Abstract
Extremely thermophilic microorganisms have been sources of thermostable and thermoactive enzymes for over 30 years. However, information and insights gained from genome sequences, in conjunction with new tools for molecular genetics, have opened up exciting new possibilities for biotechnological opportunities based on extreme thermophiles that go beyond single-step biotransformations. Although the pace for discovering novel microorganisms has slowed over the past two decades, genome sequence data have provided clues to novel biomolecules and metabolic pathways, which can be mined for a range of new applications. Furthermore, recent advances in molecular genetics for extreme thermophiles have made metabolic engineering for high temperature applications a reality.
Introduction
Microbial life in terrestrial hot springs, such as those present in Yellowstone National Park (USA), has been observed and studied in earnest since at least as early as the 1960s [1-3]. However, the discovery of microbial life in deep sea thermal vents and shallow marine seeps in volcanic regions of the world in the late 1970s and early 1980s [4-6] led to the realization that so the so called extreme thermophiles (Topt ≥ 70°C) were more phylogenetically, physiologically and geographically diverse than first thought. Many novel genera and species, both archaea and bacteria, were isolated and described, including several that became model microorganisms because of their interesting metabolic features and relative ease of cultivation in laboratory settings (see Table 1). Three archaea, Pyrococcus furiosus, a marine fermentative anaerobe (Topt 98-100°C) [5,7], Thermococcus kodakaerensis (Topt 85°C) also a marine fermentative anaerobe [8,9], and Sulfolobus solfataricus, a terrestrial heterotrophic acidophilic aerobe (Topt 80°C, pHopt 3.5) [10,11], and one bacterium, Thermotoga maritima, a marine fermentative anaerobic bacterium (Topt 80°C) [6,12,13], became the focus of most fundamental and biotechnological studies addressing life at high temperatures. In fact, much of what is known about extreme thermophile physiology and enzymology to date has been based on the study of these “model” microorganisms (sub-classified as hyperthermophiles because of their Topt ≥ 80°C). Recently, as discussed below, genetic systems have been established for the three archaea, further enhancing their value as model systems.
Table 1. Model Extreme Thermophiles.
| Organism | Isolation site | Topt (°C) | Domain | Genome size (Kb) | Growth physiology | PubMed Citations (May, 2012) |
|---|---|---|---|---|---|---|
| Sulfolobus solfataricus P2 | solfatara, Naples, Italy | 80°C | Crenarchaea | 2,992,245 | Aerobic, extreme thermoacidophile | 1252 |
| Thermotoga maritima MSB8 | shallow marine sediments, Vulcano Island, Italy | 80°C | Bacteria | 1,860,725 | Fermentative anaerobe, facultative S° reducer | 1148 |
| Thermococcus kodakaerensis KOD1 | solfatara, Kodakara Island, Japan | 85°C | Euryarchaea | 2,088,737 | Fermentative anaerobe, facultative S° reducer | 156 |
| Pyrococcus furiosus | shallow marine sediments, Vulcano Island, Italy | 98°C | Euryarchaea | 1,908,255 | Fermentative anaerobe, facultative S° reducer | 1047 |
Difficulties in isolating extreme thermophiles because of the harsh and often inaccessible environments from which they come, and subsequently cultivating these microorganisms in laboratory settings, initially presented significant challenges to their study and, consequently, to associated biotechnological applications. In the early 1990s, however, successful attempts to clone and express genes from hyperthermophiles in mesophilic recombinant hosts (e.g., Escherchia coli) facilitated efforts to produce specific enzymes for characterization and application [14,15]. Furthermore, since 1995, when the genome sequence of the hyperthermophile Methano(caldo)coccus jannaschii was reported [16], related efforts for many high temperature microorganisms have enabled and accelerated projects to not only identify promising biocatalysts for single-step biotransformations but also to discover metabolic pathways, cellular features, and biological phenomena that are relevant to biotechnology. Now, armed with virtually unlimited access to genome sequence data, and aided by molecular genetics and ‘omics’ tools, the prospects for biotechnology at elevated temperatures have never been more promising, and poised to go beyond single-step biocatalysts (i.e., the use of a single enzyme for a single biotransformation).
Isolation of novel extreme thermophiles
For the most part, the isolation of many currently known and studied extremely thermophilic microorganisms happened primarily in the late 1970s through the early ‘90s [17,18], although isolation of several Sulfolobus species was reported in the early 1970s [19]. Description of new isolates continues to appear in the literature, but most often reside within genera of previously studied extreme thermophiles. As such, discovery of significantly different extremely thermophilic genera and species is becoming rare. Coupled to this is the fact that the criteria for designating a new isolate as “novel” have become more stringent, given the availability of genome sequences and related quantitative measures for differentiating among microorganisms. Thus, new reports of truly novel extreme thermophiles based on more than marginal differences in 16S rRNA phylogeny and subtle variations in growth physiology are infrequent.
Nonetheless, interesting isolates continue to be reported. In the last several years, several new fermentative anaerobes have been isolated (Table 2). For example, Acidilobus saccharovorans [20,21], a terrestrial thermoacidophiic crenarchaeon (Topt 80-85°C, pHopt 3.5-4) contributes to closure of the anaerobic carbon cycle in terrestrial hot springs by complete oxidation of organic compounds (acetate, ethanol, and lactate). Also, Pyrococcus yayanosii is one of only a handful of microbes (and the only Pyrococcus species) shown to be an obligate piezophile (Topt 98°C, Popt 52 MPa); growth was not observed at atmospheric pressure, but rather between 20 and 120 MPa [22].
Table 2. Recently Described Extreme Thermophiles.
| Organism | Topt (°C) | 16S rRNA identity to closest relative | Isolation site | Metabolism | Notes | Ref |
|---|---|---|---|---|---|---|
| Aciduliprofundum boonei | 70 | 83 | Deep sea hydrothermal vent | Anaerobic fermentation | First obligate thermoacidophile from deep sea vents | [92] |
| Nanoarchaeum equitans | 70-98 | 81 | Submarine hydrothermal vent | Parasitism | Small (0.5 Mb) genome, parasite of Ignicoccus hospitalis | [90] |
| Methanocaldococcus villosus | 80 | 95 | Submarine hydrothermal vent | Chemolithoautotrophy | Unique striated cell surface pattern observed | [91] |
| Geoglobus acetivorans | 81 | 97 | Deep sea hydrothermal vent | Chemolithoautotrophy | 1st hyperthermophile enriched on acetate as e-donor | [92] |
| Thermogladius shockii | 84 | 96 | Hot spring | Anaerobic fermentation | Unlike close relatives, growth unaffected by sulfur | [93] |
| Desulfurococcus kamchatkensis | 85 | 98.1 | Hot spring | Anaerobic fermentation | Can use keratin as sole carbon and energy source | [94] |
| Pyrococcus yayanosii | 98 | 99.4 | Deep sea hydrothermal vent | Anaerobic fermentation | Obligate piezophile | [22] |
| Acidilobus saccharovorans | 80-85 | 98.1 | Hot spring | Anaerobic fermentation | Unlike A. aceticus, can use mono- and di-saccharides | [21] |
One promising approach to the isolation of novel microorganisms from high temperature environments is to consider mixed cultures and consortia. For example, a three-species archaeal consortium, capable of deconstructing crystalline cellulose, was recently described [23]. This consortium, with members related to archaea from the genera Ignisphaera, Thermofilum, and Pyrobaculum, was able to partially dissolve filter paper after incubating for 30 days. In other cases, inter-species interactions are critical for isolation of new extreme thermophiles. Along these lines, the discovery of nanoarchaea was reported, occurring as parasitic partners with the extremely thermophilic archaeon, Ignicoccus hospitalis [24]. Evidence from genome sequence data of lateral gene transfer between Nanoarchaeum equitans and I. hospitalis was linked to how these two microorganisms adapted to growth by sulfur-H2 respiration coupled to inorganic carbon and nitrogen fixation. As DNA sequencing rates increase and costs continue to decrease, metagenomes from thermal environments will be examined for hints to unusual physiologies [25], as well as clues to new enzymes and metabolic pathways, the components of which can be produced recombinantly (perhaps, in extremely thermophilic hosts) for further analysis.
Genomics of extreme thermophiles
As mentioned above, access to genome sequence information for extreme thermophiles (which has facilitated functional genomics, proteomics and other ‘omics-based’ approaches) has enabled rapid advances in our understanding of these microbes’ physiology over the past 15 years, despite the lack of genetic systems. In some cases, genome sequence information has been used to ask global questions about how thermophilic proteins fold and function at high temperatures [26]. Along these lines, lower levels of structural disorder and functional simplification determined at the level of individual genes and proteins, as well as of whole genomes, was proposed as the basis for prokaryotic thermophily [27]. Genome sequence information led to the provocative proposal that high temperature bacteria and archaea have lower spontaneous mutation rates than mesophiles [28]. The relationship between mesophiles and thermophiles has been given new perspective through comparative genomics. For example, reverse gyrase, an enzyme involved in thermophilic transcriptional processes to deal with uncoiling DNA, was once thought to be a defining feature of extreme thermophiles [29,30]. However, the genome sequence of Nautilla profoundicola (Topt 40-45°C) encodes the gene for this enzyme, likely acquired through lateral gene transfer within the submarine hydrothermal environment that it inhabits [31]. The phylogenetic lines between thermophily and mesophily may be more blurred than expected. Mesophilic members of the Order Thermotogales (or “mesotogas”) were recently identified [32], as well as related species that have very broad growth temperature ranges [33], raising questions of about the thermal direction of microbial evolution. It is also clear that extreme thermophiles from the same genus can have differences in their genome sequences that map to subtle but significant differences in their growth physiology [34]. Finally, examination of the P. furiosus and S. solfataricus proteomes, with respect to metal content, revealed a far more extensive set of metals implicated in protein structure and function, including “non-biological” metals, such as uranium and vanadium [35]. Since similar results were found for Escherchia coli, it seems that the microbial world employs more of the periodic table for biological function than previously thought.
In certain cases, genome sequences of extreme thermophiles have revealed physiological insights not previously known, despite years of microbiological study. The genome sequence of Metallosphaera sedula, an extremely thermoacidophilic archaeon (Topt 73°C, pHopt 2.0) [36], originally isolated and studied for its ability to mobilize metals from sulfidic ores [37], confirmed the presence of a novel CO2 fixation pathway (3-Hydroxypropionate/4-Hydroxybutyrate cycle) [38]. Consequently, this archaeon possesses a much more versatile growth physiology than previously thought [39]. In fact, M. sedula bioenergetics can be fueled by CO2/H2 autotrophy, heterotrophy, and metal/sulfur oxidation, separately or in combination (mixotrophy) [39].
Genome sequencing has also brought renewed interest to extreme thermophiles that had been isolated 20 years ago. For example, the genome sequences of Caldicellulosiruptor saccharolyticus [40,41] and Caldicellulosiruptor bescii [42] (formerly Anaerocellum thermophilum) shed new light on microbial mechanisms [43,44] and enzymology [45-47] of lignocellulose deconstruction. Furthermore, comparative genomics of eight Caldicellulosiruptor species revealed key determinants of lignocellulose degradation, based on the core and pan genomes of this genus [48].
Molecular genetics tools for extreme thermophiles
As mentioned above, studies of extreme thermophiles, and hence biotechnological applications, have been hampered to a certain extent by the limited availability of tools for genetic manipulation and metabolic engineering. Selection of thermostable markers, high temperature solid media, need for anaerobic conditions, and lack of defined media are among the challenges faced. Despite these obstacles, in recent years new molecular genetics systems have been developed for several extreme thermophiles, as well as being refined and expanded in cases where these had previously existed [49,50].
Initial success with molecular genetics in extreme thermophiles was achieved with the extremely thermoacidophilic archaeon S. solfataricus. The fact that this archaeon grows aerobically and can be cultivated on solid media no doubt contributed to progress in this regard. Most genetic manipulation in this species has utilized lacS mutants [51,52]. DNA is introduced to the cell via electroporation and growth on lactose (which requires lacS complementation) is used to select for successful transformation. This strategy has been used as the basis to generate deletion mutants for the study of copper response [53,54], toxin-antitoxin pairs [55], and antimicrobial proteins [56]. In addition, virus-based vectors encoding genes under the control arabinose- and heatinducible promoters have been used to over-express proteins in S. solfataricus [57,58], and reporter systems for monitoring gene expression have been developed based on β-galactosidase (lacS) [58] and β-glucuronidase (gusB) [59].
In recent years, success with molecular genetics has gone beyond Sulfolobus species. Development of genetic techniques for euryarchaeal Thermococcales has been facilitated by isolation of naturally competent strains of T. kodakaraensis and P. furiosus [60,61]. Manipulation of both species initially hinged upon pyrF, the gene encoding orotidine-5ʹ-monophosphate (OMP) decarboxylase; 5-fluoroorotic acid (5-FOA) can be used to select for pyrF mutants, because cells with the functional gene are sensitive to this compound. On the other hand, media lacking uracil can be used to select for complementation of pyrF, as the gene is required for uracil biosynthesis. Once pyrF mutants of P. furiosus and T. kodakaraensis were generated [60,62], methods were developed for performing selections in complex media [63,64], over-expressing proteins, secreting proteins [65], and generating “markerless” deletions that allow multiple manipulations [62,66].
These tools have facilitated more targeted investigation of fermentative H2 metabolism, a defining feature of Pyrococcus and Thermococcus species. One problem, central to understanding hydrogenase function, is producing sufficient amounts of active forms of the enzyme to study and evaluate. This issue was addressed for soluble hydrogenase I (SHI) from P. furiosus, which was recombinantly produced in E. coli by using an anaerobically-driven promoter native to this bacterium [67]. Furthermore, an engineered hydrogenase, consisting of two of the four native subunits, was overexpressed heterologously in the native host, P. furiosus, and found to utilize electrons directly from pyruvate ferredoxin oxidoreductase without the involvement of an intermediate electron carrier (NADPH or ferredoxin) [68]. There are biotechnological implications of this work. Hydrogen production is normally growth-associated, but an electron carrier-independent hydrogenase might partially decouple these processes, resulting in higher yields of hydrogen.
Directed gene knockouts have been pivotal in probing metabolic mechanisms related to H2 generation in P. furiosus [69] and T. kodakaerensis [70,71]. Deletion of surR, which encodes a transcriptional regulator, revealed that this gene is required for expression of the membrane-bound hydrogenase [71], the primary H2-evolving enzyme [70]. Deletion of cytosolic hydrogenases limited growth of the microbe, but increased specific H2 production rates, suggesting that re-oxidation of H2 by these enzymes is an important energy conservation mechanism in this species [70,71]. Deletion of the membrane-bound oxidoreductase complex, which is required for sulfide production [69,70], also resulted in increased specific hydrogen production [69-71].
Biocatalysis at elevated temperatures
To date, the largest impact of enzymes from extreme thermophiles have had on science and technology relates to their use in catalyzing the polymerase chain reaction (PCR) [72]. Beyond that, the first thoughts for biotechnological applications for enzymes from these extremophiles turned to thermostable and thermoactive “drop in” replacements for industrial enzymes already in use (e.g., proteases, amylases, glucose isomerases). Several insightful reviews have appeared over the past 25 years that examine scientific and biotechnological aspects of biocatalysis at elevated temperatures (for a recent excellent review see [73]). One of the opportunities afforded by biocatalysts that function at temperatures approaching and exceeding 100°C is the exploitation of the intrinsic features that underlie their unprecedented thermostability and thermoactivity. In this sense, the key strategic questions to consider are whether the biocatalytic process requires high temperatures, and, as a consequence, if high temperatures confer any strategic advantage.
The structural stability of enzymes from extreme thermophiles makes them attractive candidates for protein engineering, based on the premise that these are less susceptible to undesired consequences from genetic manipulations. The extensive protein engineering of β-glycosidases from S. solfataricus to catalyze chemo-enzymatic synthesis of oligosaccharides serves as an excellent example in this regard [74]. In another case, cofactor specificity of an alcohol dehydrogenase from P. furiosus could be modified by site-directed mutations in the co-factor binding pocket [75]. Building on the theme of enzymes from extreme thermophiles as robust protein engineering targets, the transcription factor Sso7d from S. solfataricus was used as a scaffold for creating binding partners to a variety of biomolecules [76]. The thermostability of these enzymes was also a key factor in examining the effect of microwaves on biocatalysis. Under certain conditions, the biocatalytic rates of enzymes from P. furiosus, S. solfataricus and T. maritima could be enhanced under microwave irradiation, a prospect not possible for mesophilic enzymes whose stability was significantly impacted by microwaves [77].
Key to many enzyme applications is the capability to immobilize the biocatalyst for stabilization or for a specific process strategy (e.g., re-use or localization). Enzymes from extreme thermophiles present some opportunities, as well as challenges, when it comes to immobilization [78]. For example, entrapment in a porous gel as a means of immobilization, commonly employed for mesophilic enzymes, is problematic because of the thermolability of the matrix. Alternatively, carbohydrate binding domains from extreme thermophiles can be employed in fusions with extremely thermophilic enzymes for immobilization, as was demonstrated with the chitin-binding domain from a P. furiosus chitinase and the xylose isomerase from Thermotoga neapolitana [79].
Not surprisingly, enzymes from extreme thermophiles have been examined closely for applications related to the emergence of biofuels. It has been argued that the deconstruction of lignocellulose is best done at elevated temperatures, either because thermal factors facilitate this process, or because biofuels bioprocessing already involves thermal steps for pretreatment [45]. To this point, computational studies suggest that thermal contributions to enzyme plasticity and molecular motion at high temperatures play a role in enhancing cellulose-binding domain and catalytic domain synergy in cellulose [80]. The genomes of species within the extremely thermophilic genus Caldicellulosiruptor encode a host of multi-domain glycoside hydrolases that contribute to the breakdown of crystalline cellulose and hemicellulose [43,48]. Recent work has looked at the contributions of the various domains within these enzymes to complex carbohydrate hydrolysis [46,81] and the potential role of certain multi-domain glycoside hydrolases, which also use S-layer homology domains to anchor to the cell envelope [82]. The high temperature, cellulose-degrading consortium of archaea, described above, also gave rise to the discovery of a novel hyperthermophilic cellulase, a multi-domain enzyme exhibiting optimal activity at 109°C [23].
Biocatalysis based on whole cells has also been the objective of efforts with extreme thermophiles. For example, the degradation of toxic pollutants has been demonstrated; S. solfataricus 98/2 could utilize phenol for growth in a fed-batch bioreactor [83]. The recovery of base, precious and strategic metals through whole cell bio-oxidation processes has been a long-term goal for extreme thermoacidophiles and recent efforts have focused on identifying process bottlenecks and improved processing strategies. It is becoming clear, not surprisingly, that mixed cultures will be the most effective approach to biohydrometallurgy and could be a way to overcome problems with surface passivation by jarosite and elemental sulfur by-products [84].
Metabolic engineering applications and opportunities at high temperatures
Until very recently, metabolic engineering involving enzyme and pathways from extreme thermophiles were carried out in mesophilic hosts. For example, the archaeal isoprenoid ether lipid biosynthesis pathway was reconstructed in E. coli to produce digeranylgeranylglyceryl phosphate (DGGGP) [85]. The strategic use of temperature for bioprocessing was demonstrated by cloning a hyperthermophilic α-amlyase from T. maritima into transgenic sweet potato [86]. Below 40°C, the normal growth temperature range of the plant, no significant amylase activity could be detected nor was plant growth and development impacted. But upon switching to 80°C, the hyperthermophilic enzyme was activated such that the plant storage root was rapidly hydrolyzed. This created a mechanism for first producing the sweet potato starch then using a thermal switch to convert the starch into a fermentable sugar for biofuels applications.
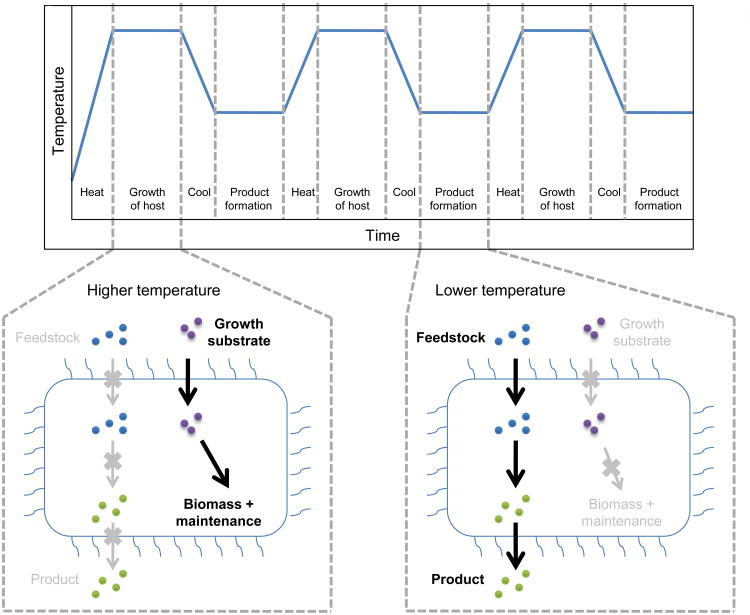
Of course, the next frontier is metabolic engineering at high temperatures. In fact, the rapid progress with molecular genetics for extreme thermophiles has given rise to the prospect of using these microorganisms as recombinant hosts for metabolic engineering. The three archaea shown in Table 1 offer the most near-term prospects in this regard. For example, S. solfatarcius P2 differs from S. solfataricus 98/2 in that the latter strain is less able grow on surfaces. However, the insertion of two genes from P2 into 98/2, encoding α-mannosidase and β-galactosidase, enabled 98/2 to mimic P2 by attaching to glass and forming static biofilms [87]. The use of thermally-driven gene regulation has been elegantly demonstrated using P. furiosus as a recombinant host. When microorganisms are employed to generate a desired product, the production pathway often competes with the microbe’s natural biosynthesis pathways for key intermediates or cofactors. In this situation, one might envision an ideal two-stage process in which biomass is generated during the first stage with minimal product formation, while cellular activity ceases and the desired product is generated during the second stage (see Figure 1). The recent proof-of-concept work of Basen et al. [63] uses temperature as the switch to halt cell growth and initiate product formation. Lactate dehydrogenase from Caldicellulosiruptor bescii (Topt = 78°C) was cloned into P. furiosus (Topt = 98°C) under the control of a P. furiosus “cold shock” promoter which is turned on at 70-75°C. In this situation P. furiosus can be cultured at 98°C until it reaches a high cell density, at which point it can be transferred to 72°C, resulting in expression of the heterologous lactate dehydrogenase and formation of lactate (a product that P. furiosus is unable to produce naturally).
Figure 1. Temperature-dependent regulation of product formation.
Two substrates are provided initially: one that can be catabolized by the extremely thermophilic host's endogenous metabolism for growth, another that cannot and is used for product formation. At higher temperature, the host grows but product formation is silenced. After sufficient cell generation has occurred, the temperature is lowered, inhibiting the host's metabolism and inducing a cold shock promoter that controls expression of heterologous enzymes active at the lower temperature. The heterologous enzymes catalyze all reactions necessary to generate the desired product from the substrate that is provided specifically for product formation. When product formation deteriorates, the temperature can be raised to rehabilitate the extremely thermophilic host and regenerate cofactors and intermediates to prepare for another round of product formation. Additional growth substrate can be provided if necessary, or the extreme thermophile may be able to subsist by catabolizing the heterologous enzymes.
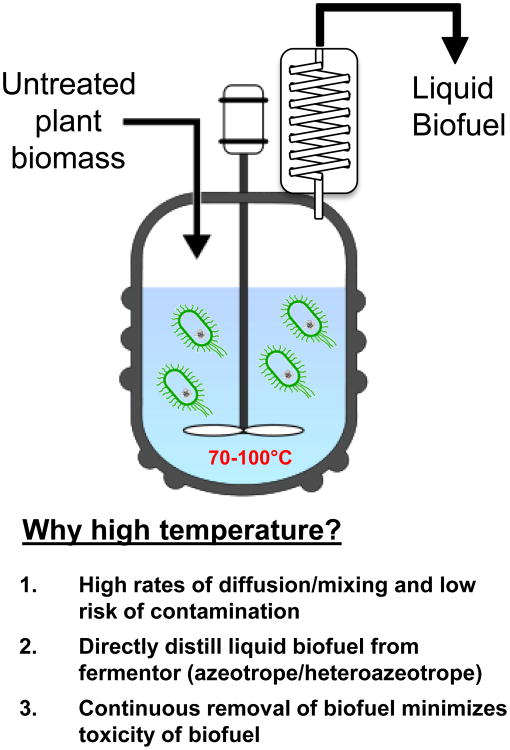
In the near future, we will likely see the first demonstrations of high temperature strains used as hosts for biotechnological applications. For example, in biofuels production, the ultimate goal is to create metabolically-engineered extreme thermophiles that breakdown lignocellulose and convert fermentable sugars to liquid biofuels (socalled “consolidated bioprocessing”). At high temperatures, there is the possibility that biofuels can be recovered directly through direct evaporation and distillation (see Figure 2). Other strategic uses of high temperatures will likely emerge as tools for molecular genetics in extreme thermophiles become more firmly established.
Figure 2. Consolidated bioprocessing at high temperatures for biofuels production.
Metabolic engineering tools for extreme thermophiles could be used to create trains capable of deconstruction of plant biomass and conversion to volatile liquid biofuels that can be recovered directly by distillation from fermentation broths.
Conclusions
Although discovered more than 40 years ago, in many ways extremely thermophilic microorganisms are just at the beginning when it comes to biotechnological applications. Virtually any enzyme that is identified in a mesophile has a homologous version in an extreme thermophile, typically with significantly higher levels of thermostabilty, if not thermoactivity. As genetic systems for extreme thermophiles become more widely used and more tractable, the challenge will be to exploit elevated temperatures to improve upon existing bioprocessing strategies, or even better, make possible novel multi-step, biotransformations. Ideas along these lines have already been proposed [88], and it is only a matter of time before these process concepts are demonstrated and put into practice.
Table 3. Considerations for Metabolic Engineering Bioprocesses at High Temperatures.
| Advantages | Disadvantages |
|---|---|
| Reduced recalcitrance of plant biomass for biofuels applications | Possible energy burden of heating reactor contents |
| Reduced risk of contamination | Lower yields of cellular biomass |
| Use of temperature regulation to optimize product formation | Genetic stability of thermophilic recombinant hosts unknown |
| Facilitated recovery of volatile products | Lower gas solubilities |
| Lower risk of release of viable genetically modified organisms | Substrate, product lability at elevated temperatures |
| Higher temperatures more consistent with chemical processes | Limited to genes encoding themostable proteins/enzymes |
| Higher mass transfer rates | Less known about microbial physiology |
| Improved solubility of carbohydrates, amino acids | Genetics tools are in infancy |
Highlights.
Current Opinion in Chemical Engineering
New developments in the use of extremely thermophilic microorganisms and enzymes are discussed.
New discoveries, particularly of mixed communities of extreme thermophiles, have been reported.
New genetics tools have opened up the possibility of metabolic engineering at high temperatures.
Acknowledgments
The authors acknowledge support from the U.S. Department of Energy ARPA-E Program (DE-AR0000081), U.S. Department of Energy GTL Program (DG-FG02-08ER64687), U.S. National Institute of Health (R01GM90209), and the U.S. Defense Threat Reduction Agency (HDTRA1-09-1-0030). AD Frock acknowledges support from an U.S. National Institutes of Health Biotechnology Traineeship (T32 GM008776-06).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Brock TD. Life at high temperatures. Evolutionary, ecological, and biochemical significance of organisms living in hot springs is discussed. Science. 1967;158:1012–1019. doi: 10.1126/science.158.3804.1012. [DOI] [PubMed] [Google Scholar]
- 2.Brock TD. Micro-organisms adapted to high temperatures. Nature. 1967;214:882–885. doi: 10.1038/214882a0. [DOI] [PubMed] [Google Scholar]
- 3.Bott TL, Brock TD. Bacterial growth rates above 90 degrees C in Yellowstone hot springs. Science. 1969;164:1411–1412. doi: 10.1126/science.164.3886.1411. [DOI] [PubMed] [Google Scholar]
- 4.Jones WJ, Leigh JA, Mayer F, Woese CR, Wolfe RS. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch Microbiol. 1983;136:254–261. [Google Scholar]
- 5.Fiala G, Stetter KO. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 6.Huber R, Langworthy TA, Konig H, Thomm M, Woese CR, Mayer F, Sleytr UB, Stetter KO. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90C. Arch Microbiol. 1986;144:324–333. [Google Scholar]
- 7.Robb FT, Maeder DL, Brown JR, DiRuggiero J, Stump MD, Yeh RK, Weiss RB, Dunn DM. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 2001;330:134–157. doi: 10.1016/s0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- 8.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005;15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl Environ Microbiol. 1994;60:4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zillig W, Stetter KO, Wunderl S, Schulz W, Priess H, Scholz I. The Sulfolobus-“Caldariella” group: Taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch Microbiol. 1980;125:269–269. [Google Scholar]
- 12.Conners SB, Mongodin EF, Johnson MR, Montero CI, Nelson KE, Kelly RM. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol Rev. 2006;30:872–905. doi: 10.1111/j.1574-6976.2006.00039.x. [DOI] [PubMed] [Google Scholar]
- 13.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 14.Fabry S, Lehmacher A, Bode W, Hensel R. Expression of the glyceraldehyde-3-phosphate dehydrogenase gene from the extremely thermophilic archaebacterium Methanothermus fervidus in E. coli. Enzyme purification, crystallization, and preliminary crystal data. FEBS Lett. 1988;237:213–217. doi: 10.1016/0014-5793(88)80204-7. [DOI] [PubMed] [Google Scholar]
- 15.Moracci M, La Volpe A, Pulitzer JF, Rossi M, Ciaramella M. Expression of the thermostable beta-galactosidase gene from the archaebacterium Sulfolobus solfataricus in Saccharomyces cerevisiae and characterization of a new inducible promoter for heterologous expression. J Bacteriol. 1992;174:873–882. doi: 10.1128/jb.174.3.873-882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 17.Stetter KO. Hyperthermophiles in the history of life. Philos Trans R Soc Lond B Biol Sci. 2006;361:1837–1842. doi: 10.1098/rstb.2006.1907. discussion 1842-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stetter KO. History of discovery of the first hyperthermophiles. Extremophilesi. 2006;10:357–362. doi: 10.1007/s00792-006-0012-7. [DOI] [PubMed] [Google Scholar]
- 19.Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Microbiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 20.Mardanov AV, Svetlitchnyi VA, Beletsky AV, Prokofeva MI, Bonch-Osmolovskaya EA, Ravin NV, Skryabin KG. The genome sequence of the crenarchaeon Acidilobus saccharovorans supports a new order, Acidilobales, and suggests an important ecological role in terrestrial acidic hot springs. Appl Environ Microbiol. 2010;76:5652–5657. doi: 10.1128/AEM.00599-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prokofeva MI, Kostrikina NA, Kolganova TV, Tourova TP, Lysenko AM, Lebedinsky AV, Bonch-Osmolovskaya EA. Isolation of the anaerobic thermoacidophilic crenarchaeote Acidilobus saccharovorans sp. nov. and proposal of Acidilobales ord. nov., including Acidilobaceae fam. nov. and Caldisphaeraceae fam. nov. Int J Syst Evol Microbiol. 2009;59:3116–3122. doi: 10.1099/ijs.0.010355-0. [DOI] [PubMed] [Google Scholar]
- 22.Birrien JL, Zeng X, Jebbar M, Cambon-Bonavita MA, Querellou J, Oger P, Bienvenu N, Xiao X, Prieur D. Pyrococcus yayanosii sp. nov., an obligate piezophilic hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2011;61:2827–2831. doi: 10.1099/ijs.0.024653-0. [DOI] [PubMed] [Google Scholar]
- 23••.Graham JE, Clark ME, Nadler DC, Huffer S, Chokhawala HA, Rowland SE, Blanch HW, Clark DS, Robb FT. Identification and characterization of a multidomain hyperthermophilic cellulase from an archaeal enrichment. Nat Commun. 2011;2:375. doi: 10.1038/ncomms1373. Deconstruction of crystalline cellulose at 90°C, the highest temperature to date, was observed by a consortium of three hyperthermophilic archaea. A multi-domain cellulase was identified with optimal activity at 109°C. [DOI] [PubMed] [Google Scholar]
- 24.Podar M, Anderson I, Makarova KS, Elkins JG, Ivanova N, Wall MA, Lykidis A, Mavromatis K, Sun H, Hudson ME, et al. A genomic analysis of the archaeal system Ignicoccus hospitalis-Nanoarchaeum equitans. Genome Biol. 2008;9:R158. doi: 10.1186/gb-2008-9-11-r158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, Richardson TH, Macur RE, Hamamura N, Jennings R, Fouke BW, et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One. 5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawle L, Ghosh K. How do thermophilic proteins and proteomes withstand high temperature? Biophys J. 2011;101:217–227. doi: 10.1016/j.bpj.2011.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burra PV, Kalmar L, Tompa P. Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PLoS One. 2010;5:e12069. doi: 10.1371/journal.pone.0012069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drake JW. Avoiding dangerous missense: thermophiles display especially low mutation rates. PLoS Genet. 2009;5:e1000520. doi: 10.1371/journal.pgen.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouthier de la Tour C, Portemer C, Nadal M, Stetter KO, Forterre P, Duguet M. Reverse gyrase, a hallmark of the hyperthermophilic archaebacteria. J Bacteriol. 1990;172:6803–6808. doi: 10.1128/jb.172.12.6803-6808.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 31.Campbell BJ, Smith JL, Hanson TE, Klotz MG, Stein LY, Lee CK, Wu D, Robinson JM, Khouri HM, Eisen JA, et al. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 2009;5:e1000362. doi: 10.1371/journal.pgen.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Nesbo CL, Kumaraswamy R, Dlutek M, Doolittle WF, Foght J. Searching for mesophilic Thermotogales bacteria: “mesotogas” in the wild. Appl Environ Microbiol. 2010;76:4896–4900. doi: 10.1128/AEM.02846-09. Mesophilic Thermotogales have been detected in mesophilic, hydrocarbon-rich environments across the world, suggesting that they originated in thermophilic oil reservoirs, adapted to lower temperatures as the reservoirs cooled, and may have spread relatively recently. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swithers KS, DiPippo JL, Bruce DC, Detter C, Tapia R, Han S, Goodwin LA, Han J, Woyke T, Pitluck S, et al. Genome sequence of Kosmotoga olearia strain TBF 19.5.1, a thermophilic bacterium with a wide growth temperature range, isolated from the Troll B oil platform in the North Sea. J Bacteriol. 2011;193:5566–5567. doi: 10.1128/JB.05828-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frock AD, Gray SR, Kelly RM. Hyperthermophilic Thermotoga species differ with respect to specific carbohydrate transporters and glycoside hydrolases. Appl Environ Microbiol. 2012;78:1978–1986. doi: 10.1128/AEM.07069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL, 2nd, Jenney FE, Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, et al. Microbial metalloproteomes are largely uncharacterized. Nature. 2010;466:779–782. doi: 10.1038/nature09265. Novel metalloproteins were identified, in some cases incorporating novel metals (i.e. molybdenum), by a metals-based approach using liquid chromatography and mass spectrometry, suggesting that metalloproteomes are much more extensive than previously realized. [DOI] [PubMed] [Google Scholar]
- 36.Auernik KS, Maezato Y, Blum PH, Kelly RM. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl Environ Microbiol. 2008;74:682–692. doi: 10.1128/AEM.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber G, Spinnler C, Gambacorta A, Stetter KO. Metallosphaera sedula gen. and sp. nov. represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst Appl Microbiol. 1989;12:38–47. [Google Scholar]
- 38.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 39•.Auernik KS, Kelly RM. Physiological versatility of the extremely thermoacidophilic archaeon Metallosphaera sedula supported by transcriptomic analysis of heterotrophic, autotrophic, and mixotrophic growth. Appl Environ Microbiol. 2010;76:931–935. doi: 10.1128/AEM.01336-09. Candidate genes of the 3-hydroxypropionate/4-hydroxybutyrate pathway wereidentified by analyzing transcription during autotrophic, heterotrophic, andmixotrophic growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rainey FA, Donnison AM, Janssen PH, Saul D, Rodrigo A, Bergquist PL, Daniel RM, Stackebrandt E, Morgan HW. Description of Caldicellulosiruptor saccharolyticus gen. nov., sp. nov: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol Lett. 1994;120:263–266. doi: 10.1111/j.1574-6968.1994.tb07043.x. [DOI] [PubMed] [Google Scholar]
- 41.van de Werken HJ, Verhaart MR, VanFossen AL, Willquist K, Lewis DL, Nichols JD, Goorissen HP, Mongodin EF, Nelson KE, van Niel EW, et al. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol. 2008;74:6720–6729. doi: 10.1128/AEM.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Dam P, Kataeva I, Yang SJ, Zhou F, Yin Y, Chou W, Poole FL, 2nd, Westpheling J, Hettich R, Giannone R, et al. Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacteria Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 2011;39:3240–3254. doi: 10.1093/nar/gkq1281. The genome of Caldicellulosiruptor bescii contains a high duplication ofcarbohydrate active domains and is enriched with genes encoding multi-domain, multi-functional carbohydrate active proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Blumer-Schuette SE, Lewis DL, Kelly RM. Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl Environ Microbiol. 2010;76:8084–8092. doi: 10.1128/AEM.01400-10. Of seven Caldicellulosiruptor species examined, four hydrolyzed crystallinecellulose and secreted a prominent S-layer protein, which is likely involved incell-substrate interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MW. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol. 2009;75:4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol. 2008;19:210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 46.VanFossen AL, Ozdemir I, Zelin SL, Kelly RM. Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng. 2011;108:1559–1569. doi: 10.1002/bit.23093. [DOI] [PubMed] [Google Scholar]
- 47.VanFossen AL, Verhaart MR, Kengen SM, Kelly RM. Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl Environ Microbiol. 2009;75:7718–7724. doi: 10.1128/AEM.01959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Blumer-Schuette SE, Giannone RJ, Zurawski JV, Ozdemir I, Ma Q, Yin Y, Xu Y, Kataeva I, Poole FL, 2nd, Adams MW, et al. Caldicellulosiruptor core and pan genomes reveal determinants for non-cellulosomal thermophilic deconstruction of plant biomass. J Bacteriol. 2012 doi: 10.1128/JB.00266-12. A comprehensive analysis of eight Caldicellulosiruptor species involving microbiological, genomic, and proteomic methods revealed GH 9 and GH 48 containing multi-domain cellulases and a novel adhesin that are key for deconstruction of plant biomass in these species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor MP, van Zyl L, Tuffin IM, Leak DJ, Cowan DA. Genetic tool development underpins recent advances in thermophilic whole-cell biocatalysts. Microb Biotechnol. 2011;4:438–448. doi: 10.1111/j.1751-7915.2010.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Leigh JA, Albers SV, Atomi H, Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev. 2011;35:577–608. doi: 10.1111/j.1574-6976.2011.00265.x. The authors review the significant progress that has been made in the development of genetic tools for manipulation of various groups of archaea. [DOI] [PubMed] [Google Scholar]
- 51.Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol. 2004;186:427–437. doi: 10.1128/JB.186.2.427-437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worthington P, Hoang V, Perez-Pomares F, Blum P. Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2003;185:482–488. doi: 10.1128/JB.185.2.482-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villafane A, Voskoboynik Y, Ruhl I, Sannino D, Maezato Y, Blum P, Bini E. CopR of Sulfolobus solfataricus represents a novel class of archaeal-specific copper-responsive activators of transcription. Microbiology. 2011;157:2808–2817. doi: 10.1099/mic.0.051862-0. [DOI] [PubMed] [Google Scholar]
- 54.Vollmecke C, Drees SL, Reimann J, Albers SV, Lubben M. Both ATPases CopA and CopB contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology. 2012 doi: 10.1099/mic.0.055905-0. [DOI] [PubMed] [Google Scholar]
- 55•.Maezato Y, Daugherty A, Dana K, Soo E, Cooper C, Tachdjian S, Kelly RM, Blum P. VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus. RNA. 2011;17:1381–1392. doi: 10.1261/rna.2679911. Gene disruption of the vapBC6 toxin-antitoxin pair resulted in increasedsensitivity to heat shock. Evidence was presented that vapC6 regulates geneexpression by exhibiting ribonucleolytic activity against specific transcripts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Ellen AF, Rohulya OV, Fusetti F, Wagner M, Albers SV, Driessen AJ. The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J Bacteriol. 2011;193:4380–4387. doi: 10.1128/JB.05028-11. Antimicrobial proteins were identified in Sulfolobus species which require twoproteins for their membrane-vesicle associated activity against closely relatedspecies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albers SV, Jonuscheit M, Dinkelaker S, Urich T, Kletzin A, Tampe R, Driessen AJ, Schleper C. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol. 2006;72:102–111. doi: 10.1128/AEM.72.1.102-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jonuscheit M, Martusewitsch E, Stedman KM, Schleper C. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol. 2003;48:1241–1252. doi: 10.1046/j.1365-2958.2003.03509.x. [DOI] [PubMed] [Google Scholar]
- 59.Honarbakhsh M, Villafane AA, Ruhl I, Sannino D, Bini E. Development of a thermostable beta-glucuronidase-based reporter system for monitoring gene expression in hyperthermophiles. Biotechnol Bioeng. 2012 doi: 10.1002/bit.24432. [DOI] [PubMed] [Google Scholar]
- 60•.Lipscomb GL, Stirrett K, Schut GJ, Yang F, Jenney FE, Jr, Scott RA, Adams MW, Westpheling J. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol. 2011;77:2232–2238. doi: 10.1128/AEM.02624-10. A naturally competent mutant was used to generate markerless single- and double-deletion mutants in P furiosus for the first time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato T, Fukui T, Atomi H, Imanaka T. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol. 2003;185:210–220. doi: 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato T, Fukui T, Atomi H, Imanaka T. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol. 2005;71:3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Basen M, Sun J, Adams MW. Engineering a hyperthermophilic archaeon for temperature-dependent product formation. MBio. 2012;3 doi: 10.1128/mBio.00053-12. P. furiosus was engineered to produce a non-native product (lactate) under conditions (low temperature) where cell growth is neglible. This was done by inserting the lactate hydrogenase from C. bescii into P. furiosus under the control of a P. furiosus “cold shock” promoter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Santangelo TJ, Cubonova L, Reeve JN. Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl Environ Microbiol. 2010;76:1044–1052. doi: 10.1128/AEM.02497-09. Genetic tools for T. kodakaraensis were expanded to allow for multiple deletions, selection on complex media, and improvement of a reporter gene system basedon a beta-glycosidase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Takemasa R, Yokooji Y, Yamatsu A, Atomi H, Imanaka T. Thermococcus kodakarensis as a host for gene expression and protein secretion. Appl Environ Microbiol. 2011;77:2392–2398. doi: 10.1128/AEM.01005-10. T. kodakaraensis was engineered to efficiently secrete overexpressed proteinsby fusing the signal sequence from a protease to a chitinase and placing expression of the fusion protein under the control of a strong constitutive promoter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Farkas J, Stirrett K, Lipscomb GL, Nixon W, Scott RA, Adams MW, Westpheling J. The recombinogenic properties of the Pyrococcus furiosus COM1 strainenable rapid selection of targeted mutants. Appl Environ Microbiol. 2012 doi: 10.1128/AEM.00936-12. A method was developed for generation of markerless deletion mutants in P. furiosus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Sun J, Hopkins RC, Jenney FE, McTernan PM, Adams MW. Heterologous expression and maturation of an NADP-dependent [NiFe]-hydrogenase: a key enzyme in biofuel production. PLoS One. 2010;5:e10526. doi: 10.1371/journal.pone.0010526. The challenge of producing significant amounts of oxygen-sensitive P. furiosus hydrogenase was overcome by expressing all the structural and maturation proteins necessary under the control of an anaerobic-inducible promoter in E. coli. It was found that some of E. coli hydrogenase maturation proteins could substitute for the P. furiosus versions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hopkins RC, Sun J, Jenney FE, Jr, Chandrayan SK, McTernan PM, Adams MW. Homologous expression of a subcomplex of Pyrococcus furiosus hydrogenase that interacts with pyruvate ferredoxin oxidoreductase. PLoS One. 2011;6:e26569. doi: 10.1371/journal.pone.0026569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bridger SL, Clarkson SM, Stirrett K, DeBarry MB, Lipscomb GL, Schut GJ, Westpheling J, Scott RA, Adams MW. Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. J Bacteriol. 2011;193:6498–6504. doi: 10.1128/JB.05445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanai T, Matsuoka R, Beppu H, Nakajima A, Okada Y, Atomi H, Imanaka T. Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol. 2011;193:3109–3116. doi: 10.1128/JB.01072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71•.Santangelo TJ, Cubonova L, Reeve JN. Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol Microbiol. 2011;81:897–911. doi: 10.1111/j.1365-2958.2011.07734.x. Targeted gene disruptions in T. kodakaraensis demonstrated that the membrane-bound hydrogenase (MBH) is primarily responsible for hydrogen production, the SurR regulator is required for MBH expression, and specific hydrogen productioncould be increased by removing the cytosolic hydrogenases (which primarilyconsume hydrogen) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 73••.Atomi H, Sato T, Kanai T. Application of hyperthermophiles and their enzymes. Curr Opin Biotechnol. 2011;22:618–626. doi: 10.1016/j.copbio.2011.06.010. Review of hyperthermophiles as sources of thermostable enzymes and potential hosts for high-temperature cell engineering and biofuel production. [DOI] [PubMed] [Google Scholar]
- 74.Cobucci-Ponzano B, Perugino G, Rossi M, Moracci M. Engineering the stability and the activity of a glycoside hydrolase. Protein Eng Des Sel. 2011;24:21–26. doi: 10.1093/protein/gzq085. [DOI] [PubMed] [Google Scholar]
- 75•.Campbell E, Wheeldon IR, Banta S. roadening the cofactor specificity of a thermostable alcohol dehydrogenase using rational protein design introduces novel kinetic transient behavior. Biotechnol Bioeng. 2010;107:763–774. doi: 10.1002/bit.22869. Analysis of mesophilic and thermophilic hydrogenases was used to guide directed mutations of P. furiosus alcohol dehydrogenase, resulting in improved activity, broadened cofactor specifity, and a cofactor concentration-dependent binding mechanism. [DOI] [PubMed] [Google Scholar]
- 76•.Gera N, Hussain M, Wright RC, Rao BM. Highly stable binding proteins derived from the hyperthermophilic Sso7d scaffold. J Mol Biol. 2011;409:601–616. doi: 10.1016/j.jmb.2011.04.020. Random mutagenesis of a small hyperthermophilic protein was used to generate stable, high affinity, highly specific binding proteins for a wide variety of model targets. [DOI] [PubMed] [Google Scholar]
- 77.Young DD, Nichols J, Kelly RM, Deiters A. Microwave activation of enzymatic catalysis. J Am Chem Soc. 2008;130:10048–10049. doi: 10.1021/ja802404g. [DOI] [PubMed] [Google Scholar]
- 78.Cowan DA, Fernandez-Lafuente R. Enhancing the functional properties of thermophilic enzymes by chemical modification and immobilization. Enzyme Microb Technol. 2011;49:326–346. doi: 10.1016/j.enzmictec.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 79.Harris JM, Epting KL, Kelly RM. N-terminal fusion of a hyperthermophilic chitin-binding domain to xylose isomerase from Thermotoga neapolitana enhances kinetics and thermostability of both free and immobilized enzymes. Biotechnol Prog. 2010;26:993–1000. doi: 10.1002/btpr.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80•.Batista PR, Costa MG, Pascutti PG, Bisch PM, de Souza W. High temperatures enhance cooperative motions between CBM and catalytic domains of a thermostable cellulase: mechanism insights from essential dynamics. Phys Chem Chem Phys. 2011;13:13709–13720. doi: 10.1039/c0cp02697b. According to computational simulations, the substrate binding and catalytic domains of a thermostable cellulase were observed to move cooperatively athigh temperatures, and two key “hinge” amino acid residues were identified. [DOI] [PubMed] [Google Scholar]
- 81.Su X, Mackie RI, Cann IK. Biochemical and mutational analyses of a multidomain cellulase/mannanase from Caldicellulosiruptor bescii. Appl Environ Microbiol. 2012;78:2230–2240. doi: 10.1128/AEM.06814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ozdemir I, Blumer-Schuette SE, Kelly RM. S-layer homology domain proteins Csac_0678 and Csac_2722 are implicated in plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol. 2012;78:768–777. doi: 10.1128/AEM.07031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christen P, Davidson S, Combet-Blanc Y, Auria R. Phenol biodegradation by the thermoacidophilic archaeon Sulfolobus solfataricus 98/2 in a fed-batch bioreactor. Biodegradation. 2011;22:475–484. doi: 10.1007/s10532-010-9420-6. [DOI] [PubMed] [Google Scholar]
- 84.Zhu W, Xia JL, Yang Y, Nie ZY, Zheng L, Ma CY, Zhang RY, Peng AA, Tang L, Qiu GZ. Sulfur oxidation activities of pure and mixed thermophiles and sulfur speciation in bioleaching of chalcopyrite. Bioresour Technol. 2011;102:3877–3882. doi: 10.1016/j.biortech.2010.11.090. [DOI] [PubMed] [Google Scholar]
- 85.Lai D, Lluncor B, Schroder I, Gunsalus RP, Liao JC, Monbouquette HG. Reconstruction of the archaeal isoprenoid ether lipid biosynthesis pathway in Escherichia coli through digeranylgeranylglyceryl phosphate. Metab Eng. 2009;11:184–191. doi: 10.1016/j.ymben.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Santa-Maria MC, Yencho CG, Haigler CH, Thompson WF, Kelly RM, Sosinski B. Starch self-processing in transgenic sweet potato roots expressing ahyperthermophilic alpha-amylase. Biotechnol Prog. 2011;27:351–359. doi: 10.1002/btpr.573. A hyperthermophilic alpha-amylase was inserted into sweet potato without affecting growth and development of the plant so that the plant could be “self-processed” by incubating at an elevated temperature that activates the thermostable enzyme. [DOI] [PubMed] [Google Scholar]
- 87.Koerdt A, Jachlewski S, Ghosh A, Wingender J, Siebers B, Albers SV. Complementation of Sulfolobus solfataricus PBL2025 with an alpha-mannosidase: effects on surface attachment and biofilm formation. Extremophiles. 2012;16:115–125. doi: 10.1007/s00792-011-0411-2. [DOI] [PubMed] [Google Scholar]
- 88.Hawkins AS, Han Y, Lian H, Loder AJ, Menon AL, Iwuchukwu IJ, Keller M, Leuko TT, Adams MW, Kelly RM. Extremely thermophilic routes to microbial electrofuels. ACS Catalysis. 2011;1:1043–1050. [Google Scholar]
- 89.Reysenbach AL, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, Tivey MK, Von Damm KL, Voytek MA. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–447. doi: 10.1038/nature04921. [DOI] [PubMed] [Google Scholar]
- 90.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 91.Bellack A, Huber H, Rachel R, Wanner G, Wirth R. Methanocaldococcus villosus sp. nov., a heavily flagellated archaeon that adheres to surfaces and forms cell-cell contacts. Int J Syst Evol Microbiol. 2011;61:1239–1245. doi: 10.1099/ijs.0.023663-0. [DOI] [PubMed] [Google Scholar]
- 92.Slobodkina GB, Kolganova TV, Querellou J, Bonch-Osmolovskaya EA, Slobodkin AI. Geoglobus acetivorans sp. nov., an iron(III)-reducing archaeon from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2009;59:2880–2883. doi: 10.1099/ijs.0.011080-0. [DOI] [PubMed] [Google Scholar]
- 93.Osburn MR, Amend JP. Thermogladius shockii gen. nov. sp. nov., a hyperthermophilic crenarchaeote from Yellowstone National Park, USA. Arch Microbiol. 2011;193:45–52. doi: 10.1007/s00203-010-0639-8. [DOI] [PubMed] [Google Scholar]
- 94.Kublanov IV, Bidjieva S, Mardanov AV, Bonch-Osmolovskaya EA. Desulfurococcus kamchatkensis sp. nov., a novel hyperthermophilic protein-degrading archaeon isolated from a Kamchatka hot spring. Int J Syst Evol Microbiol. 2009;59:1743–1747. doi: 10.1099/ijs.0.006726-0. [DOI] [PubMed] [Google Scholar]