Abstract
We describe a novel, simple, rapid, and highly sensitive method to detect single-nucleotide polymorphisms (SNPs) in Mycobacterium tuberculosis and other organisms. Amplification refractory mutation (ARMS) SNP assays were modified by converting the SNP-detecting linear primers in the ARMS assay to hairpin-shaped primers (HPs) through the addition of a 5′ tail complementary to the 3′ end of the linear primer. The improved ability of these primers to detect SNPs in M. tuberculosis was compared in a real-time PCR with SYBR-I green dye. Linear primers resulted in incorrect or indeterminate allele designation for 6 of the 13 SNP alleles tested in seven different SNP assays, while HPs determined the correct SNP in all cases. We compared the cycle threshold differences (ΔCt) between the reactions containing primer-template matches and the reactions containing primer-template mismatches (where a larger ΔCt indicates a more robust assay). The use of HPs dramatically improved the mean ΔCt values for the SNP assays (7.6 for linear primers and 11.2 for HPs). We designed 98 different HP assays for SNPs previously associated with resistance to the antibiotic isoniazid to test the large-scale utility of the HP approach. Assay design was successful in 72.4%, 83.7%, 88.8%, and 92.9% of the assays after one to four rounds of assay design, respectively. HP SNP assays are simple, sensitive, robust, and inexpensive. These advantages favor the application of this technique for SNP assays of M. tuberculosis and other organisms.
Single-nucleotide polymorphism (SNP) analysis is becoming increasingly important for studies of drug resistance, evolution, and molecular epidemiology in Mycobacterium tuberculosis (2, 7, 9, 18, 20-22), human immunodeficiency virus (3, 10, 15), and other organisms (12, 23, 26, 31, 32). High-throughput SNP analysis can be particularly beneficial for confirming associations between specific SNPs and a phenotype of interest, such as drug resistance. The ideal SNP detection method should be simple to design, easy to perform under uniform assay conditions (so that multiple SNP assays can be performed simultaneously), easy to automate, and inexpensive. Most approaches, including DNA sequencing and its derivations (9, 22), microarrays (6), allele-specific amplification (33), mass spectrometry-based techniques (27), TaqMan probes (14), and molecular beacons (28), represent a tradeoff between cost, throughput, and flexibility.
We developed an improved SNP detection method to enable us to analyze large numbers of M. tuberculosis isolates. Most causes of drug resistance in M. tuberculosis appear to be the result of SNPs in particular target genes. However, each SNP occurs at a relatively low frequency. Therefore, in the absence of large sequencing studies, it has been difficult to establish statistically valid associations between individual SNPs and resistance to a particular drug. In the case of resistance to the antibiotic isoniazid, only mutations in codon 315 of the katG gene occur with sufficient frequency. The danger of reaching conclusions after analyzing only a limited number of M. tuberculosis isolates is illustrated by the case of SNPs in codon 463 of katG and codons 269 and 312 of the kasA gene, which were originally identified as resistance associated but later shown to be common in isoniazid-susceptible isolates (1, 2, 13, 24, 25).
The amplification refractory mutation (ARMS) (17) method is an attractive starting point for designing improved SNP assays. ARMS assays differentiate among SNPs by taking advantage of the relative inability of Taq polymerase to extend primers that are mismatched to their targets at the 3′ end. This technique uses two sets of PCR primers with identical “conserved” primers but variable “SNP-specific” primers. The SNP-specific primers are designed to be identical except for their 3′-end nucleotide. At the last 3′ nucleotide, one SNP-specific primer is designed to be complementary to the wild-type sequence, while the other SNP-specific primer is designed to be complementary to the mutant sequence. Two PCRs are performed, one with each of the two SNP-specific primers. Under the usual circumstances, amplification is more efficient in the reaction that contains the perfectly complementary primer than in the reaction that contains the 3′-mismatched primer. Thus, the reaction with the complementary primer will contain more PCR product or, in the case of real-time PCR, will be identified by an earlier cycle threshold (Ct). This technique is a simple and inexpensive method for identifying SNPs; however, it is not always reliable (4).
We postulated that the thermodynamic properties that give stem-and-loop probes enhanced abilities to distinguish among SNP alleles (most notably demonstrated with molecular beacon probes [5, 7, 18, 19, 28]) could be used to improve the ARMS approach. Here, we demonstrate that conversion of the linear SNP-specific primers used in the ARMS assays to hairpin-shaped primers (HPs) designed with molecular beacon principles and parameters (29) results in dramatic improvements in SNP-detecting ability. These improvements have made it possible for us to detect M. tuberculosis SNPs simply, rapidly, and inexpensively.
MATERIALS AND METHODS
M. tuberculosis isolates and chromosomal DNA extraction.
Chromosomal DNA was extracted with the cetyltrimethylammonium bromide method as described previously (30). The reference strain H37Rv was used as a wild-type control. Clinical isolates I-524, M-5036, and M-5455 were used as mutant controls for katGS315I, katGS315N, and katGS315T, respectively. For the remaining SNPs tested, the isolates used are indicated in Table 1.
TABLE 1.
Sequences of primers useda
| SNP | LP | 5′-end tail sequence
|
Constant primer | Amplicon lengthc (bp) | Mutant isolate | ||
|---|---|---|---|---|---|---|---|
| HP | LHP | ELHP | |||||
| katG315b | catacgTcctcgatgccgc | GCGGC | CGCCC | ATATACGCCC | ccggtaaggacgcgatcac | 40 | M-5455 |
| katGS315Tb | catacgTcctcgatgccgG | CCGGC | CGCCC | ATATACGCCC | |||
| katG315b | catacgacctcgatgccgc | GCGGC | CGCCC | ATATACGCCC | |||
| katGS315Tb | catacgacctcgatgccgG | CCGGC | CGCCC | ATATACGCCC | |||
| katG485 | cgtcCtcgttccgtgg | CCACGG | GCTCGC | GATCGCGCTCGC | aggcggatgcgacca | 58 | N/Ad |
| katGG485V | cgtcCtcgttccgtgT | ACACGG | TCTCGC | GATCGCTCTCGC | |||
| katG485 | cgtcgtcgttccgtgg | CCACGG | GCTCGC | GATCGCGCTCGC | |||
| katGG485V | cgtcgtcgttccgtgT | ACACGG | TCTCGC | GATCGCTCTCGC | |||
| kasA269 | cgattCctgggtgccg | CGGCA | GCGGA | GCGGAGCGGA | aaaggcgtccgaggtgatac | 36 | M-5279 |
| kasAG269S | cgattCctgggtgccA | TGGCA | ACGGA | GCGGAACGGA | |||
| kasA269 | cgattgctgggtgccg | CGGCA | GCGGA | GCGGAGCGGA | |||
| kasAG269S | cgattgctgggtgccA | TGGCA | ACGGA | GCGGAACGGA | |||
| inhA21 | atcatcTccgactcgtcgat | ATCGACG | ATGCACC | ACGAGCAAATGCACC | gctacccgtgcgatgtgaa | 39 | M-5502 |
| inhAI21T | atcatcTccgactcgtcgaC | GTCGACG | CTGCACC | ACGAGCAACTGCACC | |||
| inhA21 | aatcatcaccgactcgtcgat | ATCGACG | ATGCACC | ACGAGCAAATGCACC | |||
| inhAI21T | aatcatcaccgactcgtcgaC | GTCGACG | CTGCACC | ACGAGCAACTGCACC | |||
| inhA94 | ggcaAgaacccaatcga | TCGATTGGA | TGGAATGCA | GAGGCAAGTGGAATGCA | cgggcaacaagctcgac | 46 | M-5041 |
| inhAS94A | ggcaAgaacccaatcgC | GCGATTGGA | CGGAATGCA | GAGGCAAGCGGAATGCA | |||
| inhA94 | ggcatgaacccaatcga | TCGATTGGA | TGGAATGCA | GAGGCAAGTGGAATGCA | |||
| inhAS94A | ggcatgaacccaatcgC | GCGATTGGA | CGGAATGCA | GAGGCAAGCGGAATGCA | |||
| ahpC73 | gcCgtcctcgaactcgtc | GACGAGT | CACCAGA | TGCATGTCACCAGA | cggcgttcagcaagctc | 38 | M-5167 |
| ahpCD73H | gcCgtcctcgaactcgtG | CACGAGT | GACCAGA | TGCATGTGACCAGA | |||
| ahpC73 | gcggtcctcgaactcgtc | GACGAGT | CACCAGA | TGCATGTCACCAGA | |||
| ahpCD73H | gcggtcctcgaactcgtG | CACGAGT | GACCAGA | TGCATGTGACCAGA | |||
| ndh268 | ggcTccgtccggcg | CGCCGA | GGCGCA | TACGAGGGCGCA | cagaccttgcaggccgactc | 38 | I-591 |
| ndhR268H | ggcTccgtccggcA | TGCCGA | AGCGCA | TACGAGAGCGCA | |||
| ndh268 | ggcaccgtccggcg | CGCCGA | GGCGCA | TACGAGGGCGCA | |||
| ndhR268H | ggcaccgtccggcA | TGCCGA | AGCGCA | TACGAGAGCGCA | |||
Shown (left to right) are linear primer (LP) sequence, additional HP tail (HP), substituted linearized HP (LHP) tail, and substituted extended LHP (ELHP) tail. Mutations (either SNPs or secondary mutations) are shown underlined in bold capital letters.
These primers detect the same SNP shown in Fig. 3 but on the complementary strand.
5′-end tail sequences not included.
N/A, not available.
Real-time PCR.
All PCRs were performed in an Applied Biosystems 7900HT sequence detector system with the 384-well block for real-time PCR. Thermal conditions were as follows: stage 1, 95°C for 10 min, 70°C for 30 s; stage 2, 72°C for 30 s, 95°C for 20 s, 69°C for 30 s, lowering one degree in the last step for every cycle during 10 cycles; and stage 3, 72°C for 30 s, 95°C for 20 s, and 60°C for 30 s, repeated 40 times. Data were collected in the last step of stage 3 for analysis with the SDS software version 2.0a23 (Applied Biosystems). Every well was loaded with PCR cocktail containing 1× Amplitaq Gold polymerase buffer, 0.15 U of Amplitaq Gold polymerase (Perkin-Elmer), 2 mM MgCl2, 2.5 pmol of each primer, 1× SYBR green I (Molecular Probes Inc.), 1.75 ng of ROX (6-carboxy-X-rhodamine, succinimidyl ester) (Molecular Probes Inc.) (used as a reference dye), and either 0.1 ng of chromosomal DNA, 105 molecules of the artificial template, or an equal volume of water (no-DNA control), followed by sufficient water to result in a final volume of 5 μl.
HP assay design.
The primers (Invitrogen or Illumina) whose sequences are shown in Table 1 were designed with the Primer Express software version 2.0 (Applied Biosystems) to produce short amplicons (30 to 90 bp) and to anneal between 60 and 65°C. A tail was added to the 5′ end of the SNP-detecting primer in order to produce a stem with the 3′ end of the primer. The stem was designed with mfold software (http://www.bioinfo.rpi.edu/applications/mfold/old/dna/) to have a Tm of 67 to 70°C with a free energy (ΔG) of between −0.5 and −2.0. Two single-stranded artificial templates (wild type and mutant) were designed to test the discriminatory power of each primer set, and chromosomal DNA of M. tuberculosis H37Rv was used as a wild-type control.
HP high-throughput assay.
We loaded 384-well plates (Applied Biosystems) with 5 μl of the SNP-specific and constant primer mix per well with a Biomek 2000 Laboratory Automation Workstation (Beckman Coulter). Plates were completely dried overnight inside a laminar-flow cabinet and kept in air-tight plastic bags at −20°C until used. We then loaded 5 μl of the PCR cocktail containing all the components except the primers per well into the microtiter plates. The plates were vortexed and then centrifuged prior to being loaded into the robot of the sequence detector system apparatus.
Comparison of linear primers and HPs.
The abilities of linear primers and HPs to distinguish between two or more SNP alleles in seven different SNP assays were compared. The principles of the HP assay are shown in Fig. 1. First, sets of linear primers for standard ARMS assays were designed with Primer Express software. Then, a second set of primers identical to the first set except that the SNP-specific primer was modified to form a stem-and-loop structure as described above were designed. Assays containing the conventional linear primers and otherwise identical assays containing the HPs were then tested for their ability to distinguish between M. tuberculosis SNPs.
FIG. 1.
Schematic of the HP assay. Two reactions with the same DNA target are performed in parallel. (A) The HP sequence is fully complementary to the target DNA sequence. (B) The HP sequence is complementary to the target DNA sequence except for the last 3′ nucleotide of the primer, which is complementary to an alternative allelic sequence. (C) The real-time PCR fluorescence curve develops more rapidly in the well where the HP sequence is fully complementary to the target DNA sequence (earlier Ct), indicating the presence of allele A.
Seven actual drug resistance-associated SNPs present in M. tuberculosis were tested with M. tuberculosis chromosomal DNA to ensure that the results would be applicable in the subsequent investigations. Each assay was performed in quadruplicate and the average Ct values for each quadruplicate set were calculated. Corresponding linear primer and HP assays were compared on two characteristics: the ability to designate the correct SNP (versus an incorrect or indeterminate assignment) and the average cycle threshold difference (ΔCt) between the reactions containing primer-template matches and the reactions containing primer-template mismatches (where a larger ΔCt indicates a more robust assay). Assays were considered indeterminate if the ΔCt was lower than 5.
HP approach for loci containing multiple alleles.
A single codon can contain more than two SNP alleles. This is the case for position 315 in the katG gene of M. tuberculosis, which is the most common position mutated in isoniazid-resistant clinical isolates (20). The ability of the HP assay to test for four possible alleles at this position was investigated. A single wild-type HP primer and three different mutant HP primers were designed to be complementary to each katG315 allele. The HPs were then tested in assays with chromosomal M. tuberculosis DNA containing each mutation.
RESULTS
Comparison of linear primer and HP.
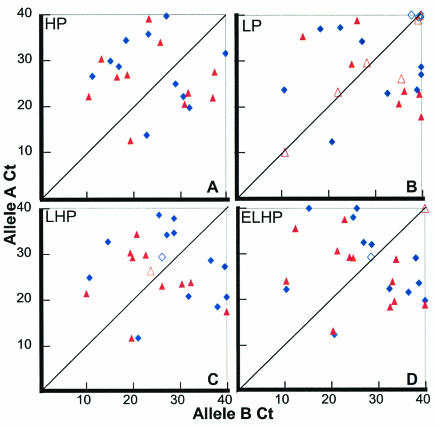
We compared the abilities of linear primers and HPs to detect seven isoniazid resistance-associated SNPs present in the chromosome of six M. tuberculosis clinical isolates. The linear primer assay identified the correct SNP in only 7 of 13 assays (producing one incorrect and five indeterminate results), while the HP assay identified the correct SNP in all 13 assays, demonstrating the superiority of this format (Fig. 2A and B, red triangles). Furthermore, the HP assay appeared to be more sensitive for SNPs, as the ΔCt values were greater in the HP assays than in the linear primer assays (Table 2).
FIG. 2.
Comparison of HP (A), linear primer (B), LHP (C), and ELHP (D) with and without secondary mutations. The Ct for each paired reaction is shown as a single point. The x axis denotes the Ct of the reaction with allele A primers, and the y axis denotes the Ct of the reaction with allele B primers. Values above the diagonal line, marking the place where Ct(B)/Ct(A) is >1, indicate that an A allele SNP should be present, and values below the diagonal line indicate that a B allele SNP should be present. Primers with secondary mutations are represented by blue diamonds, and primers without secondary mutations are represented by red triangles. Solid symbols, successful assays; open symbols, erroneous or indeterminate (ΔCt < 5) assays.
TABLE 2.
Average Cts of seven SNP assays with different primer sets on templates consisting of chromosomal DNA of M. tuberculosisa
| Primer set | Match Ct | Mismatch Ct | ΔCt | No. of rejected assays (n = 13) |
|---|---|---|---|---|
| LP | 25.0 | 32.6 | 7.6 | 6 |
| HP | 19.6 | 30.8 | 11.2 | 0 |
| LHP | 19.6 | 29.1 | 9.5 | 1 |
| ELHP | 21.1 | 32.1 | 11.0 | 1 |
See the text for a description of the isolates used. Match reaction, PCR in which the 3′ end of the primer is complementary to the target. Mismatch reaction, PCR in which the 3′ end of the primer is not complementary to the target. ΔCt, average of mismatched Cts − average of matched Cts.
HP comparison with linearized HPs and extended linearized HPs.
We postulated that the enhanced ability of the HP configuration might be due to the extended 5′ tail of the SNP-specific primer rather than to the actual stem-and-loop design. To further investigate this possibility, “linearized” HPs (LHPs) were created by mutating the 5′ tail of each HP primer so that it could no longer form a stem with the 3′ end. The LHPs were designed so that no new secondary structures were formed and they had the same GC content as the HPs. Each SNP assay was repeated with LHP sets. Figure 2C demonstrates that the performance of the LHP assays was intermediate between that of the linear primers and HPs. LHP assays produced only one indeterminate result, but the ΔCt values averaged 1.7 cycles less than those of the HP assays (Table 2).
To determine if further lengthening of the 5′ tail would continue to enhance the ability of LHPs to distinguish among SNPs, a corresponding set of “extended” LHPs (ELHPs) were created by doubling the length of each 5′ tail. The performance of assays with ELHPs was compared with those of the other primers (Fig. 2D). We found that ELHP assays produced results that were similar to those of LHP assays, with one indeterminate result out of 13 (red triangles in Fig. 2). Notably, assays with ELHPs had improved ΔCt values, with an average ΔCt that was equivalent to the average ΔCt of reactions containing HPs (Table 2). However, the ΔCt values of individual HP assays fell within a narrower range than the ΔCts of the ELHPs, suggesting that the HP-based assays remained advantageous.
Evaluation of the insertion of a secondary mismatch.
The effect of inserting additional base pair mismatches into the HPs was tested. We postulated that a secondary base pair mismatch would decrease the affinity of the mismatched primers to their target and result in a later Ct for the mismatched reactions, an observation previously reported by others for linear primers (8). A secondary mismatch was inserted into each primer either towards the center of the loop or at the end of the stem. Mismatches were designed so that total GC content was maintained. The results (blue diamonds in Fig. 2) indicate that the secondary mismatches resulted in later Cts in both match and mismatch reactions and led to ΔCt values that were improved by 1.1, 0.2, 2.1, and 0.8 cycles for the linear primer, HP, LHP, and ELHP, respectively. The number of rejected assays did not vary with the secondary mismatch except for the linear primers, where only four reactions were rejected (one incorrect and three indeterminate results) (data not shown). The incorporation of a secondary mismatch can also play an important role during HP design. The mismatch confers flexibility on the design process and can be used to avoid undesired secondary structures.
HP approach for loci containing multiple alleles.
In order to evaluate the ability of the HP assay to detect several SNP alleles at the same position, we designed HPs for four possible alleles at position 315 in the katG gene of M. tuberculosis and tested the assay with chromosomal M. tuberculosis DNAs containing each mutation. The results (Fig. 3) show that the well containing the fully complementary HP always had the earlier Ct than the wells containing the mismatched HPs.
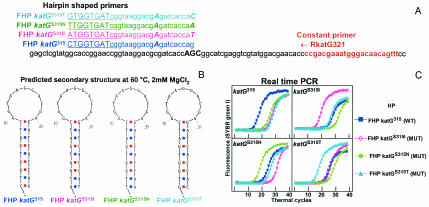
FIG. 3.
HP assay to detect four alleles at a single codon. Assays were designed to detect mutations S315I (AGC→ATC), S315N (AGC→AAC), and S315T (AGC→ACC) in the katG gene. (A) DNA sequences of the target and the HP primers. Secondary mutations and SNPs are shown in bold capitals, and 5′-end tails are underlined. The location of the constant primer is indicated in red. (B) Predicted secondary structure of each of the HP primers at 60°C, 2 mM MgCl2 (annealing conditions). (C) Real-time PCR results with chromosomal DNA from H37Rv as the wild-type control (katG315) and the mutant isolates I-524 (katGS315I), M-5036 (katGS315N), and M-5153 (katGS315T). An earlier Ct was observed for each matched reaction, indicating the correct allele. WT, wild type; MUT, mutant.
Success rate of large-scale HP assay design.
We attempted to design 98 HP assays for 207 different M. tuberculosis SNPs at 98 different polymorphic sites previously associated with resistance to isoniazid to test the utility of the HP approach in large-scale SNP analysis. Each assay was tested on chromosomal DNA from M. tuberculosis H37Rv (wild-type control) and both artificial templates. Mutant chromosomal DNA was also used when available. We succeeded in creating 91 functional HP assays (most of which included a secondary mutation) that detected 191 SNPs in the katG, kasA, ahpC, inhA, mabA, and ndh genes of M. tuberculosis. Assays that detected insertions and deletions were developed with the same parameters. Design success rates as a function of number of design attempts are shown in Table 3. We were unable to design seven (7.1%) assays to detect 16 alleles after two to four attempts.
TABLE 3.
Success rate of HP assay designa
| Attempt | Success rate, % (no. successful/no. attempted) |
|---|---|
| 1 | 72.4 (71/98) |
| 2 | 83.7 (82/98) |
| 3 | 88.8 (87/98) |
| 4 | 92.9 (91/98) |
The number of times an HP assay was redesigned until it became discriminatory is shown. The percentage of HP assays able to discriminate among SNP alleles according to our predefined criteria at a particular design round is given.
Sensitivity of the HP assay.
We investigated the sensitivity of the assays in terms of the amount of chromosomal DNA required per assay. Most assays gave consistent results with less than 0.05 ng of chromosomal DNA per well; however, we found that 0.1 ng/well resulted in smoother amplification curves (data not shown). The extreme sensitivity of our assay may be due to our design protocol, which favors very short amplicons.
DISCUSSION
HP assay offers several advantages for individual laboratories planning to perform SNP analysis on a medium scale (hundreds to thousands of assays per day). The design and setup of the assays are straightforward. SNP detection is integrated with the PCR step, experimental complexity is dramatically reduced, and it is easy to automate. Except for the cost of the real-time PCR apparatus, we found that the material costs of the technique were low. Based on 5-μl/well reactions, we calculated the reagent cost to be approximately US$0.05 per well (of which US$0.043 is attributable to the cost of TaqGold polymerase) and plasticware costs to be an additional US$0.02 per well, totaling US$0.07 per well, or US$0.14 per SNP assay. The initial investment in a real-time PCR instrument and other laboratory equipment would add considerably to these costs; however, such equipment is becoming standard in many research centers.
The HP assays enabled most alleles to be easily discriminated under a single standard condition. The efficiency of many of these assays could likely have been improved by individualizing the PCR conditions for each primer set. However, our aim was to unify conditions so that different SNP assays could be performed simultaneously. We found that ELHPs and HPs had similar performance characteristics, although HPs resulted in more consistent ΔCts and are preferred by our laboratory. It is possible that the additional nucleotides added to the 5′ end of the ELHPs result in secondary structures that mimic HP thermodynamics and that the variability of the ΔCts in ELHP assays is the result of differences in these unplanned structures. Previous reports (11, 16) have shown that HPs (which are not designed to detect SNPs) improve the specificity of PCR assays by decreasing mispriming and primer-dimer formation compared to linear primers. This feature may be an added benefit of the HP design for SNP assays. Our assays were tested with purified DNA. It is possible that HP assays would perform less well if tested on crude DNA preparations. However, the assay is likely to perform well with most common DNA purification techniques.
Acknowledgments
This research was supported by NIH grant AI 46669.
We thank Qingge Li and Sanjay Tyagi for invaluable scientific advice.
REFERENCES
- 1.Abate, G., S. E. Hoffner, V. O. Thomsen, and H. Miorner. 2001. Characterization of isoniazid-resistant strains of Mycobacterium tuberculosis on the basis of phenotypic properties and mutations in katG. Eur. J. Clin. Microbiol. Infect. Dis. 20:329-333. [DOI] [PubMed] [Google Scholar]
- 2.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameyaw, M. M., F. Regateiro, T. Li, X. Liu, M. Tariq, A. Mobarek, N. Thornton, G. O. Folayan, J. Githang'a, A. Indalo, D. Ofori-Adjei, D. A. Price-Evans, and H. L. McLeod. 2001. MDR1 pharmacogenetics: frequency of the C3435T mutation in exon 26 is significantly influenced by ethnicity. Pharmacogenetics 11:217-221. [DOI] [PubMed] [Google Scholar]
- 4.Ayyadevara, S., J. J. Thaden, and R. J. Shmookler Reis. 2000. Discrimination of primer 3′-nucleotide mismatch by taq DNA polymerase during polymerase chain reaction. Anal. Biochem. 284:11-18. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA 96:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee, M., R. Yang, E. Hubbell, A. Berno, X. C. Huang, D. Stern, J. Winkler, D. J. Lockhart, M. S. Morris, and S. P. Fodor. 1996. Accessing genetic information with high-density DNA arrays. Science 274:610-614. [DOI] [PubMed] [Google Scholar]
- 7.El-Hajj, H. H., S. A. Marras, S. Tyagi, F. R. Kramer, and D. Alland. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 39:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan, X. Y., Z. Y. Hu, F. H. Xu, Z. Q. Yan, S. Q. Guo, and Z. M. Li. 2003. Rapid detection of rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis isolates in Shanghai by using the amplification refractory mutation system. J. Clin. Microbiol. 41:993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, A. J. Brown, A. V. Savara, J. D. Hazelwood, and R. T. D'Aquila. 2000. Patterns of resistance mutations selected by treatment of human immunodeficiency virus type 1 infection with zidovudine, didanosine, and nevirapine. J. Infect. Dis. 181:904-911. [DOI] [PubMed] [Google Scholar]
- 11.Kaboev, O. K., L. A. Luchkina, A. N. Tret'iakov, and A. R. Bahrmand. 2000. PCR hot start with primers with the structure of molecular beacons (hairpin-like structure). Nucleic Acids Res. 28:E94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kota, R., R. K. Varshney, T. Thiel, K. J. Dehmer, and A. Graner. 2001. Generation and comparison of EST-derived SSRs and SNPs in barley (Hordeum vulgare L.). Hereditas 135:145-151. [DOI] [PubMed] [Google Scholar]
- 13.Lee, A. S., I. H. Lim, L. L. Tang, A. Telenti, and S. Y. Wong. 1999. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob. Agents Chemother. 43:2087-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, L. G., K. J. Livak, B. Mullah, R. J. Graham, R. S. Vinayak, and T. M. Woudenberg. 1999. Seven-color, homogeneous detection of six PCR products. BioTechniques 27:342-349. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Picado, J., K. Morales-Lopetegi, T. Wrin, J. G. Prado, S. D. Frost, C. J. Petropoulos, B. Clotet, and L. Ruiz. 2002. Selection of drug-resistant HIV-1 mutants in response to repeated structured treatment interruptions. AIDS 16:895-899. [DOI] [PubMed] [Google Scholar]
- 16.Nazarenko, I., B. Lowe, M. Darfler, P. Ikonomi, D. Schuster, and A. Rashtchian. 2002. Multiplex quantitative PCR with self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 30:E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton, C. R., A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith, and A. F. Markham. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17:2503-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations with molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 20.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 21.Ramaswamy, S. V., A. G. Amin, S. Goksel, C. E. Stager, S. J. Dou, H. El Sahly, S. L. Moghazeh, B. N. Kreiswirth, and J. M. Musser. 2000. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:326-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramaswamy, S. V., R. Reich, S. J. Dou, L. Jasperse, X. Pan, A. Wanger, T. Quitugua, and E. A. Graviss. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redd, A. J., J. Roberts-Thomson, T. Karafet, M. Bamshad, L. B. Jorde, J. M. Naidu, B. Walsh, and M. F. Hammer. 2002. Gene flow from the Indian subcontinent to Australia: evidence from the Y chromosome. Curr. Biol. 12:673-677. [DOI] [PubMed] [Google Scholar]
- 24.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338:753-760. [PMC free article] [PubMed] [Google Scholar]
- 25.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suliman-Pollatschek, S., K. Kashkush, H. Shats, J. Hillel, and U. Lavi. 2002. Generation and mapping of AFLP, SSRs and SNPs in Lycopersicon esculentum. Cell. Mol. Biol. Lett. 7:583-597. [PubMed] [Google Scholar]
- 27.Tost, J., O. Brandt, F. Boussicault, D. Derbala, C. Caloustian, D. Lechner, and I. G. Gut. 2002. Molecular haplotyping at high throughput. Nucleic Acids Res. 30:E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 29.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 30.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vignal, A., D. Milan, M. SanCristobal, and A. Eggen. 2002. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 34:275-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagenaar, T. R., V. T. Chow, C. Buranathai, P. Thawatsupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072-1081. [DOI] [PubMed] [Google Scholar]
- 33.Waterfall, C. M., and B. D. Cobb. 2001. Single tube genotyping of sickle cell anaemia with PCR-based SNP analysis. Nucleic Acids Res. 29:E119. [DOI] [PMC free article] [PubMed] [Google Scholar]