Abstract
The sequence identity of the enterovirus VP1 gene has been shown to correlate with the serotype concept. Enterovirus molecular typing methods are therefore often based on sequencing of the VP1 genomic region and monophyletic clustering of VP1 sequences of a homologous serotype. For epidemiological surveillance, 342 enterovirus samples obtained from patients with aseptic meningitis in Belgium from 1999 to 2002 were first diagnosed as being enterovirus positive by amplification of the 5′ noncoding region (5′NCR) by reverse transcription (RT)-PCR. Subsequently, samples were molecularly typed by RT-nested PCR amplification and sequencing of a portion of the VP1 gene. Phylogenetic analyses were performed to investigate enteroviral evolution and to examine the serotype and genotype correlation of the two genomic regions. Our typing results demonstrated echovirus 30, echovirus 13, echovirus 18, and echovirus 6 to be the most predominant types. Echoviruses 13 and 18 were considered to be emerging human serotypes since 2000 and 2001, respectively, as they had been rarely reported before. Several serotypes existed as multiple genotypes (subtypes) from 1999 to 2002, but genomic differences mainly resided at synonymous sites; these results strongly suggest that the subtypes exhibit similar antigenic properties. Phylogenetic analyses confirmed that VP1 is an adequate region for molecular typing. Serotype-specific clusters are not observed commonly in phylogenetic trees based on the 5′NCR, and the phylogenetic signal in the 5′NCR was found to be particularly low. However, some substructure in the 5′NCR tree made a tentative prediction of the enterovirus type possible and was therefore helpful in PCR strategies for VP1 (e.g., primer choice), provided some background knowledge on the local spectrum of enteroviruses already exists.
Within the Picornaviridae family, the Enterovirus genus represents 65 human serotypes which have been classified into five species (A to D and the polioviruses). Members of the human enterovirus B species are the most common cause of aseptic meningitis. They include the six serotypes of group B coxsackieviruses, all echoviruses, coxsackievirus A9, enterovirus 69, and the recently characterized enterovirus 73 (10, 21). Outbreaks of aseptic meningitis typically peak during the summer and early fall, and various serotypes are often associated with a single outbreak (30). The predominant enterovirus types vary from year to year, with echovirus 30, echovirus 13, and echovirus 18 being the most frequently isolated in Europe and the United States over the past few years (2, 3, 6, 7, 11, 38, 41).
Laboratory diagnosis of enterovirus infections is currently based on amplification of highly conserved regions within the enteroviral RNA genome (29). The 5′ noncoding region (5′NCR), which contains the cloverleaf and internal ribosome entry site secondary structures, seems to be the most conserved region among enteroviruses and is therefore targeted widely in enterovirus diagnostic procedures (29, 38). In addition to traditional virological methods used to identify the enterovirus serotype, multiple reverse transcription (RT)-PCR methods which are based on amplification of a portion of the VP1 capsid protein gene were recently developed (4, 5, 16, 18, 23, 38). Because VP1 is the major surface-accessible protein in the mature picornavirus virion, the VP1 gene encodes important serotype-specific neutralization epitopes; therefore, its sequence has been shown to correlate very well with the classical serotype classification (14, 23). Sequence homology criteria have been defined: a VP1 nucleotide sequence identity of more than 75% to a certain reference strain in GenBank indicates that the patient sample is of the homologous serotype, provided that the second-highest identity (next closest serotype) is less than 70% (20). Additionally, phylogenetic analyses based on VP1 sequences have demonstrated that strains of the same serotype always cluster together (4, 16, 17). Therefore, phylogenetic analyses provide an alternative method for enterovirus identification.
Two distinct mechanisms play an important role in enteroviral evolution: mutation and recombination (24, 32). Enteroviral RNA replication is characterized by a high mutation rate due to the lack of proofreading activity of the viral RNA-dependent RNA polymerase. Enteroviruses tend to have rates of spontaneous mutation of approximately one mutation per genome per replication (8). Consequently, enteroviruses exist as a dynamic mutant population termed quasispecies and corresponding to a swarm of sequence variants (9). Additionally, intratypic and intertypic forms of homologous recombination are also thought to be relatively frequent evolutionary forces for enteroviral genomes (1, 13, 24, 32). Preferential recombination breakpoints have been detected within the nonstructural P2 and P3 regions or at the 5′NCR-P1 junction, whereas the survival of offspring with recombination within the viral capsid coding region is rare (1, 13, 32).
For epidemiological surveillance, the enteroviruses in 342 cerebrospinal fluid samples (CSF) from patients hospitalized with aseptic meningitis in Belgium from 1999 to 2002 were molecularly typed by using an in-house VP1 RT-nested PCR assay. Phylogenetic analyses based on the resulting partial VP1 sequences and diagnostic partial 5′NCR sequences were carried out to investigate viral evolution and to examine whether it is possible to correlate the nucleotide sequence identity of the 5′NCR with the VP1-based serotype and genotype classification within a limited time span.
MATERIALS AND METHODS
Patients and clinical samples.
CSF samples were obtained from a total of 342 patients hospitalized with aseptic meningitis at the University Hospital Gasthuisberg Leuven (Leuven, Belgium) from January 1999 until December 2002. Patients ranged in age from 6 days to 75 years old, with a mean age of 10 years. The temporal distribution of the clinical specimens was as follows: 17 samples were collected in 1999, 150 samples in 2000, 124 samples in 2001, and 51 samples in 2002. In 2000, 82% of the samples were collected during the summer (May to September), while in 1999, 2001, and 2002, the proportions collected during the summer were only 41, 30, and 60%, respectively. The major outbreak of aseptic meningitis in Belgium in the summer of 2000 was reported previously (38), but the 122 samples collected then were included in the analyses in this study.
Extraction, amplification, and sequencing.
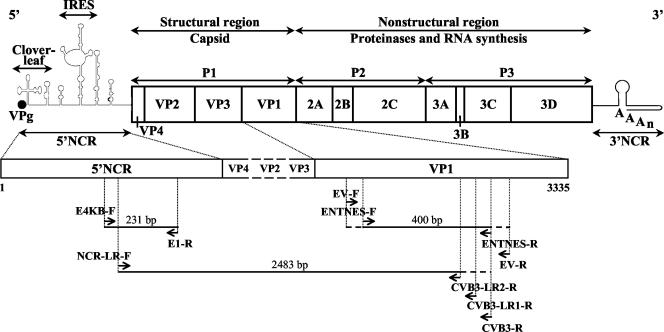
Viral RNA was extracted from CSF samples by using a QIAamp viral RNA minikit (Qiagen, Westburg, The Netherlands). Two different regions in the enteroviral genome were amplified by a one-step RT-PCR (OneStep RT-PCR kit; Qiagen) as described previously (38). Briefly, a 231-bp gene fragment in the second half of the 5′NCR was amplified with two 5′NCR-specific primers, E4KB-F and E1-R (Table 1 and Fig. 1), to confirm that the virus specimens were enterovirus positive. For typing of the virus specimens, a VP1 RT-nested PCR was carried out and generated amplicons corresponding to 388 to 400 nucleotides at the 5′ end of the VP1 gene. Degenerate outer primers EV-F and EV-R (Table 1 and Fig. 1) were used in a first amplification round. The nested PCR assay was performed either with the degenerate inner primer set (ENTNES-F and ENTNES-R) or with one of the type-specific inner primer pairs, depending on the 5′NCR clustering (Table 1 and Fig. 1). The preparation of reaction mixtures and the amplification profiles used were described previously (38). PCR products were separated by 6% polyacrylamide gel electrophoresis, stained with ethidium bromide, and visualized under UV light. The 5′NCR and VP1 amplicons were purified by using a QIAquick PCR purification kit (Qiagen) and sequenced in the forward and reverse directions by using the respective PCR primers, an ABI Prism BigDye terminator cycle sequencing reaction kit (version 3.0), and an ABI Prism 3100 DNA sequencer (Applied Biosystems, Foster City, Calif.).
TABLE 1.
Oligonucleotide primers used for 5′NCR RT-PCR, VP1 RT-nested PCR, long-range RT, and seminested PCR
| Primer | Region | Positionsa | Enterovirus specificity | Sequenceb |
|---|---|---|---|---|
| E4KB-F | 5′NCR | 416-440 | All | AAGGTGYGAAGAGYCTATTGAGCTA |
| E1-R | 5′NCR | 651-634 | All | CACCGGATGGCCAATCCA |
| NCR-LR-F | 5′NCR | 452-480 | All | TCCGGCCCCTGAATGCGGCTAATCCTAAC |
| EV-F | VP1 | 2559-2578 | Group B | GYDGARACNGGVCACACRTC |
| EV-R | VP1 | 3022-3001 | Group B | CTMATGAAHGGDATNGAYATBC |
| ENTNES-F | VP1 | 2595-2617 | Group B | GAYACWATGCARACVMGRCAYGT |
| NES6-F | VP1 | 2595-2617 | Echovirus 6 | GACACTATCCAGACACGCCATGT |
| NES9-F | VP1 | 2595-2617 | Echovirus 9 | GACACYATGCARACYAGGCACGT |
| NES13-F | VP1 | 2595-2617 | Echovirus 13 | GACACCATTCAAACCAGGTGTGT |
| NES16-F | VP1 | 2595-2617 | Echovirus 16 | GATACAATACAGACGCGCCACGT |
| NES18-F | VP1 | 2595-2617 | Echovirus 18 | GATACCATACAAACTAGGCATGT |
| NES25-F | VP1 | 2595-2617 | Echovirus 25 | GACACTATGCAGACCAGACATGT |
| NES30-F | VP1 | 2595-2617 | Echovirus 30 | GACACAATGCAGACACGACACGT |
| ENTNES-R | VP1 | 2994-2976 | Group B | GRGCAYTVCCYTCTGTCCA |
| NES6-R | VP1 | 2994-2976 | Echovirus 6 | GGGCATTTCCCTCAGTCCA |
| NES9-R | VP1 | 2994-2976 | Echovirus 9 | GRGCATTCCCTTCRGTCCA |
| NES13-R | VP1 | 2994-2976 | Echovirus 13 | GCGCGTTTCCTTCGGTCCA |
| NES16-R | VP1 | 2994-2976 | Echovirus 16 | GTGCATTCCCCTCAGTCCA |
| NES18-R | VP1 | 2994-2976 | Echovirus 18 | GAGCGTTGCCCTCTGTCCA |
| NES25-R | VP1 | 2994-2976 | Echovirus 25 | GGGCATTCCCTTCTGTCCA |
| NES30-R | VP1 | 2994-2976 | Echovirus 30 | GGGCATTACCTTCCGTCCA |
| CVB3-R | VP1 | 2994-2976 | Coxsackievirus B3 | GCGCATTGCCTTCAGTCCA |
| CVB3-LR1-R | VP1 | 2968-2936 | Coxsackievirus B3 | CTGGGATTAGTGGATGTCTGCCACACGTATGAG |
| CVB3-LR2-R | VP1 | 2934-2907 | Coxsackievirus B3 | CAACCTTATCTGGCACCGGGCCACCTGG |
Relative to the genome of echovirus 30 strain Bastianni (GenBank accession number AF311938).
Sequences are shown 5′ to 3′ with standard codes (Y is C or T; D is A, T, or G; R is A or G; N is A, C, T, or G; V is A, C, or G; M is A or C; H is A, T, or C; B is T, C, or G; and W is A or T).
FIG. 1.
Diagram showing locations of oligonucleotide primers and PCR products in the enteroviral genome (Table 1). IRES, internal ribosome entry site.
Because of the high sensitivity of RT-nested PCR assays, precautions were taken in order to prevent PCR cross-contamination. Preparation of reagents, processing of CSF samples, thermal cycling, nested PCR, and analysis of PCR products were all carried out in safety cabinets in separated laboratories and were subject to strict rules for laboratory practice. Negative controls were included in each PCR experiment.
CVB3-specific long-range RT-seminested PCR.
A coxsackievirus B3 (CVB3)-specific long-range RT-seminested PCR assay was developed to assess recombination. CVB3 cDNA was first synthesized by using an Expand reverse transcriptase kit (Roche Applied Science, Vilvoorde, Belgium) with a CVB3 VP1-specific reverse primer, CVB3-R (Table 1 and Fig. 1). With CVB3 cDNA as a template, a long-range seminested PCR assay was next carried out with an enterovirus 5′NCR-specific long-range forward primer, NCR-LR-F, two CVB3 VP1-specific long-range reverse primers, CVB3-LR1-R (outer primer; Table 1 and Fig. 1) and CVB3-LR2-R (inner primer; Table 1 and Fig. 1), and an Expand long-template PCR system kit (Roche). CVB3-specific long-range outer and inner PCR assays were both carried out with 5 μl of cDNA or a long-range outer PCR product in a 50-μl reaction volume containing 5 μl of 10× Expand long-template buffer 3, 500 μM each sodium deoxynucleoside triphosphate, 0.75 μl of Expand long-template enzyme mix, 300 nM concentrations of the respective primers, and ultrapure water to 50 μl. The same thermal cycling profile was used for both PCR assays, which were carried out by using a GeneAmp PCR system 9700 thermal cycler (Applied Biosystems) with an initial denaturation at 93°C for 2 min, a three-step cycle (10 s at 93°C, 30 s at 62°C, and 2 min at 68°C) for 10 cycles, another three-step cycle (10 s at 93°C, 30 s at 62°C, and 2 min [plus 20 s of cycle elongation for each successive cycle] at 68°C) for 30 cycles, and a final extension at 68°C for 7 min. The CVB3-specific long-range PCR products were separated on a 2% agarose gel, stained with ethidium bromide, visualized under UV light, excised from the gel, purified by using a QIAquick gel extraction kit (Qiagen), and sequenced in both directions as described above.
Sequence analysis and phylogenetic analysis.
Chromatogram sequencing files were inspected with Chromas 2.22 (Technelysium Pty Ltd., Tewantin, Queensland, Australia), and contigs were prepared by using SeqMan II (DNASTAR, Madison, Wis.). The obtained VP1 consensus sequences of 346 to 358 bp were compared to all of the corresponding enterovirus sequences available in GenBank (release 131.0-135.0) by using FASTA analysis (26) in order to identify the enterovirus type. Serotypes were designated according to the sequence homology rules proposed by Oberste et al. (20), which defined a VP1 sequence identity limit of 75% for heterologous serotypes. Pairwise nucleotide sequence alignments were calculated by using BLAST 2 Sequences (37). Multiple sequence alignments were prepared by using CLUSTAL W (39) or CLUSTAL X (40) and were manually edited with the GeneDoc alignment editor (15). Phylogenetic analyses were conducted by using MEGA version 2.1 (12). Genetic distances were calculated by using the Kimura two-parameter model with gamma-distributed rate variability among sites (alpha parameters of 0.23 and 0.33 for 5′NCR and VP1, respectively). Phylogenetic trees were constructed by using the neighbor-joining method. Pairwise distance matrices showing the number of nucleotide differences were also calculated by using MEGA version 2.1.
To analyze the phylogenetic signals of the VP1 and 5′NCR data sets, likelihood mapping analysis was performed by using Tree-puzzle version 5.0 (36). Briefly, 10,000 quartets of sequences are randomly selected, and the likelihood for three possible unrooted topologies is calculated. The resulting likelihoods for each quartet are summarized as a single dot in an equilateral triangle. The triangle is partitioned into different regions. The center region represents star-like evolution, the three corner regions represent a well-resolved phylogeny, and the three remaining regions reflect the situation in which it is difficult to distinguish between two of the three trees.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank sequence database under accession numbers AY342614 to AY343053. Previously determined sequences with accession numbers AF521310 to AF521351 and AF521353 to AF521555 were included in the analyses (38).
RESULTS
Molecular typing by sequence analysis and phylogenetic analysis.
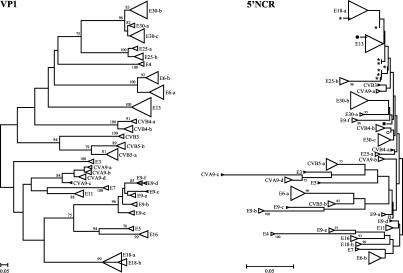
Using two distinct RT-PCR assays targeting the enterovirus 5′NCR and the VP1 gene, 342 Belgian patient samples collected over a period of 4 years were diagnosed as being enterovirus positive and subsequently were molecularly typed for epidemiological surveillance. Only a single one-step RT-PCR was needed to produce 185-bp sequences from the highly conserved 5′NCR. A phylogenetic tree was constructed from the aligned partial 5′NCR sequences by using the neighbor-joining method (Fig. 2). Distinct clusters observed in the 5′NCR tree seemed to represent a single enterovirus subtype.
FIG. 2.
VP1 phylogram (left) and 5′NCR phylogram (right) based on 342 partial VP1 sequences (346 to 358 bp) and 342 partial 5′NCR sequences (185 bp), respectively. Both neighbor-joining trees were evaluated with 100 bootstrap pseudoreplicates; only bootstrap values of over 75% are shown. The scale bar represents the genetic distance (nucleotide substitutions per site). Distinct clusters representing strains of the same (sub)type are shown as triangles (E, echovirus; CV, coxsackievirus). An illustration of the detailed trees demonstrating the positions of the individual samples can be found at the following website: http://www.kuleuven.ac.be/rega/mvr/Figure_trees_detailed_JCM.pdf. Symbols used in the 5′NCR tree: *, E13 taxon; •, CVB3 taxon; ▪, CVB4-b taxon; ○, E30-c taxon; □, E9-a taxon.
To evaluate the predictive value of the 5′NCR for subtype assignment and for surveillance purposes, a VP1 RT-nested PCR assay was carried out and revealed unambiguously the enterovirus serotypes in all samples. Nested inner primers were chosen on the basis of the 5′NCR phylogenetic grouping. For most samples, the degenerate ENTNES-F and ENTNES-R primers were able to amplify the VP1 fragment; type-specific primers were used for samples that were not amplified with this degenerate primer set (Table 1 and Fig. 1). The 346- to 358-bp VP1 sequences obtained were subjected to FASTA analysis, and each sample was assigned the type that gave the highest VP1 identity score. These assignments were further confirmed by phylogenetic analysis of the partial VP1 sequences. A tree was constructed on the basis of a nucleotide alignment of all identified patient sample VP1 fragments, including homologous VP1 reference sequences from GenBank which showed high sequence similarity to the patient sample sequences in the FASTA analysis (Fig. 2). The VP1 tree showed that strains of the same serotype (assigned by FASTA analysis) were indeed monophyletic and clustered together with their respective GenBank reference sequences, conclusions that were supported by high bootstrap values. There was consequently no conflict between the FASTA serotype assignment and the tree serotype assignment.
Distribution of enterovirus serotypes and subtypes.
The distribution of serotypes identified in Belgium from 1999 to 2002 is shown in Table 2. Sixteen different enterovirus serotypes were identified, with echovirus 30, echovirus 13, echovirus 18, and echovirus 6 being the most prevalent serotypes encountered (in 21.3, 20.2, 17.8, and 12.6% of 342 samples, respectively). Coxsackievirus B5 (7%), echovirus 9 (5%), and coxsackievirus A9 (4.1%) were detected sporadically, whereas echoviruses 3, 4, 5, 7, 11, 16, and 25 and coxsackieviruses B3 and B4 represented rather rare serotypes in our study. In 1999, echovirus 11 was predominant, while in 2000, echoviruses 30, 13, and 6 and coxsackievirus B5 behaved codominantly. In 2001, echovirus 18 peaked together with echovirus 13; echovirus 18 was again the most prevalent serotype in 2002 (Table 2).
TABLE 2.
Enterovirus serotypes identified from 1999 to 2002
| Enterovirus serotype | No. of samples collected in:
|
Total no. (%) of samples | No. (%) of single taxaa | |||
|---|---|---|---|---|---|---|
| 1999 | 2000 | 2001 | 2002 | |||
| Echovirus 3 | 0 | 0 | 0 | 1 | 1 (0.3) | 1 (0.3) |
| Echovirus 4 | 0 | 0 | 3 | 0 | 3 (0.9) | 2 (0.6) |
| Echovirus 5 | 0 | 0 | 0 | 1 | 1 (0.3) | 1 (0.3) |
| Echovirus 6 | 3 | 25 | 9 | 6 | 43 (12.6) | 33 (9.6) |
| Echovirus 7 | 0 | 1 | 2 | 0 | 3 (0.9) | 3 (0.9) |
| Echovirus 9 | 0 | 0 | 11 | 6 | 17 (5) | 16 (4.7) |
| Echovirus 11 | 5 | 0 | 0 | 3 | 8 (2.3) | 8 (2.3) |
| Echovirus 13 | 0 | 45 | 22 | 2 | 69 (20.2) | 43 (12.6) |
| Echovirus 16 | 0 | 7 | 0 | 0 | 7 (2) | 7 (2) |
| Echovirus 18 | 0 | 2 | 46 | 13 | 61 (17.8) | 42 (12.3) |
| Echovirus 25 | 1 | 0 | 7 | 0 | 8 (2.3) | 8 (2.3) |
| Echovirus 30 | 2 | 49 | 15 | 7 | 73 (21.3) | 52 (15.2) |
| Coxsackievirus A9 | 3 | 0 | 7 | 4 | 14 (4.1) | 12 (3.5) |
| Coxsackievirus B3 | 0 | 0 | 1 | 1 | 2 (0.6) | 2 (0.6) |
| Coxsackievirus B4 | 2 | 1 | 1 | 4 | 8 (2.3) | 8 (2.3) |
| Coxsackievirus B5 | 1 | 20 | 0 | 3 | 24 (7) | 18 (5.3) |
| Total no. (%) | 17 (5) | 150 (43.9) | 124 (36.3) | 51 (14.9) | 342 | 256 (74.9) |
Patient samples generating both identical 5′NCR sequences and identical VP1 sequences are grouped together and represent a single taxon in the trees.
In the VP1 tree, several serotypes showed subclustering into various genotypes or subtypes (Fig. 2). Two distinct genotypes were observed for echovirus 6 (E6-a and E6-b), echovirus 18 (E18-a and E18-b), echovirus 25 (E25-a and E25-b), coxsackievirus B4 (CVB4-a and CVB4-b), and coxsackievirus B5 (CVB5-a and CVB5-b). Echovirus 30 (E30-a, E30-b, and E30-c), coxsackievirus A9 (CVA9-a, CVA9-b, CVA9-c, and CVA9-d), and echovirus 9 (E9-a, E9-b, E9-c, E9-d, E9-e, and E9-f) demonstrated three, four, and six subclusters, respectively. The other enterovirus serotypes (echoviruses 3, 4, 5, 7, 11, 13, and 16 and coxsackievirus B3) existed as a single genotype throughout our study period.
Molecular evolution of enteroviruses.
The VP1 nucleotide sequence divergence within each subtype was not higher than 10%, with an overall mean of 2.1% (mean of the VP1 means; Table 3). Between the subtypes of a particular serotype, the mean VP1 sequence divergence was 13%. VP1 genomic differences, however, resided mainly at synonymous sites (i.e., no amino acid change). There was consequently little amino acid variation between the subtypes of a particular serotype and most likely no change in antigenic properties. For the three E30 subtypes, the amino acid variation ranged from 0 to 3.4%, while for the E18 strains, the amino acid sequence divergence varied between 0 and 1.7%. The CVB5-a and CVB5-b subtype strains showed an amino acid sequence similarity of >97%. Among all E6, E25, CVB4, E9, and CVA9 subtype strains, the amino acid sequence identities averaged 98, 97, 99, 97, and 98%, respectively. The existence of several subtype clusters appeared to be season dependent; e.g., E30-a, E30-b, and E30-c comprised samples obtained in 1999, 2000, and 2001 to 2002, respectively, with only three exceptions (Fig. 2). CVB5-b was found only in 2000, E6-b was found only in 2001 to 2002, and E18-b was found only in 2002. E9-a, E9-b, E9-c, and E9-d comprised samples from 2001, while E9-e and E9-f comprised samples from 2002, with only one exception.
TABLE 3.
Nucleotide sequence divergence within the indicated genotypes
| Genotype | No. of taxa per genotype | % Nucleotide sequence divergencea for:
|
|||
|---|---|---|---|---|---|
| VP1 (346-358 bp)
|
5′NCR (185 bp)
|
||||
| Range | Mean | Range | Mean | ||
| Echovirus 3 | 1 | NA | NA | NA | NA |
| Echovirus 4 | 2 | 0.3 | 0.3 | 4.9 | 4.9 |
| Echovirus 5 | 1 | NA | NA | NA | NA |
| Echovirus 6-a | 22 | 0.0-6.5 | 1.9 | 0.0-2.2 | 0.9 |
| Echovirus 6-b | 11 | 0.3-3.7 | 2.2 | 0.0-2.7 | 1.2 |
| Echovirus 7 | 3 | 1.1-2.8 | 2.2 | 0.5-1.1 | 0.7 |
| Echovirus 9-a | 3 | 0.6-1.1 | 0.9 | 0.0-0.5 | 0.4 |
| Echovirus 9-b | 4 | 0.6-4.2 | 2.4 | 0.0-1.6 | 0.8 |
| Echovirus 9-c | 1 | NA | NA | NA | NA |
| Echovirus 9-d | 1 | NA | NA | NA | NA |
| Echovirus 9-e | 4 | 0.0-2.0 | 1.1 | 0.0 | 0.0 |
| Echovirus 9-f | 3 | 1.7 | 1.7 | 0.0-0.5 | 0.4 |
| Echovirus 11 | 8 | 0.6-4.2 | 2.4 | 0.0-1.6 | 0.8 |
| Echovirus 13 | 43 | 0.0-4.2 | 1.5 | 0.0-4.3 | 0.9 |
| Echovirus 16 | 7 | 0.3-1.1 | 0.8 | 0.0-1.6 | 0.5 |
| Echovirus 18-a | 38 | 0.0-3.9 | 1.2 | 0.0-2.2 | 0.8 |
| Echovirus 18-b | 4 | 0.3-0.8 | 0.6 | 0.0 | 0.0 |
| Echovirus 25-a | 3 | 0.8-9.9 | 6.8 | 0.5-3.8 | 2.5 |
| Echovirus 25-b | 5 | 0.8-2.3 | 1.5 | 0.0-0.5 | 0.2 |
| Echovirus 30-a | 2 | 0.3 | 0.3 | 0.0 | 0.0 |
| Echovirus 30-b | 31 | 0.0-3.1 | 1.3 | 0.0-2.7 | 0.9 |
| Echovirus 30-c | 19 | 0.0-3.6 | 1.9 | 0.0-3.2 | 1.2 |
| Coxsackievirus A9-a | 2 | 1.1 | 1.1 | 0.0 | 0.0 |
| Coxsackievirus A9-b | 5 | 2.8-9.6 | 7.8 | 1.1-3.8 | 2.7 |
| Coxsackievirus A9-c | 1 | NA | NA | NA | NA |
| Coxsackievirus A9-d | 4 | 0.6-1.7 | 1.0 | 0.5-1.6 | 0.9 |
| Coxsackievirus B3 | 2 | 5.2 | 5.2 | 2.7 | 2.7 |
| Coxsackievirus B4-a | 1 | NA | NA | NA | NA |
| Coxsackievirus B4-b | 7 | 0.6-7.6 | 3.7 | 0.0-3.2 | 1.1 |
| Coxsackievirus B5-a | 14 | 0.0-3.4 | 1.8 | 0.0-3.2 | 1.4 |
| Coxsackievirus B5-b | 4 | 0.0-0.9 | 0.4 | 0.0-1.1 | 0.5 |
NA, not applicable (genotype has only one representative).
Analysis of the serotype and genotype correlation of VP1 and the 5′NCR.
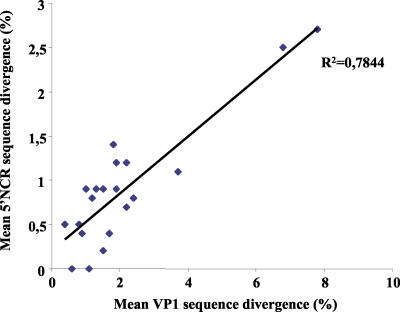
Because of the diagnostic purpose of the 5′NCR RT-PCR, the 5′NCR sequences were in fact obtained before the VP1 sequences, and information about the former sequences was used in the decision on which primers to use in the VP1 RT-nested PCR. The phylogenetic properties of the 5′NCR fragment used here were investigated by comparing the 5′NCR sequences with VP1 sequences that are used for reliable molecular typing. Compared with samples in the VP1 tree, samples in the 5′NCR tree did not cluster by serotype (Fig. 2). However, samples in the 5′NCR tree clustered together in a subtype-specific fashion. Most subclusters belonging to a particular serotype in the VP1 tree were also observed as distinct clusters in the 5′NCR tree, but they did not appear as one major serotype-specific cluster in this tree. In the 5′NCR tree, the clustering was not highly supported by bootstrap analysis, but the nucleotide sequence divergence in the 5′NCR within each subtype did not exceed 5% and the overall mean in this region was 1% (Table 3). Our results demonstrated that the VP1 mean sequence divergence values within the subtypes were all associated with low mean values in the 5′NCR (Table 3 and Fig. 3). Accordingly, closely related sequences in the 5′NCR corresponded to closely related sequences in VP1 and consequently to a particular subtype. For a few serotypes and subtypes, however, the 5′NCR clusters were not clearly distinct from one another, a finding which made prediction of the serotype from these clusters ambiguous, in particular, for E11 and E9-d, for E30-c and CVB4-b, for CVB3 and CVA9-a, for CVB3 and E13, and for E13 and E18-a (Fig. 2). However, in most instances, knowledge of the 5′NCR clustering could be used to guide the choice of primers for the VP1 RT-nested PCR.
FIG. 3.
Scatter diagram of the mean percent VP1 sequence divergence versus the mean percent 5′NCR sequence divergence within each genotype, excluding the genotypes with only one or two representatives. R2 represents the coefficient of determination of the trend line.
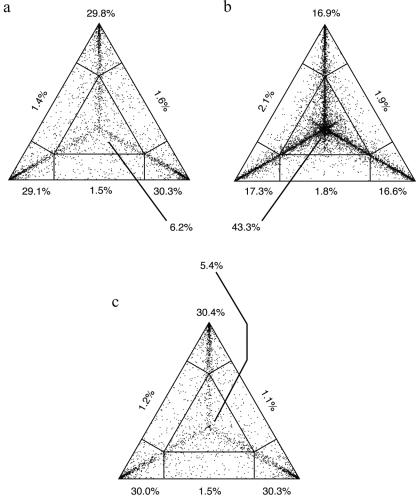
To investigate whether the ambiguous clustering in the 5′NCR was due to a low phylogenetic signal, we performed a comparative likelihood mapping analysis. Figure 4 demonstrates that the VP1 and 5′NCR sequence data clearly showed distinct patterns. While the VP1 data set had only about 6% of dots in the center of the triangle, representing phylogenetic noise, the 5′NCR data had a considerable number of quartets with uncertain topologies. The results for a concatenated alignment of both genomic regions showed a pattern similar to that of the VP1 data (Fig. 4c). In agreement with the clustering analysis, this analysis indicates that the 5′NCR data did not add contradictory or conflicting information, as would be expected if there occurred frequent recombination between the regions, but simply contributed little phylogenetic information.
FIG. 4.
Likelihood mapping results for VP1 (a), 5′NCR (b), and a concatenated alignment (c).
CVB3-specific long-range PCR.
The two CVB3 specimens clustered with different serotypes in the 5′NCR tree; this finding could be the consequence of a previous recombination event, or the partial CVB3 5′NCR and VP1 sequences could be derived from two different viruses present in a coinfected sample (virus mixture). A long-range RT-seminested PCR assay was therefore developed to amplify the entire region ranging from the second half of the 5′NCR to the first half of VP1 in the viral genome (2,483 nucleotides) (Fig. 1). However, sequencing analysis of the CVB3-specific long-range fragment showed that for both specimens, the initially determined 5′NCR and VP1 sequences were derived from a single CVB3 genome fragment, excluding the possibility of coinfection. Recombination could be an option, although the two CVB3 5′NCR sequences showed a nucleotide sequence divergence of only 2.7%. This divergence value was even lower than several 5′NCR sequence divergence values for other genotypes that clearly formed distinctive clusters in the 5′NCR tree (Table 3 and Fig. 2).
DISCUSSION
Two distant regions in the enteroviral genome were compared for their serotype-specific genetic information content in an epidemiological survey involving 342 Belgian patients with aseptic meningitis. Fragments from the second half of the highly conserved 5′NCR were generated following an initial diagnostic RT-PCR, whereas partial sequences corresponding to the 5′ end of VP1 were obtained with a VP1 RT-nested PCR assay. Both assays were developed in a previous study with the aim of accomplishing nearly universal enterovirus diagnosis (5′NCR PCR) and adequate molecular typing of members of the enterovirus B species directly from clinical specimens (VP1 PCR) (38). Based on the VP1 sequence data, the enterovirus serotypes were unambiguously identified in all patient samples. In general, from 1999 to 2002, echovirus 30, echovirus 13, echovirus 18, and echovirus 6 were identified most frequently.
The epidemiological nature of echovirus 30 has been extensively studied over the past few years (2, 19, 25, 33). It has been suggested that echovirus 30 follows an epidemic mode of transmission, causing large outbreaks followed by periods of apparent quiescence, rather than an endemic mode with a constant isolation frequency (19). In the same context, Palacios et al. (25) reported that the molecular epidemiology of echovirus 30 can be compared to influenza virus epidemiology, with a single lineage appearing to circulate worldwide at a time. Phylogenetic analysis of our echovirus 30 VP1 data (subtypes E30-a, E30-b, and E30-c) and the entire echovirus 30 VP1 data set in GenBank confirmed that circulating E30 strains at present belong to the same lineage (data not shown). On the basis of our VP1 tree, it seemed that the E30-c subtype might have evolved from E30-a (Fig. 2). We also observed a displacement from E30-b to E30-c in October 2000. The E30 genetic variants described in our study appeared to be quite similar antigenically. Genomic differences resided mainly at synonymous sites or resulted in structurally conservative amino acid changes. Displacements between these E30 variants or evolution from one E30 subtype to another was consequently not driven by host immune escape (antigenic drift).
Echovirus 13 and echovirus 18, which were rarely reported previously, emerged in Belgium as predominant serotypes in 2000 and 2001, respectively. Many reports about recent outbreaks of echoviruses 13 and 18 indeed showed the emergence of echovirus 13 in Europe in 2000 and in the United States in 2001, while echovirus 18 emerged in 2001 both in Europe and in the United States (3, 6, 7, 11, 28, 41). Echovirus 13 existed in Belgium as a single genotype—most likely the same genotype that is responsible for all recent echovirus 13 outbreaks worldwide (Fig. 2 and Table 3). Two echovirus 18 genotypes (E18-a and E18-b) were observed; they showed a VP1 nucleotide sequence divergence of approximately 9%. Again, as for the E30 subtypes, the E18 subtypes were quite similar at the amino acid level; therefore, E18-b probably did not arise as an antigenic variant. The same observations were made for the other enterovirus serotypes encountered in our study—which existed as multiple genotypes or subtypes. Echovirus 6, coxsackievirus B5, echovirus 25, coxsackievirus B4, coxsackievirus A9, and echovirus 9 all had high levels of intraserotypic genetic variations, but these variations generally resulted in minor amino acid variations, with most amino acid substitutions being phenotypically neutral. These findings are in agreement with the previous suggestion that RNA viruses evolve rather conservatively and change more than they adapt (31).
Echovirus 6 and coxsackievirus B5 were encountered in our study with a peak incidence in 2000. For the United States, coxsackievirus B5 was the serotype detected most commonly in 2000, whereas echovirus 6 appeared consistently among the 15 most common serotypes each year from 1993 to 2001 (6). Echovirus 11 dominated in 1999 in Belgium, consistent with recently reported rates of isolation of echovirus 11 in the United States (22).
The partial 5′NCR sequences obtained in our study corresponded to secondary structure domain V in the highly conserved 5′NCR (34). In general, human enterovirus phylogenetic analyses based on the 5′NCR have revealed only two major genetic clusters, a poliovirus-like cluster including the enterovirus C and D species and the polioviruses (5′NCR group I) and a coxsackievirus B-like cluster containing species A and B (5′NCR group II) (10, 27). While four clades exist in the rest of the enteroviral genome (A, B, C, and D), the existence of only two clades in the 5′NCR was suggested to be a consequence of strong evolutionary restrictions in this region due to the formation of secondary structures that are of major importance in viral translation and genome replication (27, 32).
The phylogenetic trees shown in Fig. 2 include only members of the enterovirus B species, which are the major cause of aseptic meningitis. The general topologies of both trees are not totally consistent, a fact which could be an indication of possible recombination events involving the 5′NCR. However, little is known about recombination events in the 5′NCR, and it would be rather hazardous to draw conclusions on this topic based on our 5′NCR tree. Our 5′NCR data reflect only a small portion of a highly conserved region, and clustering was therefore not highly supported by bootstrap analysis. The likelihood mapping analysis of a concatenated VP1-5′NCR alignment also indicated that the 5′NCR data did not add contradictory or conflicting information, as would be expected if recombination between the regions had occurred (Fig. 4).
In the 5′NCR tree, we observed a number of separate clusters which included members of a single enterovirus subtype. The 5′NCR phylogram tentatively displayed a subtype-specific clustering pattern, although subtypes of a given serotype were not monophyletic in this region. In most instances, we were able to predict the enterovirus serotype (or subtype) from the 5′NCR phylogram when the serotype was identified on the basis of VP1 for at least one member of each particular cluster, and we used this information to choose the most suitable primer set for our VP1 molecular typing assay. Recently, Siafakas et al. (35) reported the use of a simple restriction fragment length polymorphism assay for reliable enterovirus subclassification into five genetic groups on the basis of the 5′NCR. We suggest that, based on their and our observations, one should not underestimate the suggestive value of the 5′NCR with respect to serotype identification and the use of this knowledge for further investigation. However, our likelihood mapping analysis showed a particularly low phylogenetic information content in the 5′NCR (Fig. 4); such content can hardly result in the fully resolved topologies necessary for unambiguous molecular typing. For reliable serotyping, we therefore advise that serotype identification be confirmed by phylogenetic analysis with VP1 sequences. However, the phylogenetic information content of the 5′NCR may be increased by including a larger region in the analysis because the phylogenetic signal is influenced by two factors, degree of conservation and length of sequence.
In conclusion, our epidemiological survey comprised a large number of enterovirus samples obtained in Belgium from 1999 to 2002. We observed a serotype distribution that largely corresponded with observations in other European countries and the United States. In general, the level of intraserotypic genetic variation was rather high but did not result in antigenic variation. Our data confirm that reliable molecular typing can be obtained only from VP1 analyses, since the VP1 gene displays a superior phylogenetic signal compared to the 5′NCR. However, the suggestive value of the 5′NCR can be exploited in VP1 molecular typing strategies.
Acknowledgments
We thank our colleagues at the Laboratory of Clinical and Epidemiological Virology for helpful comments and critical reading of the manuscript.
I.T. was supported by a fellowship from the FWO, Brussels, Belgium. P.L. was supported by the Flemish Institute for Scientific-Technological Research in Industry. A.M.L. was supported by the Knowledge Foundation, Stockholm, Sweden. This work was also supported by the Flemish Fund for Scientific Research (grant G.0288.01).
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Bailly, J.-L., D. Brosson, C. Archimbaud, M. Chambon, C. Henquell, and H. Peigue-Lafeuille. 2002. Genetic diversity of echovirus 30 during a meningitis outbreak, demonstrated by direct molecular typing from cerebrospinal fluid. J. Med. Virol. 68:558-567. [DOI] [PubMed] [Google Scholar]
- 3.Böttner, A., S. Daneschnejad, W. Handrick, V. Schuster, U. G. Liebert, and W. Kiess. 2002. A season of aseptic meningitis in Germany: epidemiologic, clinical and diagnostic aspects. Pediatr. Infect. Dis. J. 21:1126-1132. [DOI] [PubMed] [Google Scholar]
- 4.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ′serotyping' of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 5.Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Enterovirus surveillance—United States, 2000-2001. Morb. Mortal. Wkly. Rep. 51:1047-1049. [PubMed] [Google Scholar]
- 7.Chomel, J.-J., D. Antona, D. Thouvenot, and B. Lina. 2003. Three echovirus serotypes responsible for outbreak of aseptic meningitis in Rhone-Alpes region, France. Eur. J. Clin. Microbiol. Infect. Dis. 22:191-193. [DOI] [PubMed] [Google Scholar]
- 8.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland, J., and E. Domingo. 1998. Origin and evolution of viruses. Virus Genes 16:13-21. [DOI] [PubMed] [Google Scholar]
- 10.Hyypiä, T., T. Hovi, N. J. Knowles, and G. Stanway. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Kirschke, D. L., T. F. Jones, S. C. Buckingham, A. S. Craig, and W. Schaffner. 2002. Outbreak of aseptic meningitis associated with echovirus 13. Pediatr. Infect. Dis. J. 21:1034-1038. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 13.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 14.Minor, P. D. 1990. Antigenic structure of picornaviruses. Curr. Top. Microbiol. Immunol. 161:121-154. [DOI] [PubMed] [Google Scholar]
- 15.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. Embnet. News 4:1-4. [Google Scholar]
- 16.Norder, H., L. Bjerregaard, and L. O. Magnius. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable for typing. J. Med. Virol. 63:35-44. [PubMed] [Google Scholar]
- 17.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberste, M. S., K. Maher, M. L. Kennett, J. J. Campbell, M. S. Carpenter, D. Schnurr, and M. A. Pallansch. 1999. Molecular epidemiology and genetic diversity of echovirus type 30 (E30): genotypes correlate with temporal dynamics of E30 isolation. J. Clin. Microbiol. 37:3928-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberste, M. S., D. Schnurr, K. Maher, S. al-Busaidy, and M. A. Pallansch. 2001. Molecular identification of new picornaviruses and characterization of a proposed enterovirus 73 serotype. J. Gen. Virol. 82:409-416. [DOI] [PubMed] [Google Scholar]
- 22.Oberste, M. S., W. A. Nix, D. R. Kilpatrick, M. R. Flemister, and M. A. Pallansch. 2003. Molecular epidemiology and type-specific detection of echovirus 11 isolates from the Americas, Europe, Africa, Australia, southern Asia and the Middle East. Virus Res. 91:241-248. [DOI] [PubMed] [Google Scholar]
- 23.Oberste, M. S., W. A. Nix, K. Maher, and M. A. Pallansch. 2003. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26:375-377. [DOI] [PubMed] [Google Scholar]
- 24.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 25.Palacios, G., I. Casas, D. Cisterna, G. Trallero, A. Tenorio, and C. Freire. 2002. Molecular epidemiology of echovirus 30: temporal circulation and prevalence of single lineages. J. Virol. 76:4940-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pöyry, T., L. Kinnunen, T. Hyypiä, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 28.Quirk, M. 2001. Echovirus to be considered in meningitis diagnosis. Lancet Infect. Dis. 1:220. [Google Scholar]
- 29.Romero, J. R. 1999. Reverse-transcription polymerase chain reaction detection of the enteroviruses. Arch. Pathol. Lab. Med. 123:1161-1169. [DOI] [PubMed] [Google Scholar]
- 30.Rotbart, H. A. 1995. Meningitis and encephalitis, p. 271-289. In H. A. Rotbart (ed.), Human enterovirus infections. ASM Press, Washington, D.C.
- 31.Sala, M., and S. Wain-Hobson. 2000. Are RNA viruses adapting or merely changing? J. Mol. Evol. 51:12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santti, J., T. Hyypiä, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savolainen, C., T. Hovi, and M. N. Mulders. 2001. Molecular epidemiology of echovirus 30 in Europe: succession of dominant sublineages within a single major genotype. Arch. Virol. 146:521-537. [DOI] [PubMed] [Google Scholar]
- 34.Siafakas, N., P. Markoulatos, and G. Stanway. 2002. Molecular classification of coxsackie A viruses on the basis of the 5′-UTR: structural and evolutionary aspects. J. Mol. Evol. 55:638-652. [DOI] [PubMed] [Google Scholar]
- 35.Siafakas, N., P. Markoulatos, C. Vlachos, G. Stanway, G. Tzanakaki, and J. Kourea-Kremastinou. 2003. Molecular sub-grouping of enterovirus reference and wild type strains into distinct genetic clusters using a simple RFLP assay. Mol. Cell. Probes 17:113-123. [DOI] [PubMed] [Google Scholar]
- 36.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 38.Thoelen, I., P. Lemey, I. Van der Donck, K. Beuselinck, A. M. Lindberg, and M. Van Ranst. 2003. Molecular typing and epidemiology of enteroviruses identified from an outbreak of aseptic meningitis in Belgium during the summer of 2000. J. Med. Virol. 70:420-429. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trallero, G., I. Casas, A. Avellón, C. Pérez, A. Tenorio, and A. De La Loma. 2003. First epidemic of aseptic meningitis due to echovirus type 13 among Spanish children. Epidemiol. Infect. 130:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]