Abstract
Viral RNA was amplified by reverse transcription-PCR from a patient suffering from hemorrhagic fever with renal syndrome (HFRS) in Germany. The virus strain could be assigned to the Dobrava hantavirus (DOBV). This is the first molecular identification of human infection by DOBV in central Europe and the first proof that a virus strain related to the DOBV-Aa lineage, carried by Apodemus agrarius rodents, is able to cause HFRS.
Members of the genus Hantavirus (family Bunyaviridae) are rodent-borne “emerging viruses” that are known to cause two human zoonoses, hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS). In Europe and Asia, the Hantavirus species Puumala virus (PUUV), Dobrava virus (DOBV), Hantaan virus (HTNV), and Seoul virus (SEOV) cause HFRS, while Sin Nombre virus (SNV), Andes virus (ANDV), and other related viruses represent the causative agents of HCPS in the Americas (8, 13, 14).
DOBV seems to be the most virulent European hantavirus. The severity of DOBV-associated HFRS can reach fatality rates of up to 12%, as reported from southeast Europe (3, 11). Members of the DOBV species are hosted by different rodent species, Apodemus flavicollis (yellow necked mouse) and Apodemus agrarius (striped field mouse) (for review, see reference 12), as well as, probably, Apodemus sylvaticus (wood mouse) (E. Tkachenko, T. Dzagurova, A. Dekonenko, L. Ivanov, A. Yampolskiy, R. Brudniy, and C. S. Schmaljohn, Abstr. 5th Int. Conf. HFRS, HPS, Hantaviruses, abstr. 19, 2001).
The first DOBV strain was isolated from lungs of A. flavicollis mice captured in a natural focus of HFRS in Slovenia, southeast Europe (2), and now represents the prototypical DOBV strain (named Slovenia, or Slo/Af) from A. flavicollis. Subsequently, related virus strains were isolated from A. agrarius animals captured on Saaremaa Island, Estonia, northeast Europe (10), and Slovakia, central Europe (B. Klempa, H. A. Schmidt, R. Ulrich, M. Schutt, M. Labuda, B. Hjelle, H. Meisel, and D. H. Kruger, Abstr. XIIth Int. Conf. Negative Strand Viruses, abstr. 289, 2003).
It has been proposed that the DOBV species forms at least two genetic branches, DOBV-Af and DOBV-Aa, that are found in A. flavicollis and A. agrarius hosts, respectively (7, 16). Whereas in southeast Europe, highly related DOBV-Af nucleotide sequences could be identified in both A. flavicollis animals and HFRS patients (1, 9, 11, 19), the situation in central Europe remained rather unclear. In this geographical region, DOBV nucleotide sequences were predominantly found in A. agrarius (but also in A. flavicollis) rodents, and neutralizing antibodies in the sera of various HFRS patients were typed as DOBV specific (see reference 16 and studies quoted therein). However, the genetic characterization of viral gene sequences from HFRS patients, enabling molecular classification of the virus strain and providing direct evidence of whether DOBV causes HFRS in central Europe, was still missing. Here, we describe the first HFRS case from central Europe associated with the DOBV-Aa lineage from A. agrarius.
A 19-year-old man (patient H169) from Ratzeburg, north Germany, was admitted to a local hospital because of increasing appendicitis-like abdominal pain. At admission, he reported a 5-day history of fever (up to 39°C) followed by nausea, vomiting, and diarrhea. According to the cardinal symptom of abdominal pain, he was scheduled for appendectomy, until acute renal failure was noticed within the next 3 days and hemodialysis became necessary because of uremic symptoms. Therefore, he was transferred to the University Hospital of Lübeck. Here, the first clinical examinations revealed hepatosplenomegaly and hypertension (systolic blood pressure, 150 mm Hg), while laboratory data showed mild thrombocytopenia and highly elevated levels of creatinine, urea (blood urea nitrogen), and C-reactive protein in serum. Urine analysis demonstrated profound proteinuria and hematuria. At day 9 after onset, a bedside test (POC Hanta test; Erilab, Ltd., Kuopio, Finland) demonstrated antihantavirus immunoglobulin M (IgM) antibodies. The patient underwent three courses of hemodialysis until the renal failure recovered spontaneously. Five days after the last hemodialysis treatment (day 16 after onset of disease), he was discharged from the hospital in good physical condition with normal laboratory values.
We examined acute-phase serum taken at day 9 after onset by in-house enzyme-linked immunosorbent assays (ELISAs). These ELISAs use recombinant DOBV and PUUV nucleocapsid protein antigens to detect the presence of specific IgM, IgA, and IgG antibodies. The titers of IgM, IgA, and IgG to DOBV antigen were determined to be 1:25,600, 1:12,800, and 1:1,200, respectively, whereas PUUV-specific antibodies could not be detected. By in-house immunofluorescence assay, an antibody titer of 1:6,400 was found against DOBV (strain Slo/Af)-infected Vero E6 cells.
RNA for reverse transcription-PCR (RT-PCR) was extracted from the patient's serum, urine, EDTA-blood, and blood stored in AVL buffer by using a QIAamp viral RNA Mini kit (Qiagen, GmbH, Hilden, Germany). A nested RT-PCR specific for the DOBV S segment generated a DNA band of expected size (599 nucleotides [nt], nt 357 to 955) only for the AVL-blood sample. The nucleotide sequence of this fragment was determined (557 nt, nt 378 to 934, excluding the primer sequences), and the new strain was designated H169. Independently, corresponding partial S segment sequences of DOBV strains Esl/29Aa/01, Esl/33Aa/01, Esl/81Aa/01, and Esl/197Aa/01 (from A. agrarius mice trapped in Slovakia, Central Europe) were amplified and determined.
Sequence comparisons showed a clear uniqueness of the sequence from the HFRS patient (H169), sharing the highest nucleotide sequence identity (87.6 to 87.7%) with S segment sequences of DOBV-Aa strains from Slovakia. The similarity to DOBV-Af strains was slightly lower, reaching 86.3 to 87.0% nucleotide identity. Within the DOBV species, sequences associated with A. sylvaticus showed the lowest similarity to the H169 sequence (85.2% nucleotide identity). Although the sequence diversity from the other DOBV sequences was rather extended, most of the nucleotide exchanges represented silent mutations, and percent identity values of the corresponding amino acids (185 amino acids [aa], amino acid positions 127 to 311) were very high, reaching 96.2% (toward strains associated with A. sylvaticus) to 99.4% (toward DOBV-Aa strains). It should be mentioned that the H169 sequence did not encompass any known sequence from virus strains handled in our laboratory before. This fact, in addition to our adherence to standard procedures of good laboratory practice, shows that the H169 sequence did not result from a contaminated PCR amplification.
For phylogenetic analysis, the sequences were aligned on the amino acid level and then reverse translated to nucleotide sequences by using DAMBE software (18). The reliability of the alignment was checked by DotPlot analysis implemented in the BioEdit software package (5). The alignment was tested for phylogenetic information by likelihood mapping analysis, and then quartet puzzling with 10,000 puzzling steps was applied to reconstruct a maximum likelihood (ML) tree with the TREE-PUZZLE software package (15, 17). The Tamura-Nei evolutionary model was used; missing parameters were reconstructed from the data set.
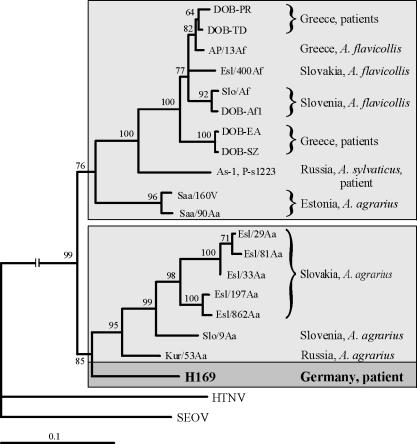
In the ML phylogenetic tree based on the 557-nt S segment sequence (nt 378 to 934), H169 clearly clustered within the DOBV species. The H169 sequence shared a common ancestor with the DOBV-Aa lineage; however, the PUZZLE support for this position of H169 was slightly below the 70% threshold limit (data not shown). The use of a smaller fragment for analysis (385 nt long, nt 378 to 762) allowed the inclusion of the only available DOBV-Aa sequence from Slovenia (Slo/9Aa) in the phylogenetic analysis (Fig. 1). When we used this alignment, including Slo/9Aa, the tree topology remained unchanged but the statistical support, particularly for position of H169, improved significantly.
FIG. 1.
ML phylogenetic tree based on the partial S segment nucleotide sequences (nt 378 to 762) of H196 and other DOBV strains, computed with TREE-PUZZLE (Tamura-Nei evolutionary model). The DOBV S segment sequence analyses included the following (accession numbers in parentheses): DOB-PR (AF060018), DOB-TD (AF060023), AP/13Af/99 (AJ410619), Esl/400Af (AY168576), Slo/Af (L41916), DOB-Af1 (AJ251996), DOB-EA (AF060020), DOB-SZ (AF060022), As-1 (AF442622), P-s1223 (AF442623), Saa/160v (AJ009773), Saa/90Aa (AJ009775), Esl/29Aa (AY533118), Esl/81Aa (AY533120), Esl33/Aa (AY533119), Esl/197Aa (AY533121), Esl/862Aa (AJ269550), Slo/9Aa (AJ251999), Kur/53Aa (AJ131673), and H169 (AY533117). HTNV strain 76-118 (M14626) and SEOV strain SR11 (M34881) were used to root the tree. Two DOBV main branches are marked by a light-gray background; the patient-associated sequence is indicated by a dark-gray box and boldface. The values at the tree branches represent the PUZZLE support values.
As shown in Fig. 1, two main branches within the DOBV species could be determined. The first group was formed by DOBV-Af sequences originating from A. flavicollis rodents (Slovenia, Greece, and Slovakia), HFRS patients from Greece, an A. sylvaticus rodent and an HFRS patient from Russia, and A. agrarius-derived Saaremaa virus strains from Estonia (which carry DOBV-Af-like S segments probably due to former reassortment events [see reference 7]). The second monophyletic group consisted of the DOBV-Aa strains from A. agrarius animals in Slovakia, Slovenia, and Russia and the new sequence, H169, from our HFRS patient.
Analysis of the H169 partial sequence clearly revealed that this strain from an HFRS patient in central Europe belongs to the DOBV species. The putative amino acid sequence encoded by the analyzed genome fragment showed 99.4% identity to the DOBV-Aa virus sequences from Slovakia, central Europe. The molecular phylogenetic analysis exhibited clustering of the H169 strain with sequences of the DOBV-Aa lineage, suggesting that a DOBV strain originating from A. agrarius was responsible for the HFRS case described. We were not able to amplify M segment sequences from our patient's rather limited material (probably because of suboptimal matching of standard primers with the M sequence, which is generally known to be more variable than the S sequence). Although our recent studies in central Europe (sympatric occurrence of DOBV-Af and DOBV-Aa strains without detection of reassortants) do not suggest it, one cannot completely exclude the possibility that H169 has a reassortant origin, because this was recently shown for an A. agrarius-derived strain from northeast Europe (7).
In additional experiments, we used a convalescent-phase serum from the patient (taken 6 months after onset of symptoms) for serotyping of neutralizing antibodies. By chemiluminescence focus reduction neutralization test (c-FRNT) (6), reciprocal neutralizing titers as low as <40 were obtained with PUUV (strain Sotkamo), Tula virus (TULV, strain Moravia), SEOV (strain 80-39), and HTNV (strain 76-118). For DOBV, two virus isolates were tested. The DOBV-Af strain Slovenia (3) and the DOBV-Aa strain Slovakia (B. Klempa, H. A. Schmidt, R. Ulrich, M. Schutt, M. Labuda, B. Hjelle, H. Meisel, and D. H. Kruger, Abstr. XIIth Int. Conf. Negative Strand Viruses, abstr. 289, 2003) both exhibited identical reciprocal titers of 640.
The results of fine serotyping by c-FRNTs are in line with the molecular genetic findings. A more-than-fourfold-higher neutralizing antibody titer was found against DOBV compared with titers against PUUV, TULV, SEOV, and HTNV. No difference in titers could be detected when the representatives of DOBV-Af and DOBV-Aa lineages (Slovenia virus and Slovakia virus, respectively) were used in neutralization assays with the patient's serum. This can be explained by the fact that the antigenicities of these virus isolates are very similar. It has previously been shown that a substantial number of human DOBV-positive sera did not exhibit clear differences in titer toward Slovenia versus Slovakia (our unpublished data) or Slovenia versus Saaremaa virus isolates (4).
In summary, we demonstrate for the first time not only serological but direct molecular genetic evidence that DOBV causes HFRS in central Europe. The sequence originating from a blood sample of an HFRS patient from Germany is clearly distinct from DOBV-Af sequences derived from patients or A. flavicollis animals in southeast Europe and is closely related to DOBV strains found in A. agrarius mice.
Nucleotide sequence accession numbers.
The S segment partial sequences (557 nt) of the following DOBV strains have been deposited in the GenBank sequence database under the following accession numbers: H169/02, AY533117; Esl/29Aa/01, AY533118; Esl33Aa/01, AY533119; Esl/81Aa/01, AY533120; Esl/197Aa/01, AY533121.
Acknowledgments
We thank Deutsche Forschungsgemeinschaft (grant KR1293/2-3) and Humboldt University Medical School for support of our work.
We are grateful to Brigitte Pohl and Heike Lerch for excellent technical assistance.
REFERENCES
- 1.Antoniadis, A., A. Stylianakis, A. Papa, S. Alexiou-Daniel, A. Lampropoulos, S. T. Nichol, C. J. Peters, and C. F. Spiropoulou. 1996. Direct genetic detection of Dobrava virus in Greek and Albanian patients with hemorrhagic fever with renal syndrome. J. Infect. Dis. 174:407-410. [DOI] [PubMed] [Google Scholar]
- 2.Avsic-Zupanc, T., S. Y. Xiao, R. Stojanovic, A. Gligic, G. van der Groen, and J. W. Leduc. 1992. Characterization of Dobrava virus: a hantavirus from Slovenia, Yugoslavia. J. Med. Virol. 38:132-137. [DOI] [PubMed] [Google Scholar]
- 3.Avsic-Zupanc, T., M. Petrovec, P. Furlan, R. Kaps, F. Elgh, and A. Lundkvist. 1999. Hemorrhagic fever with renal syndrome in the Dolenjska region of Slovenia—a 10-year survey. Clin. Infect. Dis. 28:860-865. [DOI] [PubMed] [Google Scholar]
- 4.Brus Sjolander, K., I. Golovljova, V. Vasilenko, A. Plyusnin, and A. Lundkvist. 2002. Serological divergence of Dobrava and Saaremaa hantaviruses: evidence for two distinct serotypes. Epidemiol. Infect. 128:99-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 6.Heider, H., B. Ziaja, C. Priemer, A. Lundkvist, J. Neyts, D. H. Kruger, and R. Ulrich. 2001. A chemiluminescence detection method of hantaviral antigens in neutralisation assays and inhibitor studies. J. Virol. Methods 96:17-23. [DOI] [PubMed] [Google Scholar]
- 7.Klempa, B., H. A. Schmidt, R. Ulrich, S. Kaluz, M. Labuda, H. Meisel, B. Hjelle, and D. H. Kruger. 2003. Genetic interaction between distinct Dobrava hantavirus subtypes in Apodemus agrarius and A. flavicollis in nature. J. Virol. 77:804-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruger, D. H., R. Ulrich, and A. Lundkvist. 2001. Hantavirus infections and their prevention. Microbes Infect. 3:1129-1144. [DOI] [PubMed] [Google Scholar]
- 9.Markotic, A., S. T. Nichol, I. Kuzman, A. J. Sanchez, T. G. Ksiazek, A. Gagro, S. Rabatic, R. Zgorelec, T. Avsic-Zupanc, I. Beus, and D. Dekaris. 2002. Characteristics of Puumala and Dobrava infections in Croatia. J. Med. Virol. 66:542-551. [DOI] [PubMed] [Google Scholar]
- 10.Nemirov, K., O. Vapalahti, A. Lundkvist, V. Vasilenko, I. Golovljova, A. Plyusnina, J. Niemimaa, J. Laakkonen, H. Henttonen, A. Vaheri, and A. Plyusnin. 1999. Isolation and characterization of Dobrava hantavirus carried by the striped field mouse (Apodemus agrarius) in Estonia. J. Gen. Virol. 80:371-379. [DOI] [PubMed] [Google Scholar]
- 11.Papa, A., A. M. Johnson, P. C. Stockton, M. D. Bowen, C. F. Spiropoulou, S. Alexiou-Daniel, T. G. Ksiazek, S. T. Nichol, and A. Antoniadis. 1998. Retrospective serological and genetic study of the distribution of hantaviruses in Greece. J. Med. Virol. 55:321-327. [DOI] [PubMed] [Google Scholar]
- 12.Plyusnin, A., D. H. Kruger, and A. Lundkvist. 2001. Hantavirus infections in Europe. Adv. Virus Res. 57:105-136. [DOI] [PubMed] [Google Scholar]
- 13.Schmaljohn, C. S., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmaljohn, C. S., and S. T. Nichol (ed.). 2001. Hantaviruses. Current topics in microbiology and immunology, vol. 256. Springer, New York, N.Y. [DOI] [PubMed]
- 15.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 16.Sibold, C., R. Ulrich, M. Labuda, A. Lundkvist, H. Martens, M. Schutt, P. Gerke, K. Leitmeyer, H. Meisel, and D. H. Kruger. 2001. Dobrava hantavirus causes hemorrhagic fever with renal syndrome in central Europe and is carried by two different Apodemus mice species. J. Med. Virol. 63:158-167. [PubMed] [Google Scholar]
- 17.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia, X., and Z. Xie. 2001. DAMBE: data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 19.Xiao, S. Y., G. Diglisic, T. Avsic-Zupanc, and J. W. Leduc. 1993. Dobrava virus as a new hantavirus: evidenced by comparative sequence analysis. J. Med. Virol. 39:152-155. [DOI] [PubMed] [Google Scholar]