex yields a high percentage of uniquely mappable reads and proximal peaks.
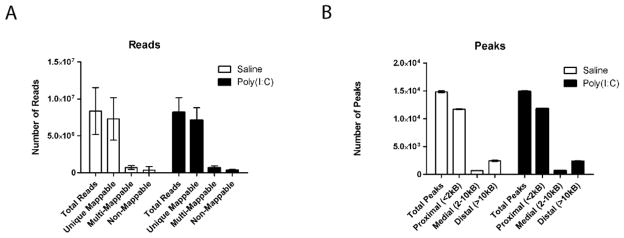
(A) A high percentage (87%) of reads from H3K4me3 libraries generated from cortex of adult mice are uniquely mappable to mouse genome (version mm9) for both saline and poly(I:C). A smaller percentage (9%) map to multiple locations and 4% are not mappable. (B) The majority of peaks called by MACS are positioned within 2Kb from an annotated transcription start site (TSS); additionally, 5% of peaks are positioned 2–10Kb from the next TSS, and 16–17% are further removed from a TSS. N = 6 mice/group.
Figure 4. Adult E17.5 poly IC offspring show subtle, non-significant H3K4me3 alterations in cortex.
The heatmap depicts the 36 genes for which DESeq detected significantly different numbers of tags between groups within 500bp from the transcription start site of the corresponding gene. None of the p-values survived correction with the false discovery rate. Fold-change compared to control group (N=6 animals from different litters, 3 male and 3 female/group as indicated) (range): 1.25 to 1.61-fold for 17 genes with increased H3K4me3 in poly IC animals;-1.26 to -2.47-fold for six genes with decreased H3K4me3 in poly IC animals.
Acute activation of IL-6 signaling alters H3K4me3 at many TSS in brain tissue
As mentioned above, poly IC administration to the pregnant dams in this study was associated with a large rise—approximately two or three orders of magnitude— in circulating IL-6 levels, a finding which is consistent with the critical role of this and related cytokines for defective neurodevelopment, and other medical sequelae in the affected offspring (Hsiao and Patterson, 2011; Pacheco-Lopez et al., 2011; Samuelsson et al., 2006; Smith et al., 2007). In human brain, IL-6R immunoreactivity has been reported in both undifferentiated neurons during earlier stages of pregnancy as well as in cortical pyramidal neurons at later stages of pregnancy and in the adult (Dame and Juul, 2000). Furthermore, the gp130 protein is expressed in neurons and glia of rat and human brain (Ringheim et al., 1998; Watanabe et al., 1996), which is of interest given that the IL-6 cytokine family signals via glycoprotein 130 (gp130); when IL-6 binds to its cognate receptor, IL-6R, then there is homodimerization with two gp130 subunits (Taga and Kishimoto, 1997). Therefore, we asked whether IL-6 was capable of modulating H3K4me3 on an acute time-scale in a model of the central nervous system. To this end, we exposed primary culture of rat embryonic forebrain to IL-6 and its cognate receptor, IL-6R, and measured levels of H3K4me3 genome-wide employing ChIP-Seq.
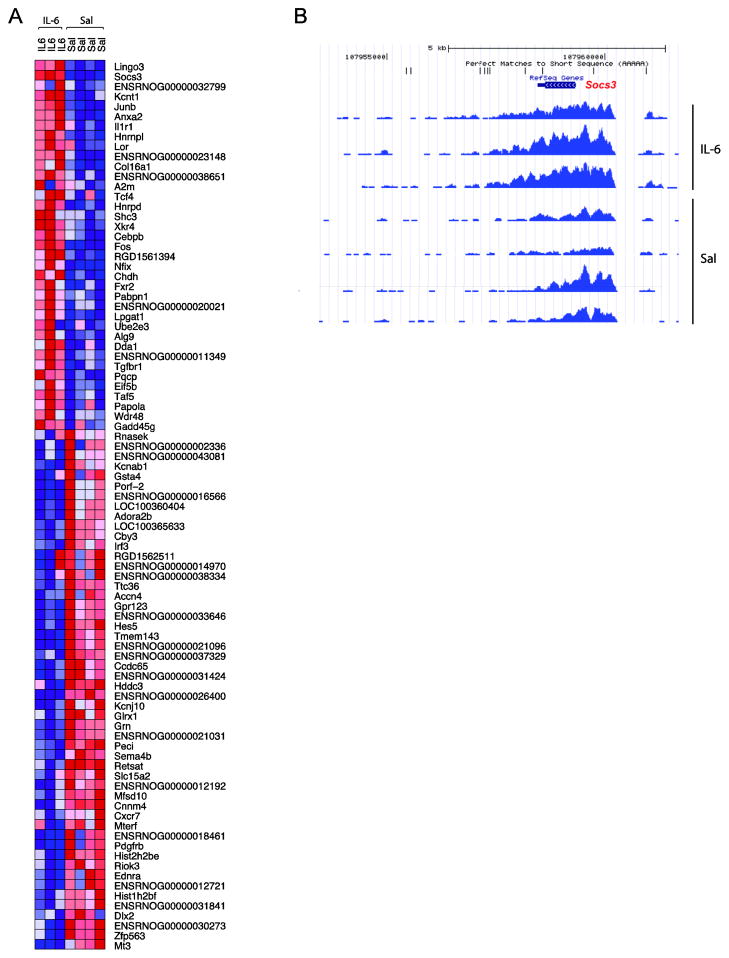
To establish the dose range of IL-6 and IL-6R that has an effect on transcription in the rat forebrain culture system, cells were treated with 5, 10, 50, 100, or 150 ng/mL IL-6 and IL-6R for 12 hours and the expression of a positive control gene—Gfap—measured with qRT-PCR and western blot. Transcript and protein levels demonstrated a concentration-dependent increase in both undifferentiated and differentiated cells compared to control. Fold-changes of Gfap mRNA in undifferentiated cells ranged from 6 to 245-fold and from 0.49 to 8-fold in differentiated cells. Co-treatment with a constant dose of IL-6 neutralizing antibody completely blocked the IL-6/IL-6R-induced increase of Gfap mRNA levels at lower doses and partially blocked the increase at the higher doses (Supplemental Figure S3). The 12 hr treatment had no discernable effect on cell viability at any dose. Based on these findings, the 100 ng/mL dose was selected for use in further experiments. Indeed, treatment for 12 hrs with IL-6 / IL-6R resulted in increased H3K4me3 levels at 33 genes ≥ 1.5-fold (149 genes ≥ 1.2-fold), while 38 genes had decreased H3K4me3 ≤-1.5-fold (60 genes ≤-1.2-fold) (Fig. 6). The heatmap in Fig. 6A depicts all genes with fold-changes of 1.5 or greater in either direction. The largest IL-6 induced H3K4me3 increase (3.2-fold; p = 1.94−20) was observed for the Suppressor of cytokine signaling 3 (Socs3) gene (Fig. 6B). There was no overlap with the list of genes showing subtle alterations in adult cortex of poly IC-exposed animals, which would suggest that at some genomic loci, the effects of maternal immune activation on histone methylation levels on the prenatal brain are transient but without evidence for persistent dysregulation in adulthood.
Figure 6. 12 hour IL-6 treatment alters H3K4me3 levels at many gene transcription start sites in rat forebrain culture.
(A) Following 12 hrs of IL-6 treatment, 33 genes demonstrated increased H3K4me3 levels ≥ 1.5-fold (149 ≥ 1.2-fold); 38 genes showed decreased levels ≤-1.5-fold (60 ≤-1.2-fold). The heatmap illustrates all IL-6 induced histone methylation changes ≥ 1.5-fold (red = increase relative to saline; blue = decrease). (B) The H3K4me3 profile from the UCSC genome browser for the Socs3 gene, which demonstrated the most robust and significant increase in methylation following 12 hrs of IL-6 treatment (fold-change = 3.2; p = 1.94−20). Note the atypical pattern of H3K4me3, which encompasses the entire Socs3 gene and surrounding region.
DISCUSSION
The goal of the present study was to explore potential changes in transcriptomes and epigenomes of the adult cerebral cortex after exposure to maternal immune activation via the viral nucleic acid mimic, poly IC. Consistent with earlier reports, we observed that acute poly IC administered at E12.5 and E17.5 elicits (1) robust changes in serum cytokine levels in at least some of the dams and (2), for litters exposed at E17.5, significant deficits in working memory in adult animal(Kneeland and Fatemi, 2012). However, in the adult cerebral cortex, the molecular signature of prenatal poly IC was surprisingly subtle, because no significant differences from controls were observed for the transcriptome and the H3K4me3 epigenome. Likewise, changes in the H3K4me3 were overall subtle, with only 24 annotated transcriptions start sites showing a significant difference between poly IC and saline exposed animals prior to correction for multiple comparison, and none survived the correction. Like, Smith and colleagues (Smith et al., 2007) recently reported that the transcriptome from rostral cerebral cortex (adult) exposed to E12.5 poly IC shows significant changes for fewer than 60 gene transcripts. Taken together, these findings clearly demonstrate that prenatal poly IC results in reproducible behavioral deficits in the adult, while the molecular pathology, at least in the cerebral cortex, remains surprisingly subtle. However, this conclusion may not be too surprising. Indeed, there is ample evidence that even severe cortical dysfunction in the context of neurological or neuropsychiatric disease is not necessarily associated with robust changes in gene expression. To mention just one additional example, both inhibitory (Chao et al., 2010) and excitatory (Dani et al., 2005) neurotransmission are severely compromised in the cortex of methyl-CpG-binding protein 2 (Mecp2, Rett Syndrome, OMIM 312750) deficient mice yet changes on the transcriptome level are minimal and with the exception of one or two genes, without significance (Tudor et al., 2002). It remains to be determined whether prenatal poly IC elicits a stronger molecular alterations in brain regions other than cerebral cortex, with the hippocampus as one of the candidate regions for poly IC-related molecular and structural pathology(Fatemi et al., 2009).
The findings reported here do not imply that cerebral cortex operates properly after prenatal poly IC. Indeed, there is ample evidence that adult cerebral cortex is functionally impaired. For example, there is widespread disruption of proper neuronal synchronization in prefrontal-hippocampal networks (Dickerson et al., 2010), and deficits in prefrontal dopamine D1 receptor density and inhibitory (GABAergic) interneuron neurochemistry (Meyer et al., 2008), and hippocampal loss of presynaptic marker proteins in (Oh-Nishi et al., 2010). These changes were not always associated with alterations in neuronal numbers (Oh-Nishi et al., 2010). In the absence of robust changes in gene expression, it remains to be determined whether post-transcriptional and -translational mechanisms controlling of protein expression and function contribute to the behavioral and neurological sequels of prenatal poly IC exposure in adult offspring.
However, litter-to-litter variability in the maternal immune response to poly IC and within-litter differences, including the levels of activated cytokines and immune complexes to which each fetal brain was exposed (Fig. 1E), could also have contributed to the largely negative findings of the present study. It is now thought that poly IC-mediated upregulation of IL-6 expression in the placenta is a key event in pathophysiological cascade leading to altered neurodevelopment in offspring(Hsiao and Patterson, 2011). Therefore it remains possible that numerous factors in the fetal/maternal unit, including disruption of blood circulation and altered endocrine status (Fatemi et al., 2011; Hsiao and Patterson, 2011) could intersect with, or modulate poly IC-mediated cytokine signaling in the fetus. However, presently no molecular or genetic tools are available to determine retrospectively in adult brain tissue the extent or intensity of MIA during prenatal development, and thus the extent of potential brain-to-brain specific variabilities in MIA exposure remains unresolved. To this end, the IL-6 sensitive primary culture from embryonic rat forebrain of the present study provide an important alternative to the whole animal/in vivo system. The fact the strongest histone methylation change after IL-6 occured at Socs3, encoding a protein with a key role in suppressing cytokine pathways after their excessive activation(Babon and Nicola, 2012; Starr et al., 1997; Yang et al., 2004), speaks to the importance of epigenetic mechanisms in the fine-tuning of the neuronal and glial response to activated immune or inflammatory systems.
Finally, it is important to mention that the current study was limited to profiling of steady state levels and distribution pattern of RNA and H3K4me3. It remains possible that the molecular signatures of poly IC exposure in otherwise unchallenged offspring are grossly normal at baseline (as reported in the present study), yet become abnormal after exposure to stress, and/or in conjunction with DNA sequence variants and mutations of susceptibility genes. Indeed, prenatal poly IC exposure triggers profound behavioral and molecular alterations in mice expressing a dominant negative form of Disc1 (Abazyan et al., 2010). It is likely that in the nearby future, other examples for such poly IC-mediated ‘unmasking’ of genetic susceptibility factors will be described (Meyer and Feldon, 2011). Conversely, the epigenetic (H3K4me3) alterations at Disc1 and other loci, as observed in the poly IC exposed wildtype cortex of the present study, could confer susceptibility to an abnormal stress response or subtle deficits in cognition (including working memory) without affecting baseline levels of the corresponding gene transcripts.
Supplementary Material
Figure 5. Histone methylation peaks at the Disc1 locus.
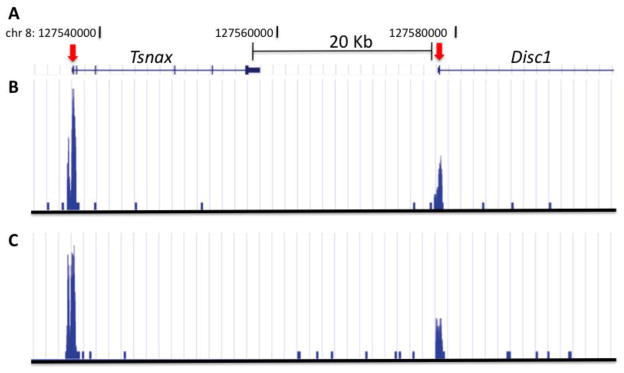
(A) University of California Santa Cruz (UCSC) genome browser track for approximately 60 Kb region on chromosome 8, surrounding Disc1 TSS as indicated. (B,C) Distribution of sequence tags from anti-H3Kme3 ChIP-seq from cerebral cortex of (B) saline and (C) poly IC exposed animal. Notice sharp peaks at TSS of Disc1 and of adjacent Tsnax gene.
Acknowledgments
The authors thank members of the Akbarian laboratory for helpful comments and technical advice, and Maria Zapp and Ellen Kittler and the UMMS Deep Sequencing Core staff. Supported by grants from the NIH, IMHRO and BBRF(NARSAD) and Autism Speaks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abazyan B, Nomura J, Kannan G, Ishizuka K, Tamashiro KL, Nucifora F, Pogorelov V, Ladenheim B, Yang C, Krasnova IN, et al. Prenatal interaction of mutant DISC1 and immune activation produces adult psychopathology. Biol Psychiatry. 2010;68:1172–1181. doi: 10.1016/j.biopsych.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babon JJ, Nicola NA. The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors. 2012 doi: 10.3109/08977194.2012.687375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boksa P. Effects of prenatal infection on brain development and behavior: a review of findings from animal models. Brain Behav Immun. 2010;24:881–897. doi: 10.1016/j.bbi.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Brown AS, Derkits EJ. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am J Psychiatry. 2010;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Patterson PH. Maternal infection and schizophrenia: implications for prevention. Schizophr Bull. 2011;37:284–290. doi: 10.1093/schbul/sbq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame JB, Juul SE. The distribution of receptors for the pro-inflammatory cytokines interleukin (IL)-6 and IL-8 in the developing human fetus. Early human development. 2000;58:25–39. doi: 10.1016/s0378-3782(00)00064-5. [DOI] [PubMed] [Google Scholar]
- Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson DD, Wolff AR, Bilkey DK. Abnormal long-range neural synchrony in a maternal immune activation animal model of schizophrenia. J Neurosci. 2010;30:12424–12431. doi: 10.1523/JNEUROSCI.3046-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Reutiman TJ, Huang H, Oishi K, Mori S. Prenatal viral infection of mice at E16 causes changes in gene expression in hippocampi of the offspring. Eur Neuropsychopharmacol. 2009;19:648–653. doi: 10.1016/j.euroneuro.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Rooney RJ, Mori S, Kornfield TE, Reutiman TJ, Kneeland RE, Liesch SB, Hua K, Hsu J, et al. The viral theory of schizophrenia revisited: Abnormal placental gene expression and structural changes with lack of evidence for H1N1 viral presence in placentae of infected mice or brains of exposed offspring. Neuropharmacology. 2011 doi: 10.1016/j.neuropharm.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Pearce DA, Brooks AI, Sidwell RW. Prenatal viral infection in mouse causes differential expression of genes in brains of mouse progeny: a potential animal model for schizophrenia and autism. Synapse. 2005;57:91–99. doi: 10.1002/syn.20162. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, Patterson PH. Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav Immun. 2011;25:604–615. doi: 10.1016/j.bbi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, Akbarian S. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, Lin CL, Straubhaar J, Martin G, Akbarian S. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneeland RE, Fatemi SH. Viral infection, inflammation and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2012 doi: 10.1016/j.pnpbp.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Feldon J. To poly(I:C) or not to poly(I:C): Advancing preclinical schizophrenia research through the use of prenatal immune activation models. Neuropharmacology. 2011 doi: 10.1016/j.neuropharm.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Meyer U, Feldon J, Fatemi SH. In-vivo rodent models for the experimental investigation of prenatal immune activation effects in neurodevelopmental brain disorders. Neurosci Biobehav Rev. 2009;33:1061–1079. doi: 10.1016/j.neubiorev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Meyer U, Nyffeler M, Yee BK, Knuesel I, Feldon J. Adult brain and behavioral pathological markers of prenatal immune challenge during early/middle and late fetal development in mice. Brain Behav Immun. 2008;22:469–486. doi: 10.1016/j.bbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Oh-Nishi A, Obayashi S, Sugihara I, Minamimoto T, Suhara T. Maternal immune activation by polyriboinosinic-polyribocytidilic acid injection produces synaptic dysfunction but not neuronal loss in the hippocampus of juvenile rat offspring. Brain Res. 2010;1363:170–179. doi: 10.1016/j.brainres.2010.09.054. [DOI] [PubMed] [Google Scholar]
- Pacheco-Lopez G, Giovanoli S, Langhans W, Meyer U. Priming of Metabolic Dysfunctions by Prenatal Immune Activation in Mice: Relevance to Schizophrenia. Schizophr Bull. 2011 doi: 10.1093/schbul/sbr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringheim GE, Szczepanik AM, Petko W, Burgher KL, Zhu SZ, Chao CC. Enhancement of beta-amyloid precursor protein transcription and expression by the soluble interleukin-6 receptor/interleukin-6 complex. Brain Res Mol Brain Res. 1998;55:35–44. doi: 10.1016/s0169-328x(97)00356-2. [DOI] [PubMed] [Google Scholar]
- Samuelsson AM, Jennische E, Hansson HA, Holmang A. Prenatal exposure to interleukin-6 results in inflammatory neurodegeneration in hippocampus with NMDA/GABA(A) dysregulation and impaired spatial learning. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1345–1356. doi: 10.1152/ajpregu.00268.2005. [DOI] [PubMed] [Google Scholar]
- Schleicher U, Hesse A, Bogdan C. Minute numbers of contaminant CD8+ T cells or CD11b+CD11c+ NK cells are the source of IFN-gamma in IL-12/IL-18-stimulated mouse macrophage populations. Blood. 2005;105:1319–1328. doi: 10.1182/blood-2004-05-1749. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007;27:10695–10702. doi: 10.1523/JNEUROSCI.2178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annual review of immunology. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Werd HJ, Rajkowska G, Evers P, Uylings HB. Cytoarchitectonic and chemoarchitectonic characterization of the prefrontal cortical areas in the mouse. Brain Struct Funct. 2010;214:339–353. doi: 10.1007/s00429-010-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D, Yoshimura R, Khalil M, Yoshida K, Kishimoto T, Taga T, Kiyama H. Characteristic localization of gp130 (the signal-transducing receptor component used in common for IL-6/IL-11/CNTF/LIF/OSM) in the rat brain. The European journal of neuroscience. 1996;8:1630–1640. doi: 10.1111/j.1460-9568.1996.tb01307.x. [DOI] [PubMed] [Google Scholar]
- Yang XP, Albrecht U, Zakowski V, Sobota RM, Haussinger D, Heinrich PC, Ludwig S, Bode JG, Schaper F. Dual function of interleukin-1beta for the regulation of interleukin-6-induced suppressor of cytokine signaling 3 expression. J Biol Chem. 2004;279:45279–45289. doi: 10.1074/jbc.M313072200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman L, Rehavi M, Nachman R, Weiner I. Immune activation during pregnancy in rats leads to a postpubertal emergence of disrupted latent inhibition, dopaminergic hyperfunction, and altered limbic morphology in the offspring: a novel neurodevelopmental model of schizophrenia. Neuropsychopharmacology. 2003;28:1778–1789. doi: 10.1038/sj.npp.1300248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.