Abstract
Background
Several germline single nucleotide polymorphisms (SNPs) have been consistently associated with prostate cancer (PCa) risk.
Objective
To determine whether there is an improvement in PCa risk prediction by adding these SNPs to existing predictors of PCa.
Design, setting, and participants
Subjects included men in the placebo arm of the randomized Reduction by Dutasteride of Prostate Cancer Events (REDUCE) trial in whom germline DNA was available. All men had an initial negative prostate biopsy and underwent study-mandated biopsies at 2 yr and 4 yr. Predictive performance of baseline clinical parameters and/or a genetic score based on 33 established PCa risk-associated SNPs was evaluated.
Outcome measurements and statistical analysis
Area under the receiver operating characteristic curves (AUC) were used to compare different models with different predictors. Net reclassification improvement (NRI) and decision curve analysis (DCA) were used to assess changes in risk prediction by adding genetic markers.
Results and limitations
Among 1654 men, genetic score was a significant predictor of positive biopsy, even after adjusting for known clinical variables and family history (p = 3.41 × 10−8). The AUC for the genetic score exceeded that of any other PCa predictor at 0.59. Adding the genetic score to the best clinical model improved the AUC from 0.62 to 0.66 (p < 0.001), reclassified PCa risk in 33% of men (NRI: 0.10; p = 0.002), resulted in higher net benefit from DCA, and decreased the number of biopsies needed to detect the same number of PCa instances. The benefit of adding the genetic score was greatest among men at intermediate risk (25th percentile to 75th percentile). Similar results were found for high-grade (Gleason score ≥7) PCa. A major limitation of this study was its focus on white patients only.
Conclusions
Adding genetic markers to current clinical parameters may improve PCa risk prediction. The improvement is modest but may be helpful for better determining the need for repeat prostate biopsy. The clinical impact of these results requires further study.
Keywords: Prostate cancer, Genetics, AUC, Detection rate, Reclassification, SNPs, Prospective study, Clinical trial
1. Introduction
Prostate cancer (PCa) is the most common solid-organ malignancy affecting American men and the second leading cause of cancer-related death [1]. Approximately 1 million prostate biopsies are performed annually in the United States, with less than half proving positive for PCa. Patients with negative biopsies have an approximately 20% incidence of PCa at repeat biopsy [2]. Novel predictors are needed to better estimate an individual’s risk for PCa.
Recently, 33 PCa risk-associated single nucleotide polymorphisms (SNPs) have been identified from genome-wide association studies (GWAS) [3–13]. These SNPs have been consistently associated with PCa risk in multiple white case-control studies [14]. Several studies have reported that a genetic score based on a combination of these risk-associated SNPs can be used to predict an individual’s risk for PCa [15–18]. However, it is unclear whether they act independent of existing clinical variables to predict PCa risk and, more importantly, whether they add value to existing clinical variables in predicting biopsy outcomes. These questions are difficult to assess in retrospective case-control studies, because many existing clinical variables, such as prostate-specific antigen (PSA), are directly or indirectly used to define cases and controls in these studies.
The Reduction by Dutasteride of Prostate Cancer Events (REDUCE) trial is the largest randomized, placebo-controlled chemoprevention trial of men following an initial negative prostate biopsy for PCa [19]. All subjects were followed systematically and had study-mandated prostate biopsies, thus affording a unique opportunity to assess the predictive performance of all existing clinical variables and genetic markers. We hypothesize that the genetic score will add to the ability of the best clinical model for PCa prediction at repeat biopsy.
2. Patients and methods
2.1. Study population
Subjects included 1654 white men in the placebo arm of the REDUCE trial who were offered (offering began partway through the study) and consented to genetic studies. The REDUCE study has been described in detail elsewhere [19]. Briefly, participants were 50–75 yr of age, with a serum PSA between 2.5 and 10 ng/ml (men 50–60 yr of age) or between 3.0 and 10 ng/ml (men >60 yr of age), and had a negative prostate biopsy (6–12 cores) within 6 mo of study enrollment. The characteristics of the participants are presented in Table 1.
Table 1.
Baseline clinical, demographic, and genetic score data of the subjects in the study
| Variables | All subjects
|
||
|---|---|---|---|
| Positive biopsies (n = 410) | Negative biopsies (n = 1244) | p | |
| Baseline clinical variables | |||
| Age | |||
| Mean (SD), yr | 63.52 (5.99) | 62.22 (6.01) | 0.0001 |
| Median (range) | 63 (50–76) | 62 (49–76) | |
| No. (%) with positive DRE | 20 (5) | 47 (4) | 0.33 |
| Prostate volume | |||
| Mean (SD)* | 44.20 (21.40) | 46.76 (16.13) | 0.03 |
| Median (range) | 41.61 (9.01–257) | 45.46 (3.66–127) | |
| Total PSA levels | |||
| Mean (SD), ml* | 5.78 (1.37) | 5.52 (1.40) | 0.01 |
| Median (range), ml | 5.7 (2.5–10.2) | 5.7 (1.8–14.2) | |
| PSA density (PSA/PV) | |||
| Mean (SD)* | 0.14 (1.67) | 0.14 (0.07) | 4.67E-06 |
| Median (range) | 0.14 (0.02–0.63) | 0.12 (0.03–0.58) | |
| Free-to-total PSA ratio | |||
| Mean (SD) | 0.16 (0.06) | 0.17 (0.06) | 0.02 |
| Median (range) | 0.16 (0.03–0.47) | 0.16 (0.03–0.47) | |
| No. of cores sampled at baseline biopsy | |||
| Mean (SD) | 8.21 (2.27) | 8.58 (2.39) | 0.004 |
| Median (range) | 8 (3–19) | 8 (2–22) | |
| Baseline genetic variables | |||
| No. (%) with positive family history | 68 (17) | 146 (12) | 0.01 |
| Genetic score based on 33 PCa risk SNPs | |||
| Mean (SD)* | 1.10 (1.83) | 0.90 (1.82) | 4.15E-09 |
| Median (range) | 1.09 (0.25–6.98) | 0.88 (0.15–13.45) | |
SD = standard deviation; DRE = digital rectal examination; PSA = prostate-specific antigen; PV = prostate volume; PCa = prostate cancer; SNP = single nucleotide polymorphism.
Mean (SD) of prostate volume, total PSA, PSA density, and genetic score is antilogarithmic.
2.2. Prostate cancer risk-associated single nucleotide polymorphisms and single nucleotide polymorphism genotyping
All 33 PCa risk-associated SNPs discovered from GWAS reported prior to December 2009 were included; each exceeded genomewide significance levels in its initial reports ( p < 10.7), and each has been replicated in independent populations (Supplementary Table 1) [3–13]. SNPs were genotyped from peripheral blood DNA using the MassARRAY platform (Sequenom, San Diego, CA, USA). One duplicated Centre d’Etude du Polymorphisme Humain (CEPH) sample and two water samples (negative controls) were blinded to technicians and included in each 96-well plate. The concordance rate between the two genotype calls of the duplicated CEPH sample was 100% for all SNPs.
2.3. Statistical analyses
A genetic score was calculated for each subject based on his genotype at the 33 SNPs and weighted by odds ratio (OR) based on an external meta-analysis for each SNP, as described previously [20,21]. For univariate analysis of biopsy outcomes, differences in binary variables (family history and digital rectal examination [DRE]) and continuous variables (age, PSA measurements, prostate volume, number of cores at prestudy entry biopsy [number of cores], and genetic score) between men with and without positive prostate biopsy were tested using a χ2 test and a two-sample t test, respectively. Total PSA levels, prostate volume, and genetic score were log-transformed to approach a normal distribution. For multivariate analysis of biopsy outcomes, predictors were modeled using logistic regression analysis.
Area under the receiver operating characteristic curve (AUC) was used to assess the ability of clinical variables and the genetic score to predict for positive prostate biopsy. This value was estimated using the entire cohort as well as in the fitting and testing of samples by four-fold cross-validation to reduce the possibility of overfitting [22]. The difference in AUC between the two models was tested using Delong’s test [23].
Reclassification of PCa risk between the two nested models (clinical parameters with or without genetic score) was calculated based on predicted values from logistic regression models. Each risk result was classified into low (1st quartile), intermediate (2nd and 3rd quartiles), or high (4th quartile) risk categories. Net reclassification improvement (NRI) was used to measure the degree to which PCa risk was appropriately reclassified [24]. The PCa detection rate (proportion of positive biopsies) in each estimated risk category was used to measure the performance of prediction models. In addition, a decision curve analysis (DCA) was performed based on the method proposed by Vickers et al. [25,26].
3. Results
Among the 1654 men in the placebo arm of the REDUCE trial, 410 (25%) had a positive biopsy for PCa from scheduled (n = 370) and “for-cause” (n = 40) biopsies over the 4-yr study period. In univariate analysis (Table 1), men with positive biopsies differed significantly (p < 0.05) from men with negative prostate biopsies for all baseline clinical and demographic variables, with the exception of DRE. Significant differences were also observed between the two groups of men for family history and the genetic score. The ability of these baseline variables to discriminate among biopsy outcomes, as measured by AUC, ranged from 0.51 to 0.59 (Table 2). The AUC was highest for the genetic score (0.59), followed by PSA density (0.58), total PSA levels (0.54), and family history (0.52). The AUC estimates obtained from the entire dataset were similar to those from testing samples by cross-validation analysis, suggesting that these estimates were stable with no evidence of overfitting.
Table 2.
Prediction performance of the clinical variables and genetic score for overall prostate cancer
| Variables and models | No. of subjects | Overall PCa
|
|||||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p* | p HL** | AUC
|
||||
| Entire cohort | Training | Testing | |||||
| Univariate analysis at baseline | |||||||
| Age (age) | 1654 | 1.04 (1.02–1.06) | 0.0002 | 0.56 | 0.56 | 0.56 | |
| DRE (DRE) | 1651 | 1.31 (0.77–2.23) | 0.33 | 0.51 | 0.51 | 0.51 | |
| Total PSA levels at baseline (PSA) | 1649 | 1.53 (1.09–2.16) | 0.014 | 0.54 | 0.54 | 0.54 | |
| Free-to-total PSA ratio (%fPSA) | 1648 | 0.09 (0.01–0.65) | 0.017 | 0.54 | 0.54 | 0.54 | |
| Prostate volume at baseline (PV) | 1630 | 0.58 (0.44–0.77) | 0.0002 | 0.56 | 0.56 | 0.56 | |
| PSA density (PSA/PV) | 1625 | 1.82 (1.43–2.32) | 1.10E-06 | 0.58 | 0.58 | 0.58 | |
| No. of cores sampled at base biopsy (Nocor) | 1652 | 0.93 (0.89–0.98) | 0.006 | 0.55 | 0.55 | 0.55 | |
| Family history at baseline (FH) | 1654 | 1.50 (1.09–2.04) | 0.0115 | 0.52 | 0.53 | 0.52 | |
| Genetic score based on 33 PCa risk SNPs (GS) | 1654 | 1.73 (1.44–2.09) | 7.5E-09 | 0.59 | 0.59 | 0.59 | |
| Best clinical model | |||||||
| Age + FH + PSA + PV + Nocor | 1624 | – | – | 0.21 | 0.62 | 0.62 | 0.60 |
| Best clinical model + genetic score | |||||||
| Age (Age) | 1624 | 1.05 (1.03–1.07) | 7.02E-06 | 0.55 | 0.66 | 0.66 | 0.64 |
| Total PSA levels at baseline (PSA) | – | 1.07 (1.01–1.13) | 0.03 | – | – | – | |
| Prostate volume at baseline (PV) | – | 0.49 (0.37–0.67) | 3.94E-06 | – | – | – | |
| No. of cores sampled at base biopsy (Nocor) | – | 0.95 (0.90–1.00) | 0.045 | – | – | – | |
| Family history at baseline (FH) | – | 1.64 (1.18–2.29) | 0.0035 | – | – | – | |
| Genetic score based on 33 PCa risk SNPs (GS) | – | 1.72 (1.42–2.08) | 3.41E-08 | – | – | – | |
PCa = prostate cancer; OR = odds ratio; CI = confidence interval; DRE = digital rectal examination; PSA = prostate-specific antigen; PV = prostate volume; SNP = single nucleotide polymorphism.
p value for OR.
p value for the Hosmer-Lemeshow goodness-of-fit test.
A multivariate analysis was performed, and the best clinical model included age, family history, total PSA level, prostate volume, and number of cores at previous biopsy. This model was well calibrated, with p = 0.21 (Hosmer-Lemeshow test). The AUC of this model was 0.62 in the entire cohort and 0.60 in the testing samples from a cross-validation analysis. The genetic score remained statistically significant when added to the clinical model (p = 3.41 × 10−8), suggesting that the genetic score is an independent predictor of biopsy outcomes. The AUC of the combined genetic and clinical models was 0.66 in the entire cohort and 0.64 in the testing samples from a cross-validation analysis. The difference in AUC between the two models was statistically significant: p < 0.001 (Table 2).
To further assess the impact of adding genetic markers to the clinical variables, we examined the reclassification of PCa risk. As shown in the cross-tabulation of risk categories, risk was reclassified in 33% of men (n = 190 + 221 + 75 + 46): 267 were downgraded (n = 221 + 46), and 265 were upgraded (n = 190 + 75; Table 3). A significantly higher percentage of men were reclassified appropriately (n = 221 + 75; 36.7%) than inappropriately (n = 190 + 46; 27.0%), with the net reclassification benefit, measured by NRI, of 10% (p = 0.002). The NRI was highest among men whose initial risk was intermediate (20%; p = 0.0001). Furthermore, the reclassified risk correlated better with the observed PCa detection rate when compared with the initial risk from the clinical model (Fig. 1a). For example, the observed detection rate was 23% among 812 subjects classified as intermediate risk based on the initial clinical model; when adding the genetic score, the observed detection rates were 14% and 35% for men whose risk was downgraded or upgraded, respectively.
Table 3.
Number of men with overall prostate cancer classified as low, middle, or high risk from predictive models with or without genetic score
| Risk categories of the model of clinical variables only | Risk categories of the model combining clinical variables and genetic score
|
NRI | p value* | |||||
|---|---|---|---|---|---|---|---|---|
| Low (1st quartile) | Middle (2nd and 3rd quartiles) | High (4th quartile) | Total | No. (%) reclassified as higher risk | No. (%) reclassified as lower risk | |||
| Low (1st quartile) | ||||||||
| No. of men | 268 | 134 | 4 | 406 | – | – | – | – |
| No. of negative | 230 | 105 | 3 | 338 | 108 (31.95) | N/A | – | – |
| No. of positive PCa | 38 | 29 | 1 | 68 | 30 (44.12) | N/A | – | – |
| 4-yr detection rate (95% CI) | 14 (10–18) | 22 (15–29) | 25 (0–67) | 17 (13–20) | – | – | 0.122 | 0.1582 |
| Middle (2nd and 3rd quartiles) | ||||||||
| No. of men | 136 | 549 | 127 | 812 | – | – | – | – |
| No. of negative | 117 | 430 | 82 | 629 | 82 (13.04) | 117 (18.6) | – | – |
| No. of positive PCa | 19 | 119 | 45 | 183 | 45 (24.59) | 19 (10.38) | – | – |
| 4-yr detection rate (95% CI) | 14 (8–20) | 22 (18–25) | 35 (27–44) | 23 (20–25) | – | – | 0.198 | 0.0001 |
| High (4th quartile) | ||||||||
| No. of men | 2 | 129 | 275 | 406 | – | – | – | – |
| No. of negative | 2 | 102 | 150 | 254 | N/A | 104 (40.94) | – | – |
| No. of positive PCa | 0 | 27 | 125 | 152 | N/A | 27 (17.76) | – | – |
| 4-yr detection rate (95% CI) | 0 | 21 (14–28) | 45 (40–51) | 37 (33–42) | – | – | 0.232 | 1.1018E-05 |
| Total | ||||||||
| No. of men | 406 | 812 | 406 | 1624 | – | – | – | – |
| No. of negative | 349 | 637 | 235 | 1221 | 190 (15.56) | 221 (18.1) | – | – |
| No. of positive PCa | 57 | 175 | 171 | 403 | 75 (18.61) | 46 (11.41) | – | – |
| 4-yr detection rate (95% CI) | 14 (11–17) | 22 (19–24) | 42 (37–47) | 25 (23–27) | – | – | 0.097 | 0.0023 |
NRI = net reclassification index; N/A = not applicable; PCa = prostate cancer; CI = confidence interval.
p value is for testing H0: NRI = 0 via the z-score.
Fig. 1.
Four-year observed detection rates of prostate cancer (PCa) among men who were classified at low (1st quartile), intermediate (2nd and 3rd quartiles), or high (4th quartile) estimated risk for PCa in two sequential prediction models. The observed detection rates for (a) any PCa and (b) high-grade PCa (Gleason score ≥7) are shown. In both figures, the top panel presents PCa detection rates of the initial prediction model based on the best clinical model, consisting of five clinical variables: age, family history, free-to-total prostate-specific antigen ratio, number of cores at base biopsy, and prostate volume. The bottom panel presents PCa detection rates of the revised prediction model by adding the genetic score estimated from 33 single nucleotide polymorphisms to the best clinical model. In subsets of men, PCa risk categories either remained the same (dotted bars) or were reclassified (hatched bars). The reclassified risk correlated better with the observed PCa detection rates.
PCa = prostate cancer.
The results based on the DCA are presented in Figure 2. The net benefit, based on true positive and false positive at various threshold probabilities, was generally higher for the model containing the best clinical variables and the genetic score than the clinical variables alone. Again, as with NRI, the difference in net benefit between the two models was larger in men with intermediate estimated risk (threshold probabilities between 17% and 31%). A 10 000-bootstrap analysis was used to internally correct for the overfit of the DCA. The bias was estimated to be small, for example, −0.0009 and −0.0026 for the threshold probabilities of 31% for models with or without the genetic score, respectively.
Fig. 2.
Decision curves for predicting prostate cancer at prostate biopsy. The y-axis represents the net benefit calculated using the methods proposed by Vickers et al. [25,26]. The x-axis represents the threshold probability (percentage) estimated from prediction models. The two vertical lines represent the 25th and 75th percentiles of estimated risk (intermediate risk) in the population, corresponding to 17% and 31% of threshold probabilities, respectively. The dotted line represents the prediction model based on the best clinical model, consisting of five clinical variables: age, family history, free-to-total prostate-specific antigen ratio, number of cores at base biopsy, and prostate volume. The dashed line represents the prediction model based on the best clinical model plus the genetic score.
GS = genetic score.
We also examined the impact of genetic scores on predicting high-grade PCa (Table 4). Among the 1624 men who had complete clinical data, 122 (7.5%) and 9 (0.5%) were diagnosed with Gleason scores ≥7 or ≥8, respectively, during the 4-yr follow-up period. Because of the small number of Gleason scores ≥8 in this study, we defined Gleason score ≥7 PCa as high-grade PCa in the analysis [19]. The best clinical model for predicting high-grade PCa included the same five variables described in Table 2. When the genetic score was added to the model, it remained significant (p = 0.003). The AUC for discriminating high-grade PCa from the clinical model to the combined clinical and genetic score model increased from 0.69 to 0.71 in the entire cohort and from 0.67 to 0.69 in the testing samples of the cross-validation analysis. The difference in AUC between the two models in the entire cohort was statistically significant (p = 0.03). Furthermore, adding the genetic score led to a reclassification of risk categories in 18% of these men, with a higher but not statistically significant percentage of men being appropriately reclassified (NRI: 3%; p = 0.50). However, a stronger benefit in net reclassification was found in each specific risk category: The NRI was 15% (p = 0.047) for the intermediate-risk group (Table 5). The reclassified risk also correlated better with the observed detection rate for high-grade PCa when compared with the initial risk from the best clinical model (Fig. 1b). For example, the detection rate of high-grade PCa was 6% among 812 men who had intermediate risk based on the best clinical model; when the genetic model was added, the detection rate lowered to 4% and increased to 14% for those downgraded to low or upgraded to high risk, respectively.
Table 4.
Prediction performance of the clinical variables and genetic score for high-grade prostate cancer
| Variables and models | No. of subjects | Overall PCa
|
|||||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p* | p HL** | AUC
|
||||
| Entire cohort | Training | Testing | |||||
| Univariate analysis at baseline | |||||||
| Age (Age) | 1654 | 1.07 (1.04–1.10) | 3.7E-05 | – | 0.61 | 0.61 | 0.61 |
| DRE (DRE) | 1651 | N/A | N/A | – | – | – | – |
| Total PSA levels at baseline (PSA) | 1649 | 2.74 (1.53–4.92) | 0.0007 | – | 0.59 | 0.59 | 0.58 |
| Free-to-total PSA ratio (%fPSA) | 1648 | 0.02 (0.0–0.73) | 0.0325 | – | 0.57 | 0.57 | 0.56 |
| Prostate volume at baseline (PV) | 1630 | 0.48 (0.31–0.74) | 0.0009 | – | 0.59 | 0.59 | 0.59 |
| PSA density (PSA/PV) | 1625 | 2.74 (1.86–4.02) | 2.81E-07 | – | 0.62 | 0.63 | 0.62 |
| No. of cores sampled at base biopsy (Nocor) | 1652 | 0.90 (0.83–0.98) | 0.0166 | – | 0.57 | 0.57 | 0.57 |
| Family history at baseline (FH) | 1654 | 1.69 (1.06–2.71) | 0.0284 | – | 0.54 | 0.54 | 0.53 |
| Genetic score based on 33 PCa risk SNPs (GS) | 1654 | 1.65 (1.23–2.12) | 9.0E-04 | – | 0.59 | 0.59 | 0.59 |
| Best clinical model | |||||||
| Age + FH + PSA + PV + Nocor | 1624 | – | – | 0.25 | 0.69 | 0.70 | 0.67 |
| Best clinical model + genetic score | |||||||
| Age (Age) | 1624 | 1.09 (1.05–1.12) | 6.45E-05 | 0.26 | 0.71 | 0.72 | 0.69 |
| Total PSA levels at baseline (PSA) | – | 2.99 (1.63–5.49) | 0.0004 | – | – | – | – |
| Prostate volume at baseline (PV) | – | 0.38 (0.24–0.60) | 3.42E-05 | – | – | – | – |
| No. of cores sampled at base biopsy (Nocor) | – | 0.92 (0.85–1.00) | 0.062 | – | – | – | – |
| Family history at baseline (FH) | – | 2.20 (1.34–3.61) | 0.0019 | – | – | – | – |
| Genetic score based on 33 PCa risk SNPs (GS) | – | 1.61 (1.18–2.19) | 0.0027 | – | – | – | – |
PCa = prostate cancer; AUC = area under the curve; OR = odds ratio; CI = confidence interval; DRE = digital rectal examination; N/A = not applicable; PSA = prostate-specific antigen; PV = prostate volume; SNP = single nucleotide polymorphism.
p value for OR.
p value for the Hosmer-Lemeshow goodness-of-fit test.
Table 5.
Number of men with high-grade* tumor classified as low, middle, or high risk from predictive models with or without genetic score
| Risk categories of the model of clinical variables only | Risk categories of the model combining clinical variables and genetic score
|
NRI | p value** | |||||
|---|---|---|---|---|---|---|---|---|
| Low (1st quartile) | Middle (2nd and 3rd quartiles) | High (4th quartile) | Total | No. (%) reclassified as higher risk | No. (%) reclassified as lower risk | |||
| Low (1st quartile) | ||||||||
| No. of men | 334 | 72 | 0 | 406 | – | – | – | – |
| No. of negative | 327 | 70 | 0 | 397 | 70 (17.63) | N/A | – | – |
| No. of positive PCa | 7 | 2 | 0 | 9 | 2 (22.22) | N/A | – | – |
| 4-yr detection rate (95% CI) | 2 (1–4) | 3 (31 to 7) | 0 | 2 (1–4) | – | – | 0.046 | 0.772 |
| Middle (2nd and 3rd quartiles) | ||||||||
| No. of men | 72 | 666 | 74 | 812 | – | – | – | – |
| No. of negative | 69 | 630 | 64 | 763 | 64 (8.39) | 69 (9.04) | – | – |
| No. of positive PCa | 3 | 36 | 10 | 49 | 10 (20.41) | 3 (6.12) | – | – |
| 4-yr detection rate (95% CI) | 4 (0–9) | 5 (4–7) | 14 (6–21) | 6 (4–8) | – | – | 0.149 | 0.047 |
| High (4th quartile) | ||||||||
| No. of men | 0 | 74 | 332 | 406 | – | – | – | – |
| No. of negative | 0 | 68 | 274 | 342 | N/A | 68 (19.88) | – | – |
| No. of positive PCa | 0 | 6 | 58 | 64 | N/A | 6 (9.38) | – | – |
| 4-yr detection rate (95% CI) | 0 | 8 (2–14) | 17 (13–22) | 16 (12–19) | – | – | 0.105 | 0.413 |
| Total | ||||||||
| No. of men | 406 | 812 | 406 | 1624 | – | – | – | – |
| No. of negative | 396 | 768 | 338 | 1502 | 134 (8.92) | 137 (9.12) | – | – |
| No. of positive PCa | 10 | 44 | 68 | 122 | 12 (9.84) | 9 (7.38) | – | – |
| 4-yr detection rate (95% CI) | 2 (1–4) | 5 (4–7) | 17 (13–20) | 8 (6–9) | – | – | 0.027 | 0.497 |
NRI = net reclassification index; N/A = not applicable; PCa = prostate cancer; CI = confidence interval.
High grade is defined as Gleason grade ≥7.
p value is for testing H0: NRI = 0 via the z-score.
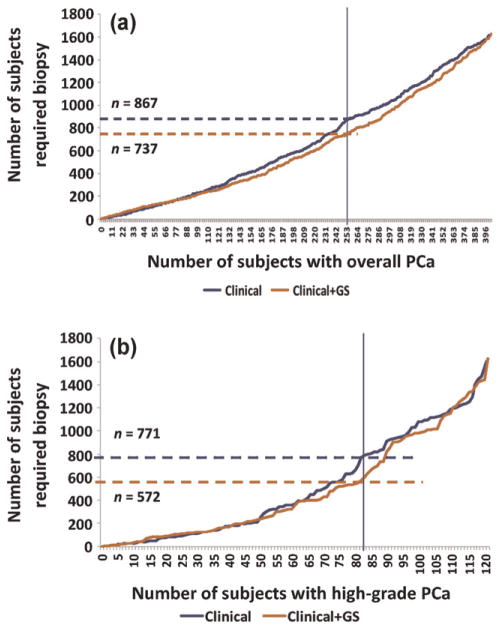
Finally, adding the genetic score to existing clinical predictors allows for fewer biopsies to identify the same number of overall and high-grade PCa patients (Fig. 3a and 3b, respectively). For example, to detect two-thirds of the overall PCa (n = 253) in this cohort, we would have to biopsy 867 men based on the best clinical model and only 737 after adding the genetic score. This difference represents a 15% reduction in the number of biopsies needed to identify the same number of cancers. Similarly, if we want to identify two-thirds of high-grade PCa patients (n = 82) in this cohort, we would need to biopsy 771 men based on the best clinical model but only 572 after adding the genetic score, representing a 25% reduction in unnecessary biopsies.
Fig. 3.
Curves demonstrating the number of men needed to undergo prostate biopsy to identify a given number of (a) overall or (b) high-grade disease based on the clinical model alone (blue) or with the genetic score (orange). Horizontal hatched lines represent the number of biopsies required to identify two-thirds of cancers (represented by the vertical blue line) based on the clinical model alone (blue) or with the genetic score (orange).
PCa = prostate cancer; GS = genetic score.
4. Discussion
Based on a clinical trial population, this study provides evidence that genetic score, calculated from inherited PCa risk-associated markers, is an independent predictor of PCa risk and can provide added value to existing clinical variables in better predicting the outcomes of repeat biopsies. The first finding has been demonstrated in multiple case-control studies [15–18]. The second finding was similar to the results of a recent observational prospective study from Sweden, in which 5241 men underwent for-cause prostate biopsy and had a genetic score calculated based on 27 PCa risk-associated SNPs [25]. In that study, genetic score significantly predicted biopsy outcome, independent of clinical variables such as age and total PSA levels. The AUC for predicting positive biopsy increased from 0.64 for the combined clinical model to 0.67 for the combined clinical and genetic score; the difference in AUCs was statistically significant (p = 0.01). The current study builds on this “real-world” example by demonstrating the genetic score’s value in the setting of a clinical trial whereby subjects, unlike in the clinic, underwent study-mandated biopsy, thus overcoming the confounding issue of PSA-driven biopsy.
The clinical utility of these findings remains to be seen. The added value of genetic markers as assessed by AUC was modest. However, because the current clinical predictors for positive biopsy are poor (AUC of 0.60 for the combined clinical model in our study, where AUC is 0.50 for variables without any predictive ability), the added value is appreciable in relative terms. The 4% increase in AUC represents a 40% [(0.64–0.60)/(0.60–0.50)] improvement. For the millions of patients and their treating urologists who are struggling to make a decision regarding the need for biopsy, this additional information may be helpful.
The potential added value of genetic markers was also demonstrated in the NRI and DCA analyses, particularly in men with intermediate risk. In the REDUCE trial, these men had low but concerning levels of PCa risk based on clinical parameters only (23% for any PCa and 6% for high-grade PCa). After adding the genetic score, the lowered observed PCa detection rate (14% for any PCa and 4% for high-grade PCa) among those downgraded and the increased PCa detection rate (35% for any PCa and 14% for high-grade PCa) among those upgraded provides additional information that may help guide care. In contrast, among men in the low-risk or high-risk group as defined by existing clinical parameters, the genetic score may have limited clinical impact; their PCa detection rates were already low enough (17% for any PCa and 2% for high-grade PCa) to consider forgoing repeat biopsy or sufficiently high (37% for any PCa and 16% for high-grade PCa) to consider repeat biopsy. However, the decision to undergo repeat biopsy is complex and depends on many factors, affecting the risk perception of urologists and their patients.
A further demonstration of the benefit of the genetic score is a reduction in the number of biopsies needed to detect the same number of cancers. Given the morbidity, cost, and even low but measurable mortality from prostate biopsy, this benefit may be of clinical and public health importance.
The interpretation of the genetic score is similar to family history but has several advantages. Genetic score has a better AUC (0.59) than family history (0.52) in discriminating prostate biopsy outcome. It can be measured objectively and does not change over time, while family history depends on the number and ages of male relatives and can change from negative to positive. All 33 SNPs can be genotyped in a single panel using a noninvasive saliva sample, with a cost comparable to a PSA test. Finally, SNPs can be used together to improve the risk estimate.
There are several caveats in this study. First, because of the small number of men of other races in the REDUCE population, our analyses were restricted to white men. Thus, it is necessary to replicate these findings in men of other racial groups. Second, although we have shown that the genetic score adds value to clinical variables for the assessment of overall and high-grade PCa risk, we did not test its ability to distinguishing risk for indolent as opposed to aggressive PCa. Multiple studies demonstrate that these SNPs do not distinguish between these two forms of PCa [18,27]. Finally, it is acknowledged that the predictive performance of the combined genetic and clinical model remains less than ideal. Other emerging biomarkers, such as urine PCA3, TMPRESS-ERG, and endorectal coil magnetic resonance imaging need to be incorporated in future studies, as these data were not available for most subjects in this study [28–30].
5. Conclusions
In the context of a clinical trial, we provide evidence that the genetic score is an independent predictor of PCa and adds to current clinical parameters for predicting biopsy outcomes. The improvement in risk prediction is modest but may be helpful for the millions of men with an initial negative prostate biopsy.
Supplementary Material
Acknowledgments
Funding/Support and role of the sponsor: This study is partially supported by a National Cancer Institute RC2 grant (CA148463) to Dr Xu and a research contract by GlaxoSmithKline to Dr Xu.
The authors thank the patients enrolled in REDUCE who provided the consent and genetic samples that enabled this study and the clinicians who contributed their expertise in recruiting study patients for the REDUCE clinical study. Dave Pulford, Jennifer Aponte, Jon Charnecki, and Mary Ellyn Volk participated in consent reconciliation and sample management to enable genetic sample selection for inclusion and genotype determination. Karen King provided data management support for this project. We thank Silviu-Alin Bacanu and Matt Nelson for multiple discussions on power determination, method limitations, and results. We appreciate the assistance of Lauren Marmor in coordinating the support of the Avodart Collaborative Research Team.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.eururo.2012.05.006.
Footnotes
Author contributions: Jianfeng Xu had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Xu, Condreay, Kader, Spraggs, Sun.
Acquisition of data: Sun, Zheng, Kim, Condreay.
Analysis and interpretation of data: Kader, Sun, Reck, Newcombe, Kim, Hsu, D’AgostinoJr, Tao, Zhang, Turner, Platek, Spraggs, Whittaker, Lane, Bleecker, Meyers, Torti, McConnell, Isaacs, Zheng, Condreay, Rittmaster, Xu.
Drafting of the manuscript: Xu, Kader.
Critical revision of the manuscript for important intellectual content: Xu, Kader, Condreay, Rittmaster, Lane, D’Agostino Jr, Bleecker, Meyers, Torti, McConnell, Isaacs.
Statistical analysis: Sun, Spraggs, Newcombe, D’Agostino Jr, Kim, Hsu, Tao.
Obtaining funding: Xu.
Administrative, technical, or material support: Zheng.
Supervision: Xu, Condreay.
Other: Rittmaster (study oversight), Zheng (genotyping).
Financial disclosures: Jianfeng Xu certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: A patent application has been filed by Drs. Xu, Zheng, Kader, and Sun, the Wake Forest University School of Medicine, and Johns Hopkins University School of Medicine to preserve patent rights for the technology.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–9. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Presti JC., Jr Repeat prostate biopsy—when, where, and how. Urol Oncol. 2009;27:312–4. doi: 10.1016/j.urolonc.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 3.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 5.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 7.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 8.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11. 22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 11.Yeager M, Chatterjee N, Ciampa J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–7. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eeles RA, Kote-Jarai Z, Al Olama AA, et al. UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators; PRACTICAL Consortium. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–21. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–15. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 15.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Sun J, Kader AK, et al. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69:1565–72. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salinas CA, Koopmeiners JS, Kwon EM, et al. Clinical utility of five genetic variants for predicting prostate cancer risk and mortality. Prostate. 2009;69:363–72. doi: 10.1002/pros.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald LM, Kwon EM, Koopmeiners JS, Salinas CA, Stanford JL, Ostrander EA. Analysis of recently identified prostate cancer susceptibility loci in a population-based study: associations with family history and clinical features. Clin Cancer Res. 2009;15:3231–7. doi: 10.1158/1078-0432.CCR-08-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andriole GA, Bostwick D, Brawley OW. The influence of dutasteride on the risk of biopsy-detectable prostate cancer: outcomes of the REduction by DUtasteride of Prostate Cancer Events (REDUCE) study. N Engl J Med. 2010;362:1192–202. doi: 10.1056/NEJMoa0908127. [DOI] [PubMed] [Google Scholar]
- 20.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 21.Kim ST, Cheng Y, Hsu FC, et al. Prostate cancer risk-associated variants reported from genome-wide association studies: meta-analysis and their contribution to genetic variation. Prostate. 2010;70:1729–38. doi: 10.1002/pros.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–34. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 23.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 24.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statist Med. 2008;27:157–72. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 25.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kader AK, Sun J, Isaacs SD, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69:1195–205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Kok JB, Verhaegh GW, Roelofs RW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–8. [PubMed] [Google Scholar]
- 29.Aubin SM, Reid J, Sarno MJ, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol. 2010;184:1947–52. doi: 10.1016/j.juro.2010.06.098. [DOI] [PubMed] [Google Scholar]
- 30.Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.