Abstract
Hepatitis C virus isolates which disclosed a novel genotype 1-associated restriction pattern by restriction fragment length polymorphism analysis were characterized. Except for a mother and child pair, the patients were unrelated. Sequence analysis showed a G→A substitution leading to a new RsaI recognition site. Phylogenetic analysis revealed that these isolates constitute a novel genetic lineage within the main cluster of genotype 1 strains.
Hepatitis C virus (HCV) is an enveloped virus which belongs to the genus Hepacivirus in the family Flaviviridae (15). It is the most important cause of posttransfusion non-A, non-B hepatitis worldwide. The genome of HCV consists of a single strand of positive RNA (≈9.5 kb), which codes for at least 10 viral proteins and is flanked by 5′- and 3′-end noncoding regions.
The 5′-end untranslated region (5′UTR) of HCV is the most highly conserved portion of the viral genome and has been used to develop sensitive assays for RNA detection as well as for genotyping. According to current recommendations, HCV isolates are classified into six major clades, 1 to 6, whose nucleotide and inferred amino acid sequences differ by 35% (15, 18). Clade assignment is achieved by sequencing and aligning the HCV core, E1, or NS5B sequences with those of the prototypical strains of each clade (15). Although exact and reliable, this method cannot be easily implemented in conventional diagnostic laboratories. Consequently, several other methods to determine HCV genotype have been developed, including HCV RNA amplification followed by either reamplification with genotype-specific primers in the core region (11), hybridization with type-specific probes in the 5′UTR (16), or digestion of PCR products with restriction endonucleases that recognize genotype- and even subtype-specific sequence polymorphisms in the 5′UTR of the HCV (restriction fragment length polymorphism [RFLP]) (3, 9).
Using the RFLP method described by Davidson et al. (3), a widely used technique in our region, we reported a high prevalence of genotype 1a/c in children and infants in Argentina (6). Although rapid and simple, RFLP turned out to be unsatisfactory for the identification of HCV genotypes in isolates for five of our cases. Four of them were children and represented 14% of our pediatric population under study. Our aim was to characterize these isolates and to evaluate their diversity by means of nucleic acid sequencing and subsequent phylogenetic analysis.
Plasma samples from five patients with chronic HCV infection (four children and one adult) were analyzed. Viral RNA was reverse transcribed, and the 5′UTR was amplified as previously described (5), with Kwok and Higuchi's recommendations (8). Amplicons were digested with restriction enzymes, followed by 15% or 12% polyacrylamide gel electrophoresis to evaluate the HCV genotype or subtype, as described by Davidson et al. (3). In addition, fragments were purified and sequenced with the Big Dye terminator cycle sequencing kit, version 1.1, and the 3100 genetic analyzer (Applied Biosystems).
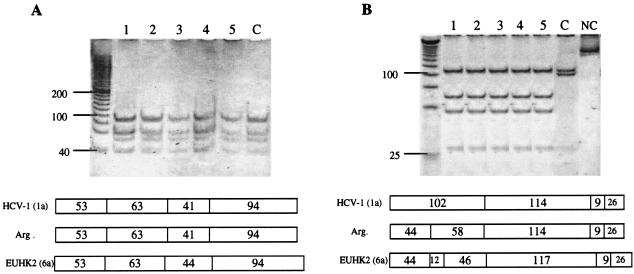
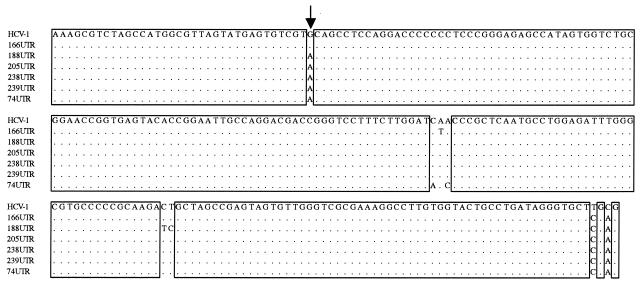
DNA digestion of the five samples with HinfI and MvaI gave a genotype 1- or 6-associated pattern (Fig. 1A), whereas they were untypeable by RsaI and HaeIII digestion (Fig. 1B). On the other hand, digestion with BstUI gave the genotype 1a/c-associated pattern (data not shown). Thus, our strains exhibit a restriction map more similar to that of genotype 1 than to any of the other genotypes. Sequence alignment showed a G→A substitution at position −235 of the 5′UTR in all untypeable isolates tested compared to a prototypical genotype 1 strain (Fig. 2). This point mutation resulted in the generation of a new recognition site for RsaI, modifying the typical pattern for HCV genotype 1. The analysis also included an Argentine isolate for which the genotype had been clearly determined as 1a/c by the same technique. This isolate does not display the above-mentioned substitution, confirming that the abnormal pattern was a consequence of this nucleotide substitution.
FIG. 1.
RFLP analysis of Argentine HCV isolates. Amplified DNA fragments were digested with HinfI and MvaI (A) or RsaI and HaeIII (B). (Top) Polyacrylamide gel electrophoresis. Lanes 1 to 5, untypeable isolates; lane C, Argentine isolate corresponding to genotype 1a/c; lane NC, negative control (251 bp). (Bottom) Restriction map of the isolates tested and prototypic HCV strains, as predicted from the DNA sequences (Webcutter program version 2.0). The numbers indicate the lengths of the restriction fragments (in base pairs). Genotypes are indicated in parentheses. HCV-1 (1a), prototypic HCV genotype 1 strain; Arg, untypeable Argentine isolates; EUHK2, prototypic HCV genotype 6 strain. The numbers on the left indicate the sizes of the standards (in base pairs).
FIG. 2.
Alignment of the amplified 5′UTR sequences of Argentine HCV isolates with the corresponding sequence of strain HCV-1 (genotype 1a). Isolates are indicated by name on the left side. Dots indicate nucleotide identity with strain HCV-1. The arrow marks position −235.
The mutation described above was not related to either the patient's age or a history of blood transfusions. This is supported by the fact that it was present in samples from children and an adult who were infected by different routes at different times. Except for one case of mother-to-child transmission, in which samples from both the mother and child were studied, the patients were unrelated to each other. This indicates that our observations depict a general phenomenon which should be taken into account when determining viral genotypes. The detection of the same mutation in the mother-and-child paired samples strongly supports the idea that this variant is adapted to the host environment. Therefore, the mutation is unlikely an intrahost substitution due to natural quasispecies dynamics.
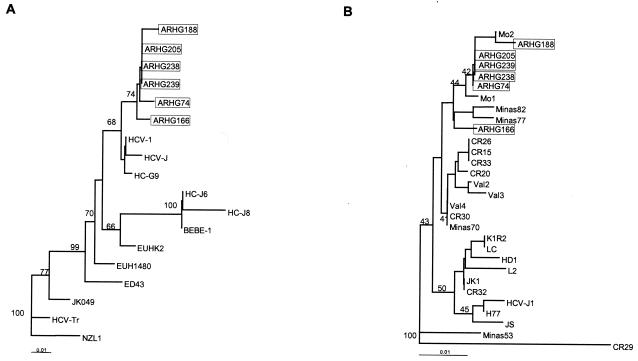
To elucidate the genetic heterogeneity of our isolates and to determine their phylogenetic relatedness to prototypic HCV strains, we applied phylogenetic analysis with the programs of the PHYLIP package (DNAdist, Neighbor, Seqboot and Consense) (4). The five untypeable isolates clustered together inside the main cluster of the genotype 1 strains, and they were segregated from HCV prototypes corresponding to subtypes a, b, and c (Table 1; Fig. 3A). We also evaluated the genetic variability of our isolates in comparison to other genotype 1 HCV isolates recently described in our region as well as in the rest of the world (Table 1; Fig. 3B). The two HCV isolates from Montevideo, Uruguay, displayed the same point mutation at position −235 and clustered with our mutated isolates. The nonmutated Argentine isolate used as a genotype 1 control and other HCV isolates from Brazil clustered apart from the untypeable isolates which have been described here.
TABLE 1.
HCV isolates used for phylogenetic analysis
| Isolate | HCV genotype | Country of origin | Accession no. |
|---|---|---|---|
| HCV-1 | 1a | USA | M62321 |
| HCV-J | 1b | Japan | D90208 |
| HC-G9 | 1c | Indonesia | D14853 |
| K1R2 | 1b | Japan | D50482 |
| LC | 1 | Taiwan | U89019 |
| HD1 | 1 | Germany | U45476 |
| L2 | 1 | Korea | U01214 |
| JK1 | 1b | Japan | X61596 |
| HCV-J1 | 1 | Japan | D10749 |
| H77 | 1a | USA | AF009606 |
| JS | 1 | Japan | D85516 |
| HC-J6 | 2a | Indonesia | D00944 |
| HC-J8 | 2b | Indonesia | D10988 |
| BEBE1 | 2c | Italy | D50409 |
| JK049 | 3 | Indonesia | D63821 |
| NZL1 | 3a | Japan | D17763 |
| HCV-Tr | 3b | Thailand | E10839 |
| ED43 | 4a | Egypt | Y11604 |
| EUH1480 | 5a | United Kingdom | Y13184 |
| EUHK2 | 6a | Hong Kong | Y12083 |
| Montevideo 1 | 1 | Uruguay | AJ012831 |
| Montevideo 2 | 1 | Uruguay | AJ012832 |
| Minas 53 | 1 | Brazil | AF077230 |
| Minas 70 | 1 | Brazil | AF077232 |
| Minas 77 | 1 | Brazil | AF077235 |
| Minas 82 | 1 | Brazil | AF077236 |
| Valdivia 2 | 1 | Chile | AJ291456 |
| Valdivia 3 | 1 | Chile | AJ291457 |
| Valdivia 4 | 1 | Chile | AJ291458 |
| CR 15 | 1 | Costa Rica | AJ437146 |
| CR 20 | 1 | Costa Rica | AJ437148 |
| CR 26 | 1 | Costa Rica | AJ437150 |
| CR 29 | 3 | Costa Rica | AJ437144 |
| CR 30 | 1 | Costa Rica | AJ437147 |
| CR 32 | 1b | Costa Rica | AJ437145 |
| CR 33 | 1 | Costa Rica | AJ437149 |
| ARHG166 | 1a/c | Argentina | AY376832 |
| ARHG188 | UTa | Argentina | AY376833 |
| ARHG205 | UT | Argentina | AY376834 |
| ARHG238 | UT | Argentina | AY376835 |
| ARHG239 | UT | Argentina | AY376836 |
| ARHG74 | UT | Argentina | AY376837 |
UT, untypeable.
FIG. 3.
Phylogenetic analysis of the amplified 5′UTR of untypeable isolates and corresponding sequences of other HCV strains. Isolates are shown by their names. The values obtained after 100 bootstrap resamplings are indicated. The horizontal branch lengths are proportional to the genetic distances. (A) Phylogenetic tree obtained with our untypeable isolates and prototypic HCV strains of all genotypes. (B) Phylogenetic tree of untypeable isolates and other genotype 1 isolates.
Interestingly, the HCV strains from Central and South America were more related to each other than to the others from the rest of the world. This further supports the idea of regional diversification of HCV. In fact, a geographic distribution of HCV genotypes has been documented; genotypes 1, 2, and 3 are the most commonly detected worldwide (18). HCV genotype 1 in particular has been extensively reported by other authors in Argentina (5, 14), Chile (10), Venezuela (12), Uruguay (2), and Brazil (7). In a recent study, Pybus et al. used a mathematical model to analyze the epidemic behavior of HCV infection. Strikingly, it seems that HCV genotypes 1a and 1b originated about 100 years ago and are evolving at a faster rate than genotypes 4 and 6 (13). This dissimilar evolution rate between HCV genotypes may account for the mutated variant described in this paper that clustered into the novel genetic lineage reported by Vega et al. (Fig. 3B) (17).
The existence of HCV variants containing an A at position −235 may affect clade determination by RFLP in conventional diagnostic laboratories. Other authors have demonstrated that, as it is the most conserved region in the HCV genome, the 5′UTR may also be useful to identify many of the different genotypes by phylogenetic analysis (1). For genotype assignment purposes, our mutated isolates may be classified as genotype 1. Nevertheless, it should be borne in mind that although the untypeable strains disclosed a subtype a/c-associated pattern of bands when digested with BstUI, phylogenetic analysis did not indicate an association between them and the prototypic genotype 1 subtype a and c strains. Thus, it may be advisable to classify the new HCV isolates showing this abnormal restriction pattern only as genotype 1, without a subtype label, until this novel genetic lineage is completely characterized.
Nucleotide sequence accession numbers. The GenBank accession numbers of the sequences reported in this work are AY376832 to AY376837.
Acknowledgments
M.I.G. was supported by a fellowship from the Fundación Argentina de Transplante Hepático (fellowship 171202/03), and M.V.P. is a member of the National Research Council (CONICET) Research Career Program.
REFERENCES
- 1.Alfonso, V., D. Flichman, S. Sookoian, V. Mbayed, and R. Campos. 2001. Phylogenetic characterization of genotype 4 hepatitis C virus isolates from Argentina. J. Clin. Microbiol. 39:1989-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colina, R., C. Azambuja, R. Uriarte, C. Mogdasy, and J. Cristina. 1999. Evidence of increasing diversification of hepatitis C viruses. J. Gen. Virol. 80:1377-1382. [DOI] [PubMed] [Google Scholar]
- 3.Davidson, F., P. Simmonds, J. Ferguson, L. Jarvis, B. Dow, E. Follet, C. Seed, T. Krusius, C. Lin, G. Medgyesi, H. Kiyokawa, G. Olim, G. Duraisamy, T. Cuypers, A. Saeed, D. Teo, J. Conradie, M. Kew, M. Lin, C. Nuchaprayoon, O. Ndimbie, and P. Yap. 1995. Survey of major genotypes and subtypes of hepatitis C using RFLP of sequences amplified from the 5′ non-coding region. J. Gen. Virol. 76:1197-1204. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 2001. PHYLIP: phylogeny inference package, version 3.6(alpha2). Department of Genetics, University of Washington, Seattle, Wash.
- 5.Gismondi, M. I., M. V. Preciado, I. Badía, A. Ferro, C. Galoppo, and S. Grinstein. 2001. Estudio y caracterización genotípica de la infección por el virus de Hepatitis C en niños. Medicina (Buenos Aires) 61:815-820. [PubMed] [Google Scholar]
- 6.Gismondi, M. I., E. I. Turazza, S. Grinstein, M. C. Galoppo, and M. V. Preciado. Hepatitis C virus infection in infants and children from Argentina. J. Clin. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 7.Krug, L. P., V. R. Lune, N. Ikuta, A. S. Fonseca, H. Cheinquer, L. S. Ozaki, and S. G. Barros. 1996. Hepatitis C virus genotypes in Southern Brazil. Braz. J. Med. Res. 29:1629-1633. [PubMed] [Google Scholar]
- 8.Kwok, S., and R. Higuchi. 1997. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 9.McOmish, F., P. Yap, and B. Dow. 1994. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J. Clin. Microbiol. 32:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munoz, G., M. Velasco, V. Thiers, C. Hurtado, J. Brahm, M. Larrondo-Lillo, A. Guglielmetti, G. Smok, C. Bréchot, and E. Lamas. 1998. Prevalence and genotypes of hepatitis C virus in blood donors and in patients with chronic liver disease and hepatocarcinoma in a Chilean population. Rev. Med. Chile 126:1035-1042. [PubMed] [Google Scholar]
- 11.Okamoto, H., K. Sugiyama, S. Okada, K. Kurai, Y. Akahane, Y. Sugai, T. Tanaka, K. Sato, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1992. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J. Gen. Virol. 73:673-679. [DOI] [PubMed] [Google Scholar]
- 12.Pujol, F. H., C. L. Loureiro, M. Devesa, L. Blitz, K. Parra, S. Beker, and F. Liprandi. 2002. Determination of genotypes of hepatitis C virus in Venezuela by restriction fragment length polymorphism. J. Clin. Microbiol. 35:1870-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pybus, O. G., M. A. Charleston, S. Gupta, A. Rambaut, E. C. Holmes, and P. H. Harvey. 2001. The epidemic behavior of the hepatitis C virus. Science 292:2323-2325. [DOI] [PubMed] [Google Scholar]
- 14.Quarleri, J., B. Robertson, V. Mathet, M. Feld, L. Espínola, M. Requeijo, O. Mandó, G. Carballal, and J. Oubiña. 2000. Genomic and phylogenetic analysis of hepatitis C virus isolates from argentine patients: a six-year retrospective study. J. Clin. Microbiol. 38:4560-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin-i, P. Simmonds, D. Smith, L. Stuyver, and A. J. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 16.Stuyver, L., R. Rossau, A. Wyseur, M. Duhamel, B. Vanderborght, H. Van Heuverswyn, and G. Maertens. 1993. Typing of hepatitis C virus isolates and characterisation of new subtypes using a line probe assay. J. Gen. Virol. 74:1093-1102. [DOI] [PubMed] [Google Scholar]
- 17.Vega, I., R. Colina, L. García, R. Uriarte, C. Mogdasy, and J. Cristina. 2001. Diversification of hepatitis C viruses in South America reveals a novel genetic lineage. Arch. Virol. 146:1623-1629. [DOI] [PubMed] [Google Scholar]
- 18.Zein, N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]