Abstract
Whole-house surveillance for healthcare-associated infection is no longer the recommended practice because of the large personnel time investment required. We developed a computer-based tracking system using microbiologic data as an aid in detecting potential outbreaks of healthcare-associated infections on a hospital-wide basis. Monthly total isolates of 25 clinically significant hospital pathogens were tallied from 1991 to 1998 to form a database for future comparison. Two different algorithm tools (based on increases of organism numbers over baseline) were applied to determine alert thresholds for suspected outbreaks using this information. The first algorithm (2SD) defined an alert as two standard deviations above the mean monthly number of isolates. The second (MI) defined an alert as either a 100% increase from the baseline organism number over 2 months or a ≥50% increase (compared to baseline) during a three-consecutive-month period. These two methods were compared to standard infection control professional surveillance (ICP) for the detection of clonal outbreaks over 12 months. Overall, a total of seven clonal outbreaks were detected during the 1-year study. Using standard methods, ICP investigated nine suspected outbreaks, four of which were associated with clonal microbes. The 2SD method signaled a suspected outbreak 15 times, of which three were clonal and ICP had detected one. The MI method signaled a suspected outbreak 30 times; four of these were clonal, and ICP had detected one. The sensitivity and specificity values for ICP, 2SD, and MI for detecting clonal outbreaks were 57, 43, and 57% and 17, 83, and 67%, respectively. Statistical methods applied to clinical microbiology laboratory information system data efficiently supplement infection control efforts for outbreak detection.
A focus in modern healthcare is to reduce the incidence of medical errors, a major source of which are healthcare-associated infections (22; M. J. Berens, Chicago Tribune, July 21, 22, and 23 [p. 1, 14-16; 1, 11; 1, 8-10], 2002). Not only do nosocomial infections add to excess morbidity and mortality, but they also lead to potentially avoidable costs, often amounting to $15,000 to $35,000 per affected patient (19, 22). In addition, healthcare organizations are faced with controlling the spread of emerging and reemerging pathogens, especially drug-resistant bacteria that are the cause of many nosocomial infections. Many years ago it was recognized that the microbiology laboratory can help in this effort by monitoring its extensive data generated from culture results for patterns of infection (4, 8, 11, 20). The laboratory holds the earliest opportunity to detect specific organism or antimicrobial resistance patterns emerging in the hospital. Efficiently doing so, followed by communication with infection control, can be a key enhancement to detection and the elimination of potential microbial outbreaks (6, 16). Each month a typical microbiology laboratory generates many billions of potentially useful pieces of information drawn from all parts of the healthcare organization. Ideally, if microbiologic data can be collected and efficiently analyzed, monthly organism totals could be compared to a threshold, with outliers noted as a “suspected outbreak(s)” and discussed with infection control for appropriate action (17). However, given the limited resources available to microbiology laboratories, such a strategy must be rapid, require little additional personnel time, and be easy to use for widespread implementation.
Our goal here was to create a monitoring system using microbiologic data that could (i) identify trends in an organism population in a timely manner in order to highlight where infection control intervention might be useful, (ii) be sensitive enough to identify most suspected outbreaks and yet specific so that positive results signaled meaningful problems, and (iii) be easily used in any microbiology laboratory with trained microbiology technologists.
MATERIALS AND METHODS
During the study period (1 January through 30 December 1999) Northwestern Memorial Hospital (NMH) was a 688-bed tertiary care teaching facility with approximately 39,000 annual admissions, with 7,000 newborn deliveries, 56,000 emergency room visits, 115,000 home care, and 265,000 outpatient visits.
Definition of terms.
Two algorithm tools were developed. The first algorithm, 2SD (described in detail below), defined an alert as two standard deviations above a mean. The second, MI (also described in detail below), defined an alert as either a 100% increase from the baseline organism number over 2 months or a ≥50% increase during a three-consecutive-month period. An “alert” was defined as any time either 2SD or MI signaled the possibility of an outbreak, as indicated by the threshold being exceeded for their predetermined algorithms. To determine the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for standard infection control practice, 2SD, and MI surveillance, results were compared to the identification of any clonal outbreak during the 12-month study period. A “clonal outbreak” is defined as a potential outbreak that subsequently was determined to be due to genetically identical microbial pathogens. A “potential outbreak” was an event identified by 2SD, MI or the Infection Control Professionals' (ICP) routine practice and was investigated by molecular DNA fingerprinting analysis of the suspected organisms. A “suspected outbreak” was an event identified by 2SD, MI, or ICP methods that was discussed as a candidate for molecular DNA fingerprinting analysis. Therefore, for the computerized methods, a total of 300 suspected outbreaks may have occurred (25 organisms times 12 months). There was no limitation on the number of suspected outbreaks that could have been identified by the ICP method. We defined an outbreak as an increase in the rate of nosocomial infection above that noted in the past (24). Since the spread of a clonal organism from one patient to another is an undesired goal, the definition of an outbreak was minimally two patients on the same nursing unit or related nursing units with a nosocomial infection due to a single clonal microbial strain.
Statistical analysis.
Sensitivity was determined by the number of clonal outbreaks detected by all three surveillance approaches; there were seven such outbreaks in all that formed the performance standard for outbreak detection. Specificity was determined by the performance of the three approaches in detecting the 7 clonal outbreaks within the group of 13 total potential outbreaks throughout the year that were investigated by genetic typing.
Creating the database.
In this hypothesis-based computer model, we selected our 25 most common hospital pathogens, purposefully including drug-resistant bacteria, for surveillance (Table 1). The data were available for these organisms in the hospital laboratory information system (Sunquest, Tucson, Ariz.) from 1991 to 1998. The isolate totals for each month of every year were derived from computer generated MIC susceptibility reports, with duplicates removed by a defined algorithm. A duplicate is defined by Sunquest as the same organism from the same patient from the same source in the same month with the identical susceptibility pattern. Slight alterations in the susceptibility pattern of an isolate will result in counting some organisms in replicate. Thus, most but not all replicates were removed. Organism totals included isolates from all inpatient units and outpatient sites. The monthly totals for each organism were entered into an Excel (Microsoft Corp., Redmond, Wash.) spreadsheet for data analysis.
TABLE 1.
Organisms selected for use in the database for the 2SD and MI algorithm tools
| Strain | Strain | |
|---|---|---|
| Acinetobacter calcoaceticus/baumannii complex | ||
| Citrobacter koseri | ||
| Citrobacter freundii | ||
| Clostridium difficile | ||
| Escherichia coli | ||
| Enterobacter aerogenes | ||
| Enterobacter cloacae | ||
| Enterococcus faecalisa | ||
| Enterococcus faecalisb | ||
| Enterococcus faeciuma | ||
| Enterococcus faeciumb | ||
| Haemophilus influenzae | ||
| Klebsiella oxytoca | ||
| Klebsiella pneumoniae | ||
| Proteus mirabilis | ||
| Pseudomonas aeruginosa | ||
| Staphylococcus epidermidisc | ||
| Staphylococcus aureusc | ||
| Serratia marcescens | ||
| Streptococcus pneumoniae | ||
| Stenotrophomonas maltophilia |
Vancomycin-susceptible and -resistant strains combined.
VRE strains only, with clinical and surveillance strains segregated.
Oxacillin-susceptible and -resistant strains segregated.
Analysis of variance (2SD) tool.
Analysis of variance (Excel; two factor analysis of variance) was used to identify significant differences (P < 0.05) in the mean organism totals by month and year (Table 2). If differences between yearly and monthly means were not statistically significant, it was assumed for the algorithm that the rate was sufficiently constant, and the threshold for a suspected outbreak was defined as the overall mean plus two standard deviations (to minimize the effect of natural variation). If the monthly mean organism totals differed significantly (P < 0.05) with similar patterns in different seasons of a single year, then the data were divided into “low months” and “high months” and separate thresholds determined for each time period by using the above method in order to achieve constancy (in the absence of an outbreak) from year to year. High-month periods were defined as the month with the highest month-specific mean organism total plus the preceding and subsequent two months. Low-month periods included the remaining seven months. When organism-specific data showed significant differences in annual means, the calculated thresholds were based on the most recent 5 years of data. This was also done to achieve constancy of the baseline data.
TABLE 2.
Example of 2SD: two-factor without replication using mean Citrobacter freundii organisms per month for 8 yearsa
| Mo | Occurrences of replication duringa:
|
Avg | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1991 | 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | ||
| Jan | 5 | 6 | 11 | 9 | 9 | 7 | 6 | 6 | 7 |
| Feb | 8 | 13 | 8 | 4 | 8 | 10 | 6 | 5 | 8 |
| Mar | 12 | 11 | 18 | 13 | 6 | 1 | 8 | 11 | 10 |
| Apr | 13 | 9 | 6 | 6 | 7 | 9 | 13 | 11 | 9 |
| May | 13 | 8 | 6 | 8 | 12 | 9 | 9 | 15 | 10 |
| Jun | 13 | 15 | 8 | 11 | 13 | 10 | 17 | 8 | 12 |
| Jul | 9 | 8 | 25 | 11 | 15 | 14 | 13 | 11 | 13 |
| Aug | 10 | 15 | 18 | 13 | 10 | 4 | 12 | 8 | 11 |
| Sep | 10 | 9 | 16 | 8 | 6 | 11 | 16 | 10 | 11 |
| Oct | 10 | 11 | 8 | 9 | 3 | 8 | 5 | 13 | 8 |
| Nov | 11 | 5 | 6 | 5 | 6 | 6 | 7 | 11 | 7 |
| Dec | 9 | 12 | 4 | 9 | 7 | 3 | 5 | 5 | 7 |
| Avg | 10 | 10 | 11 | 9 | 9 | 8 | 10 | 10 | 9 |
Values greater than the mean by two standard deviations are indicated in bold face. The upper boundary for the “high months” (May to Sept) is 19. The upper boundary for the “low months” (Oct to Apr) is 14.
Monthly increases (MI) tool.
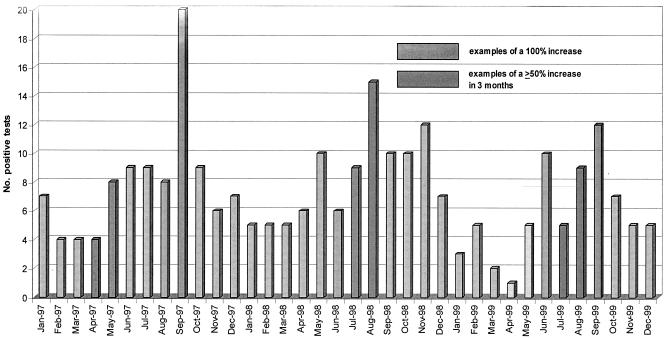
The current month's total for each organism was compared to the monthly totals of that organism for the two prior months in order to form a “rolling” 3-month analysis period. If a 100% increase occurred in the third month compared to the prior month or if a ≥50% total increase occurred over the course of a 3-month period, the organism increase triggered a suspected outbreak alert. An example of this is in Fig. 1.
FIG. 1.
Number of positive cultures per month with Acinetobacter baumannii complex analyzed using the MI algorithm (January 1997 to December 1999).
Suspected outbreak determination.
Suspected outbreak alerts by either MI or 2SD were presented to the NMH Infection Control and Prevention Department where a panel consisting of two physicians and the hospital's three infection control nurses, all experienced in infection control decided if further investigation was needed. These panelists remained constant throughout the year-long study period and were not blinded to the source of information provided; however, they were unaware a formal comparison of clonal outbreak detection was being done. Their decision regarding any need for further investigation was based on past experience of the participants, suggesting the likelihood of a nosocomial infection outbreak. Factors weighed as to the likelihood and potential importance of the microbiology results included the seriousness of the infection (e.g., Staphylococcus aureus in the blood versus S. epidermidis in surgical site infection), the organism being a multidrug-resistant pathogen (e.g., vancomycin-resistant enterococci [VRE]), and most importantly, the potential that there was probable contact spread of the detected microbe (e.g., multiple organisms of the same genus and species recovered from patients on a single nursing unit versus from several nursing units that did not share patient transfers). A decision to investigate the suspected outbreak, thus making it a potential outbreak, led to molecular typing of the target organism(s). If the panel made the decision that the suspected outbreak was unlikely to represent a true nosocomial infection problem, there was no further investigation.
Infection control initiated investigation.
Practices to determine whether hospital-associated infections may be occurring were based on current Centers for Disease Control and Prevention (CDC) recommendations for definition of a nosocomial infection (5). Nosocomial infections were detected by ongoing surveillance using trained nurses for intensive care units (ICUs), including a neonatal ICU, and postsurgery units applying standard nosocomial infection definitions. Additional data were collected through manual review of microbiology reports and patients' medical records, direct observation of medical and nursing practice, and active surveillance using rectal cultures of patients residing on nursing units caring for those with high-risk conditions. In addition, there was an evaluation of suspected nosocomial infections reported by healthcare providers throughout the hospital.
Time analysis.
For each trended organism that exceeded a 2SD or MI threshold, additional computer generated reports were done to determine whether there were any nursing unit trends, and if these appeared evident, the patient admission histories were reviewed to determine whether the positive cultures had been obtained more than 2 days after admission. If this revealed a high number of nosocomial positives, the potential for drug resistance or specimen source, then the trends were evaluated.
The use of microbiology laboratory data for development and validation of a tracking tool to enhance detection of nosocomial infections was approved by the Institutional Review Board of Northwestern University and the CDC.
RESULTS
The overall pattern of suspected and potential outbreak discovery is shown in Table 3. There were a total of 13 potential outbreaks during the study period. ICP detected nine potential outbreaks that they investigated during the 12 months of the present study, and four of these were determined to be due to clonal microorganisms. Of the 300 possible events in 1999 available to the computerized systems, 2SD signaled a suspected outbreak 15 times, and four became potential outbreaks, three of which were from clonal bacteria. One (clonal) of the fifteen was a potential outbreak already investigated by the ICP, and the other three were potential outbreaks investigated by ICP after detection by 2SD. Eleven reported suspected outbreaks were not investigated after panel discussion. For the same 300 potential events, MI signaled a suspected outbreak 30 times (Table 3), and six of these became potential outbreaks, four of which were determined to be clonal in origin. Two of these (one clonal, one nonclonal) were potential outbreaks ICP already was investigating.
TABLE 3.
Suspected outbreaks detected by the 2SD and MI algorithm tools during 1999
| Mo | Organism | DNA typing resulta | Infection control ini- tiated inves- tigation | Flagged by the:
|
|
|---|---|---|---|---|---|
| 2SD method | MI method | ||||
| Jan | Clinical VREfaecium | Clonal | No | Yes | Yes |
| Feb | S. maltophilia | Clonal | Yes | No | No |
| Feb | C. freundii group | Not tested | No | Yes | No |
| Mar | S. marcescens | Clonal | Yes | No | No |
| Mar | E. coli | Not clonal | Yes | No | No |
| Mar | E. cloacae | Not clonal | Yes | No | No |
| Mar | C. difficile | Not tested | No | No | Yes |
| Mar | H. influenzae | Not tested | No | No | Yes |
| Apr | E. aerogenes | Not tested | No | No | Yes |
| May | C. diversus | Not tested | No | No | Yes |
| Jun | MSSA | Not clonal | Yes | No | No |
| Jun | A. anitratus/baumannii | Not tested | No | No | Yes |
| Jun | C. difficile | Not tested | No | No | Yes |
| Jun | K. pneumoniae | Not tested | No | No | Yes |
| Jun | MRSA | Not tested | No | No | Yes |
| Jun | S. marcescens | Not tested | No | No | Yes |
| Jul | H. influenzae | Not tested | No | No | Yes |
| Jul | K. pneumoniae | Not tested | No | No | Yes |
| Jul | P. mirabilis | Not tested | No | No | Yes |
| Aug | S. maltophilia | Not clonal | Yes | No | Yes |
| Aug | K. pneumoniae | Not clonal | Yes | No | No |
| Aug | Clinical VREfaecalis | Not tested | No | Yes | Yes |
| Aug | E. aerogenes | Not tested | No | Yes | No |
| Aug | MRSA | Not tested | No | Yes | No |
| Aug | Total VREfaecalis | Not tested | No | Yes | Yes |
| Sep | S. maltophilia | Clonal | Yes | Yes | Yes |
| Sep | P. aeruginosa | Clonal | Yes | No | No |
| Sep | Clinical VREfaecium | Not tested | No | Yes | Yes |
| Sep | A. anitratus/baumannii | Not tested | No | No | Yes |
| Sep | C. diversus | Not tested | No | No | Yes |
| Sep | S. pneumoniae | Not tested | No | No | Yes |
| Oct | Clinical VREfaecalis | Not tested | No | Yes | Yes |
| Oct | E. faecalis | Not tested | No | Yes | Yes |
| Oct | Total VREfaecalis | Not tested | No | Yes | No |
| Oct | C. difficile | Not tested | No | No | Yes |
| Oct | H. influenzae | Not tested | No | No | Yes |
| Nov | Total VREfaecium | Clonal | No | No | Yes |
| Nov | C. freundii group | Not clonal | No | Yes | Yes |
| Nov | S. marcescens | Not tested | No | No | Yes |
| Dec | Total VREfaecium | Clonal | No | Yes | Yes |
| Dec | C. difficile | Not tested | No | Yes | No |
| Dec | Clinical VREfaecium | Not tested | No | Yes | Yes |
| Total | 15 | 30 | |||
A result indicating “clonal” or “not clonal” indicates 1 of the 13 potential outbreaks that was investigated by molecular typing.
Comparison of results from applying the 2SD and MI methods to 1999 data shows that 2SD detected three of the seven clonal outbreaks and MI detected four. In one case, MI detected a potential outbreak 1 month sooner than 2SD. However, MI signaled twice as many suspected outbreaks as 2SD, suggesting the possibility of more false-positive alerts by using the more simplistic MI approach.
The sensitivity, specificity, PPV, and NPV for the detection of clonal outbreaks by the ICP, 2SD, and MI methods involving the 13 potential outbreaks during 1999 are compared in Table 4.
TABLE 4.
Sensitivity, specificity, PPV, NPV for ICP, 2SD, and MI for detecting a clonal outbreak
| Method (no. of potential outbreaks) | % Sensitivity | % Specificity | PPV (%) | NPV (%) |
|---|---|---|---|---|
| ICP (6) | 57.1 | 16.7 | 44.4 | 25.0 |
| 2SD (4) | 42.9 | 83.3 | 75.0 | 55.6 |
| MI (6) | 57.1 | 66.7 | 66.7 | 57.1 |
Two clinical infection control interventions were initiated based on information gathered from the trend 2SD and MI analysis tools that gave an alert signal for a suspected outbreak. In January of 1999, 2SD and MI analyses signaled a suspected outbreak for clinical isolates of vancomycin-resistant Enterococcus faecium (VREF). Examination of patient room assignments indicated that one nursing unit had three patients with VREF. The molecular epidemiology laboratory typed the isolates of VREF and found that all three isolates were the same genetic type (clonal). Further investigation indicated that one of the patients had been positive with this strain type in the past, while the other two represented new acquisitions, suggesting evidence of nosocomial transmission. As per our infection control procedure, we informed the nursing unit of the results and reeducated the personnel on standard infection control practices. The following month, only one new positive patient with this strain of VREF was found on the unit, and the overall number of clinical isolates of VREF returned to baseline. This small cluster of VREF illustrates the typical pattern of nosocomial transmission at our institution, in the form of “miniclusters” (23).
In November and December 1999, MI and 2SD analyses (2SD gave an alert only in December) again signaled a suspected outbreak for both clinical and surveillance isolates of VREF. Based upon patient room assignments, three nursing units had multiple patients with VREF in November and five nursing units had multiple patients with VREF in December. Typing of these isolates revealed that three of the suspect nursing units each had three to five patients with a common genomic strain type in the course of 1 month. Importantly, each unit had three to four patients with the identical VREF clone that were positive for the first time, indicating the high likelihood that a nosocomial outbreak of VREF had been detected.
The greatest expenditure of time was the initial creation of the database. The ongoing process of retrieving statistics electronically from the Sunquest susceptibility (MIC) reports required approximately 2 h of operator time per month, plus the time needed to assess each suspected outbreak to determine whether further investigation was needed. Overall, these trend analyses used in our report can be done over the course of one 8-h working day each month, provided that the organism numbers are easily gathered from the laboratory information system using minimal operator time. During the period of the present study, there were three full-time infection control professionals assigned to the inpatient services at NMH.
DISCUSSION
There is considerable current interest in developing statistical “rules” for application to surveillance. These range from relatively simple analysis of measures of change indicator infection rates from historical data (12, 14, 21) to sophisticated association rules for the application of data mining to microbiology laboratory culture results (2). A rationale for this approach is exemplified by Mylotte, who described a diseased-based observational system for identifying possible outbreaks in long-term care (13). Mylotte's analysis was done in an attempt to provide an objective evaluation of surveillance data in order to standardize outbreak detection. Threshold testing has been suggested as a means of using trends to objectively define alerts as part of an infection control surveillance strategy (1, 7). The data from Parkhurst et al. using threshold analysis of microbiology culture reports suggest this approach detected potential problems more rapidly than did standard observational surveillance (15). Recently, surveying microbiology laboratory data was evaluated as a tool for detection of nosocomial infections in an ICU and found to be comparable to routine infection surveillance (25). However, we are unaware that algorithms such as those we describe have been applied to microbiology laboratory information system data on a hospital-wide basis and subsequently compared to standard infection control surveillance practice.
Selecting a “gold standard” for a comparison such as this is difficult because any system may miss at least some outbreaks. We chose to compare detection of clonal events to provide an objective measure of performance between the varied approaches, even though no single one may be optimal. Support to the findings of our investigation comes from the recent report by Poulakou et al., who compared analysis of microbiology information system data to two other computerized surveillance approaches and to a reference standard in which each patient in the general and cardiac surgery department was monitored for surgical-site infection (G. Poulakou, A. Chalfine, A. Ben Ali, D. Cauet, J. Gonot, F. Goldstein, and J. Carlet, Prog. Abstr. 5th Eur. Cong. Chemother. Infect., abstr. FP1.01, 2003). These researchers found that monitoring microbiology laboratory data performed the best, with a sensitivity exceeding 80%, for the detection of these infections (Poulakou et al., Prog. Abstr. 5th Eur. Cong. Chemother. Infect.). Our results suggest that the current practice of focused infection control surveillance to detect nosocomial infections can be supplemented through the use of data generated in the clinical microbiology laboratory, even when relatively rudimentary computer tools based on threshold analysis are utilized. The 2SD and, particularly, the MI methods of analysis using monthly microbiologic data have potential use as an adjunct to routine outbreak detection. These methods met most of our objectives for enhancing infection control activity by identifying trends in a timely manner, while demonstrating reasonable sensitivity and specificity. The results are comparable to the 84% sensitivity and 48% specificity suggested by Laxson et al. in 1984 when they used positive cultures from the microbiology laboratory to detect possible nosocomial infections (11). One year later Schifman and Palmer demonstrated that by using excess rates of positive cultures, on a location-specific basis, far out-performed traditional surveillance in detecting small clusters of cross-infections (20). More recently, Glenister et al. used manual review of microbiology laboratory results to detect hospital-acquired infections and found a sensitivity similar to ours, depending on whether laboratory test results were only reviewed (48%) or if there was a twice-weekly follow-up visit to the nursing unit for a discussion of potential infections (71%). These surveillance approaches required only one-sixth to one-third the amount of time as their standard surveillance method (8).
One important aspect of the computer tools we describe here is that they can be used without the need for more resources in the microbiology laboratory, other than the time for data retrieval and trend analysis that was approximately 1 day per month for our healthcare organization. Significantly, not all clonal events during the study period were detected with routine infection control surveillance methods, indicating that there is potential for using the microbiology information system to identify nosocomial outbreaks of infection that currently go undetected using current standard practices. Our concurrent evaluation of 2SD and MI methods, along with routine infection control surveillance, found four suspected outbreaks that were not detected by standard practice methods, and three of these events proved to involve clonal isolates. Furthermore, 2SD analysis detected 11, and MI analysis detected 24 suspected outbreaks that were never investigated.
Interestingly, the MI method identified one clonal outbreak a month sooner than the 2SD approach (VREF in November), which reinforces the remainder of our data suggesting that MI is the more sensitive of the tools, with similar specificity. Another benefit of MI is that organism trends are noted as the numbers are increasing and a developing problem can be seen over a period of 2 to 3 months, even before the number meets the 2SD upper boundary to signal a problem. The possible disadvantage of MI is that it signaled twice as many suspected outbreaks as 2SD (including one proven nonclonal event for which 2SD did not generate an alert), indicating potentially too much monthly variation in recovered organisms to make MI the optimal surveillance tool.
We desired a method that was sensitive enough to identify all suspected outbreaks while sufficiently specific so that a suspected outbreak is likely to represent a true nosocomial infection event. When suspected outbreaks identified by the 2SD method were compared to the clonal outbreaks investigated by the ICP approach, 2SD analysis did not signal four of the seven outbreaks that were eventually detected and thus detected 43% of the clonal outbreaks. The MI tool performed better than the 2SD method and detected four of the seven clonal outbreaks (57%) found during 1999, which was equivalent to standard infection control practice performance. This relative lack of sensitivity for the current computer tools may be due to the fact that all of the clonal outbreaks investigated during this 1999 study consisted of small numbers (three to five) of patients. Databases using hospital-wide figures may mask small outbreaks that occur on individual units. This limitation is highlighted by the report of Price et al., who detected an outbreak of bacteremia in an outpatient dialysis unit using the 2SD and MI approaches (18). Although these algorithms did recognize the potential problem that had not been detected by routine infection control surveillance, detection required some 8 months and only occurred when the bacteremias in a given month were primarily associated with a single bacterial genus (Enterococcus sp.). The sensitivity of computerized analysis may increase if unit specific databases for detecting outbreaks were created. If unit specific trends were to be monitored, the organisms chosen for the database could be selected based on the population of patients on the unit surveyed and include important drug-resistant organisms so as to increase outbreak detection sensitivity.
In conclusion, an active, integrated infection control program has a major positive medical and economic impact on a healthcare organization (10). Our data demonstrates that a fully manual surveillance approach does not capture all of the potential outbreaks in an active medical center practice. Over 25 years ago it was recognized that computer tools could help the hospital epidemiologist and infection control committee in managing healthcare-associated infectious diseases (9). Newer reports have confirmed that hypothesis (1). We found that monitoring the activity of 25 important nosocomial pathogens with the 2SD and MI methods did enhance our infection control program by detecting four potential outbreaks not investigated by the Infection Control and Prevention Department in 1999, three of which were due to the unsuspected spread of clonal organisms. The amount of additional work required for use of these two tools totaled approximately 8 h per month. New computerized tools for increased detection of potential infection control opportunities are rapidly being developed (3) and offer great potential for system-wide surveillance in detecting healthcare-associated infectious diseases (16). After this investigation the 2SD and MI tools were routinely used in our infection control program and continued to supplement outbreak detection at our medical center (18). We have recently adopted a new computer tool with considerably enhanced analysis capacity and over the next 12 months will compare the data mining system to the 2SD and MI analysis methods (16). Use of statistical methods and information technology to analyze microbiology culture result data can supplement traditional infection control outbreak detection in an efficient and cost-effective manner to enhance infection control practice.
Acknowledgments
This study was supported by Public Health Service grant UR8/CCU515081 from the Centers for Disease Control and Prevention and Excellence in Academic Medicine grant 142 from the State of Illinois.
REFERENCES
- 1.Birnbaum, D. 1984. Analysis of hospital infection surveillance data. Infect. Control 5:332-538. [DOI] [PubMed] [Google Scholar]
- 2.Brossette, S. E., A. P. Sprague, J. M. Hardin, K. B. Waites, W. T. Jones, and S. A. Moser. 1998. Association rules and data mining in hospital infection control and public health surveillance. J. Am. Med. Inform. Assoc. 5:373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brossette, S. E., A. P, Sprague, W. T. Jones, and S. A. Moser. 2000. A data mining system for infection control surveillance. Methods Infect. Med. 39:303-310. [PubMed] [Google Scholar]
- 4.Courcol, R. J., F. F. Saulnier, A. V. Durocher, F. E. Wattel, and G. R. Martin. 1987. Computerized colonization-surveillance based on antimicrobial susceptibility patterns. Eur. J. Epidemiol. 3:243-246. [DOI] [PubMed] [Google Scholar]
- 5.Emori, T. G., D. H. Culver, T. C. Horan, W. R. Jarvis, J. W. White, D. R. Olson, S. Banerjee, J. R. Edwards, W. J. Martone, R. P. Gaynes, and J. M. Hughes. 1991. National nosocomial infections surveillance system (NNIS): description of surveillance methods. Am. J. Infect. Control. 19:19-35. [DOI] [PubMed] [Google Scholar]
- 6.Emori, G. T., and R. P. Gaynes. 1983. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, R. S., J. P. Burke, D. C. Classen, R. M. Gardner, R. L. Menlove, K. M. Goodrich, L. E. Stevens, and S. L. Pestotnik. 1992. Computerized identification of patients at high risk for hospital-acquired infection. Am. J. Infect. Control 20:4-10. [DOI] [PubMed] [Google Scholar]
- 8.Glenister, H. M., L. J. Taylor, C. L. R. Bartlett, E. M. Cooke, J. A. Sedgwick, and C. A. Mackintosh. 1993. An evaluation of surveillance methods for detecting infections in hospital inpatients. J. Hosp. Infect. 23:229-242. [DOI] [PubMed] [Google Scholar]
- 9.Gooch, J. J., and D. D. Wood. 1976. Computerized system analyzes epidemiological data. Hospitals 50:91-96. [PubMed] [Google Scholar]
- 10.Hacek, D. M., T. Suriano, G. A. Noskin, J. Kruszynski, B. Reisberg, and L. R. Peterson. 1999. Medical and economic benefit of a comprehensive infection control program that includes routine determination of microbial clonality. Am. J. Clin. Pathol. 111:647-654. [DOI] [PubMed] [Google Scholar]
- 11.Laxson, L. B., M. J. Blaser, and S. M. Parkhurst. 1984. Surveillance for the detection of nosocomial infections and the potential for nosocomial outbreaks. I. Microbiology culture surveillance is an effective method of detecting nosocomial infection. Infect. Control 12:318-324. [DOI] [PubMed] [Google Scholar]
- 12.Morton, A. P., M. Whitby, M. L. McLaws, A. Dobson, S. McElwain, D. Looke, J. Stackelroth, and A. Sartor. 2001. The application of statistical process control charts to the detection and monitoring of hospital-acquired infections. J. Qual. Clin. Pract. 21:112-117. [DOI] [PubMed] [Google Scholar]
- 13.Mylotte, J. M. 1996. Analysis of infection control surveillance data in a long-term-care facility: use of threshold testing. Infect. Control Hosp. Epidemiol. 17:101-107. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien, S. J., and P. Christie. 1997. Do CuSums have a role in routine communicable disease surveillance? Public Health 111:255-258. [DOI] [PubMed] [Google Scholar]
- 15.Parkhurst, S. M., M. J. Blaser, L. B. Laxson, and W. L. Wang. 1985. Surveillance for the detection of nosocomial infections and the potential for nosocomial outbreaks. II. Development of a laboratory-based system. Am. J. Infect. Control. 13:7-15. [DOI] [PubMed] [Google Scholar]
- 16.Peterson, L. R., and S. E. Brossette. 2002. Hunting healthcare-associated infections from the clinical microbiology laboratory: passive, active, and virtual surveillance. J. Clin. Microbiol. 40:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaller, M. A., and L. A. Herwaldt. 1997. The clinical microbiology laboratory and infection control: emerging pathogens, antimicrobial resistance, and new technology. Clin. Infect. Dis. 25:858-870. [DOI] [PubMed] [Google Scholar]
- 18.Price, C. S., D. M. Hacek, G. A. Noskin, and L. R. Peterson. 2002. Outbreak of bloodstream infections in an outpatient hemodialysis center. Infect. Control Hosp. Epidemiol. 23:725-729. [DOI] [PubMed] [Google Scholar]
- 19.Roberts, R. R., R. D. Scott 2nd, R. Cordell, S. L. Solomon, L. Steele, L. M. Kampe, W. E. Trick, and R. A. Weinstein. 2003. The use of economic modeling to determine the hospital costs associated with nosocomial infections. Clin. Infect. Dis. 36:1424-1432. [DOI] [PubMed] [Google Scholar]
- 20.Schifman, R. B., and R. A. Palmer. 1985. Surveillance of nosocomial infections by computer analysis of positive culture rates. J. Clin. Microbiol. 21:493-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stern, L., and D. Lightfoot. 1999. Automated outbreak detection: a quantitative retrospective analysis. Epidemiol. Infect. 122:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stone, P. W., E. Larson, and L. N. Kawar. 2002. A systematic audit of economic evidence linking nosocomial infections and infection control interventions: 1990-2000. Am. J. Infect. Control. 30:145-152. [DOI] [PubMed] [Google Scholar]
- 23.Stosor, V., J. Kruszynski, T. Suriano, G. A. Noskin, and L. R. Peterson. 1999. Molecular epidemiology of vancomycin-resistant enterococci: a 2-year perspective. Infect. Control Hosp. Epidemiol. 20:653-659. [DOI] [PubMed] [Google Scholar]
- 24.Wenzel, R. P. 1987. Epidemics: identification and management, p. 94-108. In R. P. Wenzel (ed.), Prevention and control of nosocomial infections. The Williams & Wilkins Co., Baltimore, Md.
- 25.Zolldann, D., H. Haefner, C. Poetter, S. Buzello, D. Sohr, R. Luetticken, and S. W. Lemmen. 2003. Assessment of a selective surveillance method for detecting nosocomial infections in patients in the intensive care department. Am. J. Infect. Control 31:261-265. [DOI] [PubMed] [Google Scholar]