Abstract
Purpose
USP-11, a member of the ubiquitin-specific protease family, has emerged as an essential regulator of double-strand break (DSB) repair. Few studies have shown that silencing USP-11 led to hypersensitivity to poly(ADP-ribose) polymerase (PARP) inhibition, ionizing radiation (IR), and DNA-damaging agents. We sought to examine the predictive and prognostic relevance of USP-11 in patients treated with neoadjuvant systemic therapy (NST) for breast cancer.
Methods
56 women who were treated with NST for breast cancer between 1999 and 2004 were included in the study. The Kaplan-Meier product-limit method was used to estimate disease-free survival (DFS) and overall survival (OS) rates. Logistic regression models were fit to determine the associations between USP-11 status, pCR and survival.
Results
Sixteen (29%) patients had high USP-11 expressing tumors, and 40 (71%) patients had low USP-11 expressing tumors. No significant differences were observed in pCR rates with respect to USP-11 status. At a median follow-up of 7.4 years, 33 patients (59%) experienced a disease recurrence or death. Patients with high USP-11 expressing tumors had a higher risk of recurrence (Odds ratio [OR] = 3.87; 95% CI:1.51 to 9.93; P= 0.005) and death (OR = 6.03; 95% CI:2.00 to 18.17; P= 0.001) than those with low USP-11 expressing tumors. Patients who did not achieve a pCR had an increased risk of recurrence (OR = 5.16; 95% CI:1.16 to 23.07; P= 0.03).
Conclusions
Our data indicates that USP-11 is not a predictor of a pCR after anthracycline-taxane containing NST for breast cancer. Low USP-11 expression was independently correlated with better survival outcomes.
INTRODUCTION
Recent evidence implicates a substantial commitment of the cellular DNA damage response (DDR) machinery to the synthesis, recognition, and hydrolysis of ubiquitin chains at DNA damage sites. Ubiquitin, the highly conserved 76-amino acid protein, once linked to the target protein by a cascade of enzymes including ubiquitin-activating enzyme (E1), ubiquitin-conjugated enzyme (E2), and ubiquitin ligase (E3), can initiate a number of post-translational cellular processes [1]. The hydrolysis of the isopeptide bond connecting ubiquitin to a substrate protein is performed by deubiquitinating enzymes (DUBs), and this renders target proteins able to escape proteasome degradation and thus resume their original designated functions [2]. Two families of DUBs have been identified, all of which exhibit specificity for one type of ubiquitin chain isoform [3–5]. USP-11, located on the X chromosome [6], is a member of USP subclass of the ubiquitin-specific protease (UBP) family [7]. Proteins in the UBP family differ in length but possess conserved domains such as Cys box and His box, responsible for the catalytic activity of UBPs [1]. USP-11 is primarily localized in the nucleus of non-dividing cells, and was found throughout mitotic cells [8].
USP-11 deubiquitylates specific substrates which play key roles in correct microtubule nucleation (RanBPM) [8]; antigen-presenting cell function (RELB) [9]; inflammation, immunity, cell proliferation and apoptosis (TNFa-induced NK-kB, and IKKa/p53 signaling pathway) [10, 11]; and E7-modulated cell growth and transformation (HPV-16E7) [12]. Further, USP-11 has emerged as an essential regulator of double-strand break (DSB) repair through its interaction with BRCA2 [13], and by recruitment of a subset of DSB repair proteins including 53BP1 and RAD51 to repair foci [14]. More importantly, few studies have shown that silencing USP-11 led to spontaneous DDR activation in otherwise undamaged cells and hypersensitivity to poly(ADP-ribose) polymerase (PARP) inhibition, ionizing radiation (IR), and genotoxic agents including bleomycin, cisplatin, and mitomycin C which induce DNA damage via homologous recombination (HR)-dependent pathways [13, 14]. However, there is no clinical data on the effectiveness of neoadjuvant systemic chemotherapy (NST) in breast cancers with USP-11 low and high expression, and also if USP-11 expression is predictive of improved long-term survival in sporadic breast cancers. Thus, we postulated that tumors with low USP-11 expression would be defective in HR repair and therefore more sensitive to NST. We conducted this retrospective analysis to determine the pathologic complete response rates (pCR) following NST in breast cancer tumors with high vs. low USP-11 expression. Our secondary endpoints included disease-free survival (DFS) and overall survival (OS).
MATERIALS AND METHODS
Patient Population
The prospectively maintained Breast Cancer Management System research database of The University of Texas M.D. Anderson Cancer Center (MDACC) identified 56 consecutive women treated between 1999 and 2004, and who had prechemotherapy diagnostics core biopsy tissue available. Initial clinical stage of all patients were revised and based on the seventh edition of the American Joint Committee on Cancer (AJCC) staging criteria [15]. This study was approved by Institutional Review Board at MDACC. Patient clinical data, tumor characteristics, lymph node status, chemotherapy type, and survival data were retrieved from the clinical database and correlated with USP-11 status, pCR and survival outcomes.
Pathologic Assessment
All pathologic specimens were reviewed by designated breast pathologists at MDACC. Histologic type and tumor grade were defined according to the WHO classification system [16] and the modified Black’s nuclear grading system [17], respectively. Immunohistochemical (IHC) analysis was used to determine estrogen-receptor (ER) and progesterone-receptor (PR) status. Tumors were considered ER- or PR-positive if ≥10% of cells showed nuclear staining. HER2-positivity was defined as 3+ receptor overexpression by IHC staining and/or as gene amplification (gene copy ratio of >2.0 of HER2:CEP17) found on fluorescence in situ hybridization. pCR was defined as the absence of any invasive disease in the breast, and the absence of micrometastasis or macrometastasis in the ipsilateral axillary lymph nodes.
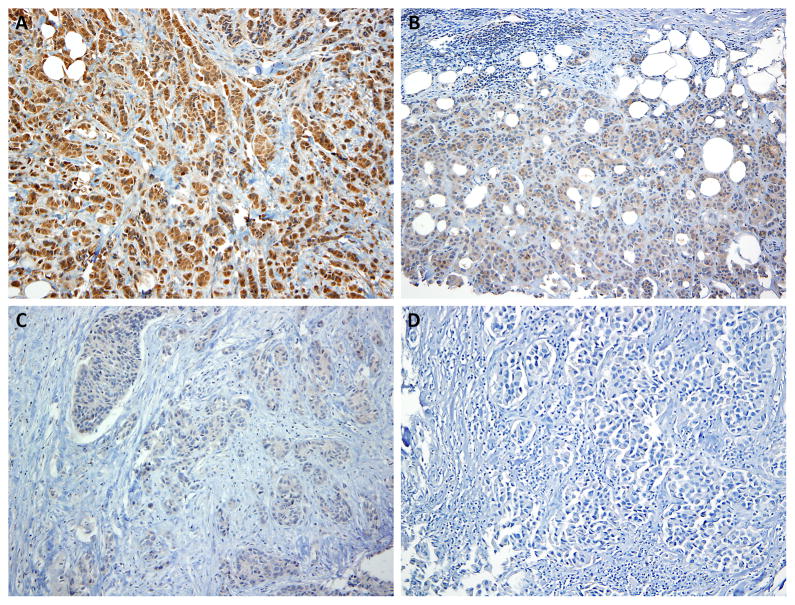
Expression of USP-11 was analyzed on pre-chemotherapy diagnostic biopsy samples using anti-USP11 (C-term) polyclonal antibody [18] by IHC. The degree of USP-11 overexpression on IHC determined how patients were grouped. Percentage of overall cytoplasmic expression in tumor cells was recorded (no nuclear staining was detected) (Figure 1). Similarly, the intensity was scored as 0 (no expression), 1 (weak expression), 2 (moderate expression), and 3 (strong expression). Percentage (P) and intensity of cytoplasmic expression were multiplied to generate a numerical score, and the resulting score was used to dichotomize the tumors. Martingale residual suggested that a cut-off of 250 is appropriate for OS. Low USP-11 expression was defined as <250 (n=40) and high USP-11 expression was defined as ≥ 250 (n=16).
Figure 1.
Representative immunostaining of USP-11 in breast cancer tumor sections. (A) strong staining, (B) moderate staining, (C) weak staining, and (D) no staining.
Treatment
All patients were treated with anthracycline plus paclitaxel based NST as per standard clinical protocols used in the institution. Thirty-six (64%) patients received sequential weekly paclitaxel 80 mg/m2 in 12 courses followed by 4–6 courses of FAC (5-FU 500 mg/m2, doxorubicin 50 mg/m2, and cyclophosphamide 500 mg/m2); 19 (34%) patients received sequential weekly paclitaxel 80 mg/m2 in 12 courses followed by 4–6 courses of FEC (5-FU 500 mg/m2, epirubicin 75 mg/m2, and cyclophosphamide 500 mg/m2). One patient received only weekly paclitaxel x 12. Of 15 patients that had HER2-positive breast cancer, 6 (40%) also received IV trastuzumab during NST.
After completion of NST, all patients underwent definitive breast surgery and axillary lymph node dissection or sentinel node dissection. Surgical intervention was breast conserving surgery for 34% of patients (n=19) and mastectomy for 66% of patients (n=37). When appropriate, locoregional radiotherapy and hormone therapy were given after surgery.
Statistical Analysis and Outcome Measures
The demographic and clinical characteristics were summarized and compared between the 2 groups, defined by USP-11 status (low vs. high expression) with the chi-square test for categorical variables and Kruskal-Wallis test or Wilcoxon rank sum test for continuous variables. Chi-square test or Fisher’s exact test was used to identify the significant factors predictive of a pCR.
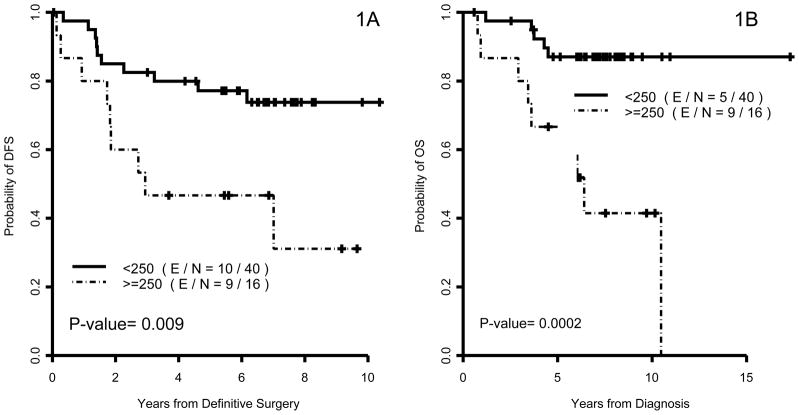
DFS was calculated from the surgery date until the first date of documented disease recurrence or death or the date of to first metastasis or death last follow-up. OS was calculated from the time of initial diagnosis until the date of death from any cause or last follow-up. Martingale residual was calculated using Cox model to define an appropriate cut-off point for dividing patients using USP-11 expression. OS and DFS were estimated using the Kaplan-Meier method and the comparisons between or among patients’ characteristics groups were assessed using log-rank test (Figure 2). Univariate Cox proportional hazard models were applied to assess the effect of covariates on survival outcomes. P-values ≤ 0.05 were considered statistically significant; all tests were two-sided. All computations were carried out in SAS 9.2 and S-plus 8.0.
Figure 2.
Kaplan–Meier estimates of disease-free survival (DFS) and overall survival (OS) by USP-11 status for all patients (1A and 1B).
RESULTS
Patient demographics and pretreatment clinical characteristics are summarized in Table 1. Patients who presented with clinical tumor stage I/II and had negative lymphovascular invasion (LVI) were more likely to have low USP-11 tumor cell expression. USP-11 expression was not linked to any other clinical-phenotypic characteristics of breast cancer tumors.
Table 1.
Patient demographics and baseline disease characteristics by USP-11 groups
| USP11 Low (N=40) | USP11 High (N=16) | P | ||
|---|---|---|---|---|
| Age | ||||
| ≤ 50 | 18 (45%) | 5 (31.3%) | 0.38 | |
| >50 | 22 (55%) | 11 (68.8%) | ||
|
| ||||
| Race | ||||
| White | 32 (80%) | 10 (62.5%) | 0.82 | |
| Black | 2 (5%) | 2 (12.5%) | ||
| Hispanic | 5 (12.5%) | 2 (12.5%) | ||
| Other | 1 (2.5%) | 2 (12.5%) | ||
|
| ||||
| Clinical tumor stage | ||||
| T1 | 7 (17.5%) | 3 (18.8%) | 0.04 | |
| T2 | 20 (50%) | 4 (25%) | ||
| T3 | 9 (22.8%) | 2 (12.5%) | ||
| T4 | 4 (10%) | 7 (43.8%) | ||
|
| ||||
| Clinical nodal stage | ||||
| N0 | 14 (35%) | 6 (37.5%) | 0.86 | |
| N1-3 | 26 (65%) | 10 (62.5%) | ||
|
| ||||
| Clinical stage | ||||
| I | 2 (5%) | 1 (6.3%) | 0.29 | |
| II | 24 (60%) | 6 (37.5%) | ||
| III | 13 (32.5%) | 9 (56.5%) | ||
| IV | 1 (2.5%) | - | ||
|
| ||||
| ER status | ||||
| Negative | 20 (50%) | 7 (43.8%) | 0.67 | |
| Positive | 20 (50%) | 9 (56.3%) | ||
|
| ||||
| PR status | ||||
| Negative | 23 (57.5%) | 8 (50%) | 0.61 | |
| Positive | 17 (42.5%) | 13 (50%) | ||
|
| ||||
| HER2 status | ||||
| Negative | 28 (70%) | 13 (81.3%) | 0.51 | |
| Positive | 12 (30%) | 3 (18.8%) | ||
|
| ||||
| Triple negative status | ||||
| No | 32 (80%) | 12 (75%) | 0.72 | |
| Yes | 8 (20%) | 4 (25%) | ||
|
| ||||
| Nuclear grade | ||||
| I–II | 9 (22.5%) | 6 (37.5%) | 0.25 | |
| III | 31 (77.5%) | 10 (62.5%) | ||
|
| ||||
| Lymphatic invasion | ||||
| Negative | 31 (77.5%) | 8 (50%) | 0.04 | |
| Positive | 9 (22.5%) | 8 (50%) | ||
|
| ||||
| Vascular invasion | ||||
| Negative | 55 (96.5%) | 8 (50%) | 0.01 | |
| Positive | 2 (3.5%) | 8 (50%) | ||
|
| ||||
| Skin involvement | ||||
| Negative | 38 (95%) | 15 (93.8%) | 1.00 | |
| Positive | 2 (5%) | 1 (6.3%) | ||
ER= estrogen receptor; PR=progesterone receptor; HER2 = human epidermal growth factor receptor-2
Response to NST
Overall, pCR was achieved by 15 patients (26%) - a response rate that is consistent with our previous experience in a larger randomized study using similar NST [19]. The pCR rates in various subgroups in relation to the various clinical and biological factors are shown in Table 2. Confirming the previous studies [20] [21–23], non-Caucasians, patients whose tumors were ER/PR-negative, triple-negative (TN), or with negative LVI appeared to be more likely to have a pCR. USP-11 status did not significantly influence the pCR rate. Eleven (27.5%) patients achieved a pCR in the low USP-11 group compared to 4 (25%) patients in the high USP-11 group (P=1.00).
Table 2.
Pathologic complete response rates by clinical characteristics
| pCR No (N=41) | pCR Yes (N=15) | P | ||
|---|---|---|---|---|
| Age | ||||
| ≤ 50 | 16 (69.6%) | 7 (31.3%) | 0.60 | |
| >50 | 25 (75.8%) | 8 (68.8%) | ||
|
| ||||
| Race | ||||
| White | 34 (81%) | 8 (19%) | 0.01 | |
| Black | 2 (50%) | 2 (50%) | ||
| Hispanic | 2 (28.6%) | 5 (71.4%) | ||
| Other | 3 (100%) | - | ||
|
| ||||
| Clinical tumor stage | ||||
| T1 | 7 (70%) | 3 (30%) | 0.56 | |
| T2 | 16 (66.7%) | 8 (33.3%) | ||
| T3 | 8 (72.7%) | 3 (27.3%) | ||
| T4 | 10 (90.9%) | 1 (9.1%) | ||
|
| ||||
| Clinical nodal stage | ||||
| N0 | 13 (65%) | 7 (35%) | 0.30 | |
| N1-3 | 28 (77.8%) | 8 (22.2%) | ||
|
| ||||
| Clinical stage | ||||
| I | 2 (66.7%) | 1 (33.3%) | 0.19 | |
| II | 19 (47.5%) | 11 (52.5%) | ||
| III | 20 (91%) | 2 (9%) | ||
| IV | - | 1 (100%) | ||
|
| ||||
| ER status | ||||
| Negative | 15 (55.6%) | 12 (44.4%) | 0.006 | |
| Positive | 26 (89.7%) | 3 (10.3%) | ||
|
| ||||
| PR status | ||||
| Negative | 19 (61.3%) | 12 (38.7%) | 0.03 | |
| Positive | 22 (88%) | 3 (12%) | ||
|
| ||||
| HER2 status | ||||
| Negative | 30 (73.2%) | 11 (26.8%) | 1.00 | |
| Positive | 11 (73.3%) | 4 (26.7%) | ||
|
| ||||
| Triple negative status | ||||
| No | 35 (79.5%) | 9 (20.5%) | 0.04 | |
| Yes | 6 (50%) | 6 (50%) | ||
|
| ||||
| Nuclear grade | ||||
| I–II | 13 (86.7%) | 2 (13.3%) | 0.30 | |
| III | 28 (68.3%) | 13 (31.7%) | ||
|
| ||||
| Lymphatic invasion | ||||
| Negative | 25 (64.1%) | 14 (35.9%) | 0.02 | |
| Positive | 16 (94.1%) | 1 (5.9%) | ||
|
| ||||
| Vascular invasion | ||||
| Negative | 26 (63.4%) | 15 (36.6%) | 0.005 | |
| Positive | 15 (100%) | - | ||
|
| ||||
| Skin involvement | ||||
| Negative | 39 (73.6%) | 14 (26.4%) | 1.00 | |
| Positive | 2 (66.7%) | 1 (33.3%) | ||
|
| ||||
| USP-11 expression | ||||
| Low | 29 (72.5%) | 11 (27.5%) | 1.00 | |
| High | 12 (75%) | 4 (25%) | ||
pCR = pathologic complete response; ER= estrogen receptor; PR= progesterone receptor; HER2 = human epidermal growth factor receptor-2
Survival estimates
Median follow-up of all patients was 7.4 years (range 0.6 to 17.3 years). Overall, 19 (34%) breast cancer recurrences or deaths were observed; 10 in the low USP-11 group vs. 9 in the high USP-11 group. The estimated 5-year DFS rates were 77.2% (95% CI: 60.7% to 87.4%) in the low USP-11 group vs. 46.7% (95% CI: 21.2% to 68.7%) in the high USP-11 group (P=0.009) (Table 3).
Table 3.
Five-year overall survival and disease-free survival estimates in patient subgroups
| 5-Year Survival Rate (95% CI) | |||||
|---|---|---|---|---|---|
|
| |||||
| OS | P | DFS | P | ||
| Age | |||||
| ≤50 | 95% (69.5%, 99.3%) | 0.04 | 81.1% (57.0%, 92.5%) | 0.07 | |
| >50 | 72.7% (54.1, 84.8%) | 60.6% (42.0%, 74.9%) | |||
|
| |||||
| Race | |||||
| Black | 25% (0.9%, 66.5%) | 0.0000 | 25% (0.9%, 66.5%) | 0.0000 | |
| White | 87.4% (72.2%, 94.5%) | 70.4% (53.7%, 82%) | |||
|
| |||||
| Clinical stage | |||||
| I/II | 93.9% (77.9%, 98.4%) | 0.008 | 81.8% (63.9%, 91.4%) | 0.02 | |
| III/IV | 61.5% (37.4%, 78.6%) | 48.5% (26.2%, 67.6%) | |||
|
| |||||
| Clinical nodal stage | |||||
| N0 | 90% (65.6%, 97.4%) | 0.59 | 75% (50%, 88.7%) | 0.40 | |
| N1-3 | 76.5% (58.4%, 87.5%) | 65.1% (46.7 %, 78.5%) | |||
|
| |||||
| ER status | |||||
| Negative | 77.8% (57.1%, 89.3%) | 0.89 | 74.1% (53.2%, 86.7%) | 0.53 | |
| Positive | 84.9% (64.5%, 94%) | 63.6% (42.8%, 78.6%) | |||
|
| |||||
| PR status | |||||
| Negative | 80% (60.8%, 90.5%) | 0.57 | 76.7% (57.2%, 88.1%) | 0.48 | |
| Positive | 83.3% (61.3%, 93.4%) | 59.1% (37.1%, 75.6%) | |||
|
| |||||
| HER2 status | |||||
| Negative | 82% (65.9%, 91%) | 0.93 | 72.4% (55.7%, 83.7%) | 0.60 | |
| Positive | 80% (50%, 93.1%) | 60% (31.8%, 79.7%) | |||
|
| |||||
| Triple negative status | |||||
| No | 83.3% (68.1%, 91.7%) | 0.66 | 67.1% (50.8%, 79%) | 0.85 | |
| Yes | 75% (40.8%, 91.2%) | 75% (40.8%, 91.2%) | |||
|
| |||||
| Nuclear grade | |||||
| I/II | 92.9% (59.1%, 99%) | 0.09 | 80% (50%, 93.1%) | 0.50 | |
| III | 77.5% (61.2%, 87.6%) | 64.8% (47.9%, 77.4%) | |||
|
| |||||
| Lymphatic invasion | |||||
| Negative | 81.3% (64.8%, 90.6%) | 0.22 | 75.9% (58.8%, 86.7%) | 0.15 | |
| Positive | 81.6% (53%, 93.7%) | 52.9% (27.6%, 73%) | |||
|
| |||||
| Vascular invasion | |||||
| Negative | 82.3% (66.4%, 91.1%) | 0.07 | 77.1% (60.5%, 87.4%) | 0.06 | |
| Positive | 79% (47.9%, 92.7%) | 46.7% (21.2%, 68.7%) | |||
|
| |||||
| Skin involvement | |||||
| No | 80.4% (66.5%, 88.9%) | 0.97 | 69% (54.5%, 79.7%) | 0.92 | |
| Yes | 100% | 66.7% (54.5%, 94.5%) | |||
|
| |||||
| USP-11 status | |||||
| Low | 87% (71.6%, 94.4%) | 0.0002 | 77.2% (60.7%, 87.4%) | 0.009 | |
| High | 66.7% (37.5%, 84.6%) | 46.7% (21.2%, 68.7%) | |||
|
| |||||
| pCR* | |||||
| No | - | 59.6% (42.7%, 73%) | 0.04 | ||
| Yes | - | 93.3% (61.3%, 99.0%) | |||
CI = confidence interval; OS: overall survival; DFS: disease-free survival; ER= estrogen receptor; PR= progesterone receptor; HER2 = human epidermal growth factor receptor-2; pCR = pathologic complete response.
Calculated from surgery date.
Overall, patients who achieved a pCR had a better DFS than patients who did not (5-year rate, 93.3% [95% CI: 61.3% to 99.0%] vs. 59.6% [95% CI: 42.7% to 73.0%]; P=0.04). In univariate analyses, age >50, black race, and clinical stage III/IV was associated with a significantly increased risk of recurrence. The multivariate Cox proportional hazards model showed that patients who did not achieve a pCR, and patients with high USP-11 expressing tumors had an increased risk of recurrence (Odds ratio [OR] = 5.16; 95% CI:1.16 to 23.07; P= 0.03; and OR = 3.87; 95% CI:1.51 to 9.93; P= 0.005, respectively) (Table 4).
Table 4.
Multivariate logistic regression model for DFS
| OR | 95% CI | P | |
|---|---|---|---|
| pCR | |||
| No (N=41) vs. Yes (N=15) | 5.16 | (1.15, 23.06) | 0.03 |
|
| |||
| USP-11 status | |||
| High (N=16) vs. low (N=40) | 3.87 | (1.51, 9.93) | 0.004 |
There were a total of 14 (25%) deaths; 5 in the low USP-11 group vs. 9 in the high USP-11 group. The 5-year OS estimates were 87.0% (95% CI: 71.6% to 94.4%) in the low USP-11 group compared to 66.7% (95% CI: 37.5% to 84.6%) in the high USP-11 group (P=0.0002). In univariate analyses, age >50, black race, and clinical III/IV stage was associated with an increased risk of death. The multivariate Cox proportional hazards model showed that clinical stage I/II was associated with a lower risk of death (OR = 0.25; 95% CI:0.08 to 0.78; P= 0.02) whereas patients with high USP-11 expressing tumors had a higher risk of death (OR = 6.03; 95% CI:2.00 to 18.17; P= 0.001) (Table 5).
Table 5.
Multivariate logistic regression model for OS
| OR | 95% CI | P | |
|---|---|---|---|
| Clinical stage: | |||
| I/II (N=33) vs. III/IV (N=23) | 0.25 | (0.08, 0.78) | 0.01 |
|
| |||
| USP-11 status | |||
| High (N=16) vs. low (N=40) | 6.025 | (1.99, 18.17) | 0.001 |
OR = Odds ratio; CI = confidence interval
DISCUSSION
Our data indicates that USP-11 is not a predictor of pCR after anthracycline-taxane containing NST for breast cancer; however this does not exclude the possibility of hypersensitivity to other DNA-damaging agents including platinums, PARP inhibitors, mitomycin, and bleomycin as suggested by previous preclinical studies [13, 14]. We show that low USP-11 expression was independently correlated with better survival outcomes following NST. Predictions of pCR have been attempted on the basis of tumor expression of proliferation and apoptosis markers [24], endocrine and growth factors, oncogenes [25], genes involved in the ubiquitin-proteasome pathway and DNA repair and cell death regulators [26]. And to achieve a greater predictive value, multiple gene expression profiles are examined simultaneously and correlated with pCR and survival [27–33]. Besides the ER/PR-negative status, lack of p53 expression, high baseline proliferative and apoptotic indices [34–37], only few additional single biomarkers (eIF4E [38], CXCR4 [39], ALDH1 [40], bcl-2 [27], aB-crystallin [41], KRT13/NFAT5 [42]) have been shown to possess a sufficient predictive/prognostic value to render them clinically useful. In this study we fail to show that USP-11 has a predictive value; however we demonstrate that USP-11 deubiquitinase can be a potential prognostic marker of subsequent survival benefit.
Transcriptional and post-translational modification and degradation of proteins by the ubiquitin-proteasome system are key regulatory events in cellular responses to various stimuli. Recently, identification of mutations in a E3 ubiquitin ligase, RNF168, as the underlying genetic disorder of the DNA repair deficiency disorder, RIDDLE syndrome, has brought to light a critical role for the ubiquitin system in regulating the cellular DSBs. The finding that RNF168 functions downstream of RNF8 to orchestrate the recruitment of repair proteins, such as BRCA1 and 53BP1, to sites of DNA damage suggests that ubiquitin cascade regulates the spatial relocalization of DSB repair proteins [43].
Although much progress has been made in characterizing enzymes that link ubiquitin to proteins, our understanding of deubiquitinating enzymes is less well developed with only few enzymes identified [44–46]. Preclinical studies suggest that deubiquitinating enzymes play an important role in maintaining the delicate balance in DDR by regulating p53 expression [11], and recruitment of DNA repair/checkpoint proteins to DSB foci [43, 47, 48] through ubiquitin conjugation/de-conjugation or ubiquitin turnover. This notion was supported by 2 observations: first, when cells are treated with proteasome inhibitor MG132, which functions to reduce the pools of free ubiquitin, the formation of IR-induced RNF168 foci was affected [49] and second, overexpression of the deubiquitylating enzyme USP3 blocked RNF168 accumulation at sites of DNA breaks [50]. It is also clear that defective ubiquitylation/de-ubiquitylation activities at DSBs significantly increases predilection to tumorigenesis, increases hypersensitivity to IR or DNA-damaging agents [51–55]. Given the DUB impact on the relative ratios of different ubiquitin linkages of DNA repair proteins found at DSBs, one would expect that cells with low USP-11 expression have increased sensitivity to NST, and therefore higher pCR rates and better survival outcomes. In this study, we failed to show any pCR difference, and intriguingly, patients with low USP-11 expression had tumors with negative LVI, presented at an earlier stage (stage I/II) and had better survival outcomes. It is tempting to speculate that USP-11 low cells with a defective DNA repair protein recruitment compensate by up-regulating the MRN/MDC1/ATM pathways [56] or BRCA1-BARD1 domain [57]. This phenomenon is supported by the fact that proteins may contain both a DUB domain and ligase domain in a single polypeptide chain. For example, BRCA1-RAP80 complex contains both BRCC36-encoded K63-specific DUB activity and BRCA1-derived K6-specific E3 ligase activity, raising the possibility that “ubiquitin editing” activities may be performed by different members of the same complex. It is also noteworthy to mention that of factors associated with low USP-11 expression (stage I/II and negative LVI), only LVI was found to be related to pCR but not the tumor stage, as we know from the previous studies that tumor stage is one of the predictive factors for pCR. Therefore, it is possible that the lack of association between USP-11 expression, tumor stage and pCR could well be related to small sample size. Future studies with larger sample size need to be conducted to delineate the predictive vs prognostic implications of USP-11 expression. If correlation between USP-11 expression and pCR or survival is confirmed, then we can explore inhibition of USPs as a novel therapeutic strategy. For example, the curcumin-like derivative HO-3867, diarylidenyl piperidone compound, has shown promising activity in pre-clinical studies on a number of cell types [58]. In ovarian carcinoma cells, HO-3867 has been shown to decrease levels of both USP2a and its targets FASN and FAK, along with a concomitant reduction in oncogenic behaviors, such as migration and invasion. Using a high-throughput activity-based assay, Tian [59] and colleagues have identified a novel and selective inhibitor of USP7 paving the way for identification and characterization of additional USP-targeted agents. Further delineation of how DUBs themselves are regulated by phosphorylation, ubiquitination and protein-protein interactions will likely identify additional strategies for clinical intervention.
BRCA1 and BRCA2 are tumor suppressor genes which play an important role in mediating the DDR of DSBs by HR [60]. BRCA1 has been shown to be required for the activation of both S- and G2/M-phase cell-cycle arrest after DNA damage, the latter being dependent on prior phosphorylation of BRCA1 by the master checkpoint kinase ATM [ataxia telangiectasia mutated] [61]. BRCA1 has also been shown to interact with multiple DNA repair/recombination proteins, including RAD51, the RAD50/MRE11/Nibrin complex, Bloom’s helicase, and the Fanconi D2 protein [62]. The major role of BRCA2 is regulation of RAD51 filament formation and activity which works as a key enzyme catalyzer in HR [63]. Cells lacking BRCA1 or BRCA2 are unable to repair DSBs by HR, and die. Therefore combining PARP (a DNA repair enzyme) inhibition with tumors that have defective BRCA1 or BRCA2 proteins exerts a “chemical synthetic lethality” [64, 65]. Likewise, in vitro studies have shown enhanced cytotoxicity to multiple DNA-damaging agents and PARP-inhibitors in USP-11 silenced cells similar to that observed when BRCA1 is silenced [13, 14]. The question as to whether USP-11 silenced tumors with BRCA1 or BRCA2 dysfunction behave in the same way as BRCA1-mutant or USP-11 silent tumors remains unresolved. The preclinical evidence suggests that hypersensitivity to DNA-damaging agents in USP-11 silenced cells are dependent on intact BRCA2 [13]. The interaction between BRCA1 and USP-11 has not been investigated yet; however a recent study reported that the DSB repair function of USP-11 were independent of the BRCA1 localization [14]. The latter study also detected that RAD51 foci had an accelerated disappearance in USP-11-silenced cells indicating an inability to maintain a sufficient RAD51 filament to facilitate HR repair via BRCA2-signaling pathway. Mass spectrometry results indicate that USP-11 also interacts with a large number of proteins including the transcriptional elongation factors TCEAL1 and 4, other DUBs including USP-7, the NRF2 regulatory protein KEAP1 [66], and p53 [11]. Therefore, it is possible that the BRCA1/2 interaction provides a mechanism to recruit USP-11 to DSBs where it can act on other substrates to exert its prosurvival functions. One therapeutically relevant intervention for future studies could be to identify the USP-11 targets mediating its repair function, and also to assess the synthetic lethality index in BRCA1/2- and USP-11-mutant breast tumors exposed to DNA-damaging agents, or in cells that are resistant to PARP inhibitors.
Several limitations must be considered when interpreting the results of our study. Our study was a retrospective analysis with small sample size. Our study does not address whether a post-treatment change in USP-11 expression would be predictive of a pCR or survival. Due to small sample size, we could not evaluate the prognostic significance of USP-11 in a separate cohort of triple-negative tumors. Additionally, USP-11 staining pertained only to cytoplasm but not to nucleus in all of the breast tumor samples, which may have resulted in differential outcomes. However it is an important pilot study in that it identifies possible in vivo biologic effects of USP-11 in the ongoing research for predictive molecular markers. It also serves to illustrate the shortcomings of cell line-based studies and the need to validate the data on human tissue studies.
In summary, we did not find a correlation between USP-11 expressions in tumor cells and pCR, which is traditionally associated with prognosis. However our study demonstrates that USP-11 has an independent prognostic impact in patients treated with NST. The lack of association of USP-11 status and pCR in our study may reflect the small sample size of the cohort. It remains to be determined if mutation of the catalytically active site of USP-11 could cause disruption of protein interactions, accounting for the DSB repair defect seen in USP-11 silenced cells. Future experimental models and clinical studies need to be conducted to investigate the in vivo biological targets of USP-11, and perhaps using USP-11 as a potential biomarker for chemosensitive phenotype in patients treated with PARP inhibitors or other DNA-damaging agents.
Acknowledgments
Research support: Partially funded by NCI 2 U10 CA045809; the Lynn Cohen Breast and Ovarian Cancer Project, and the Nelly B. Connally Breast Cancer Research Fund.
Footnotes
Disclaimers: The manuscript has never been published and is not under consideration for publication elsewhere. Authors have no financial interest to declare.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annual review of biochemistry. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual review of biochemistry. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1997;11(14):1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Ubiquitin-dependent protein degradation. Annual review of genetics. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 5.Chung CH, Baek SH. Deubiquitinating enzymes: their diversity and emerging roles. Biochemical and biophysical research communications. 1999;266(3):633–640. doi: 10.1006/bbrc.1999.1880. [DOI] [PubMed] [Google Scholar]
- 6.Swanson DA, Freund CL, Ploder L, Mcinnes RR, Valle D. A ubiquitin C-terminal hydrolase gene on the proximal short arm of the X chromosome: implications for X-linked retinal disorders. Human molecular genetics. 1996;5(4):533–538. doi: 10.1093/hmg/5.4.533. [DOI] [PubMed] [Google Scholar]
- 7.Nijman SM, Luna-Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Ideguchi H, Ueda A, Tanaka M, et al. Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. The Biochemical journal. 2002;367(Pt 1):87–95. doi: 10.1042/BJ20011851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nature cell biology. 2004;6(2):97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 10.Sun W, Tan X, Shi Y, et al. USP11 negatively regulates TNFalpha-induced NF-kappaB activation by targeting on IkappaBalpha. Cellular signalling. 2010;22(3):386–394. doi: 10.1016/j.cellsig.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi T, Kimura J, Miki Y, Yoshida K. The deubiquitinating enzyme USP11 controls an IkappaB kinase alpha (IKKalpha)-p53 signaling pathway in response to tumor necrosis factor alpha (TNFalpha) The Journal of biological chemistry. 2007;282(47):33943–33948. doi: 10.1074/jbc.M706282200. [DOI] [PubMed] [Google Scholar]
- 12.Lin CH, Chang HS, Yu WC. USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. The Journal of biological chemistry. 2008;283(23):15681–15688. doi: 10.1074/jbc.M708278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Molecular and cellular biology. 2004;24(17):7444–7455. doi: 10.1128/MCB.24.17.7444-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiltshire TD, Lovejoy CA, Wang T, Xia F, O’connor MJ, Cortez D. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. The Journal of biological chemistry. 2010;285(19):14565–14571. doi: 10.1074/jbc.M110.104745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seventh Edition of the AJCC Cancer Staging Manual and Handbook. 2010. [Google Scholar]
- 16.The world Health Organization Histological Typing of Breast Tumors. The World Organization. Am J Clin Pathol. (2) 1982;78(6):806–816. doi: 10.1093/ajcp/78.6.806. [DOI] [PubMed] [Google Scholar]
- 17.Black MM, Speer FD. Nuclear structure in cancer tissues. Surg Gynecol Obstet. 1957;105(1):97–102. [PubMed] [Google Scholar]
- 18.Sab1300069 S-aP. Anti-USP11 (C-term) antibody produced in rabbit.
- 19.Green MC, Buzdar AU, Smith T, et al. Weekly paclitaxel improves pathologic complete remission in operable breast cancer when compared with paclitaxel once every 3 weeks. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(25):5983–5992. doi: 10.1200/JCO.2005.06.232. [DOI] [PubMed] [Google Scholar]
- 20.Macgrogan G, Mauriac L, Durand M, et al. Primary chemotherapy in breast invasive carcinoma: predictive value of the immunohistochemical detection of hormonal receptors, p53, c-erbB-2, MiB1, pS2 and GST pi. British journal of cancer. 1996;74(9):1458–1465. doi: 10.1038/bjc.1996.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arun B, Bayraktar S, Liu DD, et al. Response to neoadjuvant systemic therapy for breast cancer in BRCA mutation carriers and noncarriers: a single-institution experience. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(28):3739–3746. doi: 10.1200/JCO.2011.35.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurley J, Doliny P, Reis I, et al. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. J Clin Oncol. 2006;24(12):1831–1838. doi: 10.1200/JCO.2005.02.8886. [DOI] [PubMed] [Google Scholar]
- 23.Burstein HJ, Harris LN, Gelman R, et al. Preoperative therapy with trastuzumab and paclitaxel followed by sequential adjuvant doxorubicin/cyclophosphamide for HER2 overexpressing stage II or III breast cancer: a pilot study. J Clin Oncol. 2003;21(1):46–53. doi: 10.1200/JCO.2003.03.124. [DOI] [PubMed] [Google Scholar]
- 24.Ellis PA, Smith IE, Mccarthy K, Detre S, Salter J, Dowsett M. Preoperative chemotherapy induces apoptosis in early breast cancer. Lancet. 1997;349(9055):849. doi: 10.1016/s0140-6736(05)61752-7. [DOI] [PubMed] [Google Scholar]
- 25.Chang J, Ormerod M, Powles TJ, Allred DC, Ashley SE, Dowsett M. Apoptosis and proliferation as predictors of chemotherapy response in patients with breast carcinoma. Cancer. 2000;89(11):2145–2152. [PubMed] [Google Scholar]
- 26.Sotiriou C, Powles TJ, Dowsett M, et al. Gene expression profiles derived from fine needle aspiration correlate with response to systemic chemotherapy in breast cancer. Breast cancer research : BCR. 2002;4(3):R3. doi: 10.1186/bcr433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Von Minckwitz G, Sinn HP, Raab G, et al. Clinical response after two cycles compared to HER2, Ki-67, p53, and bcl-2 in independently predicting a pathological complete response after preoperative chemotherapy in patients with operable carcinoma of the breast. Breast cancer research : BCR. 2008;10(2):R30. doi: 10.1186/bcr1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayers M, Symmans WF, Stec J, et al. Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22(12):2284–2293. doi: 10.1200/JCO.2004.05.166. [DOI] [PubMed] [Google Scholar]
- 29.Buchholz TA, Stivers DN, Stec J, et al. Global gene expression changes during neoadjuvant chemotherapy for human breast cancer. Cancer J. 2002;8(6):461–468. doi: 10.1097/00130404-200211000-00010. [DOI] [PubMed] [Google Scholar]
- 30.Chang JC, Wooten EC, Tsimelzon A, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362(9381):362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 31.Gianni L, Zambetti M, Clark K, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(29):7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 32.Hannemann J, Oosterkamp HM, Bosch CA, et al. Changes in gene expression associated with response to neoadjuvant chemotherapy in breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(15):3331–3342. doi: 10.1200/JCO.2005.09.077. [DOI] [PubMed] [Google Scholar]
- 33.Bertucci F, Finetti P, Rougemont J, et al. Gene expression profiling for molecular characterization of inflammatory breast cancer and prediction of response to chemotherapy. Cancer research. 2004;64(23):8558–8565. doi: 10.1158/0008-5472.CAN-04-2696. [DOI] [PubMed] [Google Scholar]
- 34.Chang J, Powles TJ, Allred DC, et al. Biologic markers as predictors of clinical outcome from systemic therapy for primary operable breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1999;17(10):3058–3063. doi: 10.1200/JCO.1999.17.10.3058. [DOI] [PubMed] [Google Scholar]
- 35.Stearns V, Singh B, Tsangaris T, et al. A prospective randomized pilot study to evaluate predictors of response in serial core biopsies to single agent neoadjuvant doxorubicin or paclitaxel for patients with locally advanced breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(1):124–133. [PubMed] [Google Scholar]
- 36.Kandioler-Eckersberger D, Ludwig C, Rudas M, et al. TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2000;6(1):50–56. [PubMed] [Google Scholar]
- 37.Makris A, Powles TJ, Dowsett M, et al. Prediction of response to neoadjuvant chemoendocrine therapy in primary breast carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 1997;3(4):593–600. [PubMed] [Google Scholar]
- 38.Hiller DJ, Chu Q, Meschonat C, Panu L, Burton G, Li BD. Predictive value of eIF4E reduction after neoadjuvant therapy in breast cancer. The Journal of surgical research. 2009;156(2):265–269. doi: 10.1016/j.jss.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 39.Hiller D, Chu QD. CXCR4 and axillary lymph nodes: review of a potential biomarker for breast cancer metastasis. International journal of breast cancer. 2011;2011:420981. doi: 10.4061/2011/420981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resetkova E, Reis-Filho JS, Jain RK, et al. Prognostic impact of ALDH1 in breast cancer: a story of stem cells and tumor microenvironment. Breast cancer research and treatment. 2010;123(1):97–108. doi: 10.1007/s10549-009-0619-3. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov O, Chen F, Wiley EL, et al. alphaB-crystallin is a novel predictor of resistance to neoadjuvant chemotherapy in breast cancer. Breast cancer research and treatment. 2008;111(3):411–417. doi: 10.1007/s10549-007-9796-0. [DOI] [PubMed] [Google Scholar]
- 42.Jauliac S, Lopez-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nature cell biology. 2002;4(7):540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- 43.Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 2009;8(10):1532–1538. doi: 10.4161/cc.8.10.8351. [DOI] [PubMed] [Google Scholar]
- 44.Andersen MW, Ballal NR, Goldknopf IL, Busch H. Protein A24 lyase activity in nucleoli of thioacetamide-treated rat liver releases histone 2A and ubiquitin from conjugated protein A24. Biochemistry. 1981;20(5):1100–1104. doi: 10.1021/bi00508a009. [DOI] [PubMed] [Google Scholar]
- 45.Kim JH, Park KC, Chung SS, Bang O, Chung CH. Deubiquitinating enzymes as cellular regulators. Journal of biochemistry. 2003;134(1):9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Seminars in cell & developmental biology. 2000;11(3):141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 47.Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicassio F, Corrado N, Vissers JH, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Current biology : CB. 2007;17(22):1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 49.Blundred RM, Stewart GS. DNA double-strand break repair, immunodeficiency and the RIDDLE syndrome. Expert review of clinical immunology. 2011;7(2):169–185. doi: 10.1586/eci.10.93. [DOI] [PubMed] [Google Scholar]
- 50.Doil C, Mailand N, Bekker-Jensen S, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 51.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nature reviews Molecular cell biology. 2008;9(10):759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y. DNA damage: a trigger of innate immunity but a requirement for adaptive immune homeostasis. Nature reviews Immunology. 2006;6(4):261–270. doi: 10.1038/nri1804. [DOI] [PubMed] [Google Scholar]
- 53.Demuth I, Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene. 2007;26(56):7792–7798. doi: 10.1038/sj.onc.1210876. [DOI] [PubMed] [Google Scholar]
- 54.Devgan SS, Sanal O, Doil C, et al. Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell death and differentiation. 2011;18(9):1500–1506. doi: 10.1038/cdd.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikkila J, Coleman KA, Morrissey D, et al. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 2009;28(16):1843–1852. doi: 10.1038/onc.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mochan TA, Venere M, Ditullio RA, Jr, Halazonetis TD. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer research. 2003;63(24):8586–8591. [PubMed] [Google Scholar]
- 57.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. The EMBO journal. 2006;25(10):2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selvendiran K, Tong L, Bratasz A, et al. Anticancer efficacy of a difluorodiarylidenyl piperidone (HO-3867) in human ovarian cancer cells and tumor xenografts. Mol Cancer Ther. 2010;9:1169–1179. doi: 10.1158/1535-7163.MCT-09-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian X, Isamiddinova NS, Peroutka RJ, et al. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay Drug Dev Technol. 2011;9:165–173. doi: 10.1089/adt.2010.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nature reviews. Cancer. 2004;4(9):665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 61.Xu B, Kim S, Kastan MB. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Molecular and cellular biology. 2001;21(10):3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yun MH, Hiom K. Understanding the functions of BRCA1 in the DNA-damage response. Biochemical Society transactions. 2009;37(Pt 3):597–604. doi: 10.1042/BST0370597. [DOI] [PubMed] [Google Scholar]
- 63.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. The Journal of biological chemistry. 1997;272(51):31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 64.Mccabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer research. 2006;66(16):8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 65.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(22):3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 66.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]